Discovery of Species-Specific Peptide Markers for Superseed Authentication Using Targeted LC-MS/MS Proteomics
Abstract
1. Introduction
2. Results
2.1. Initial Sample Assessment
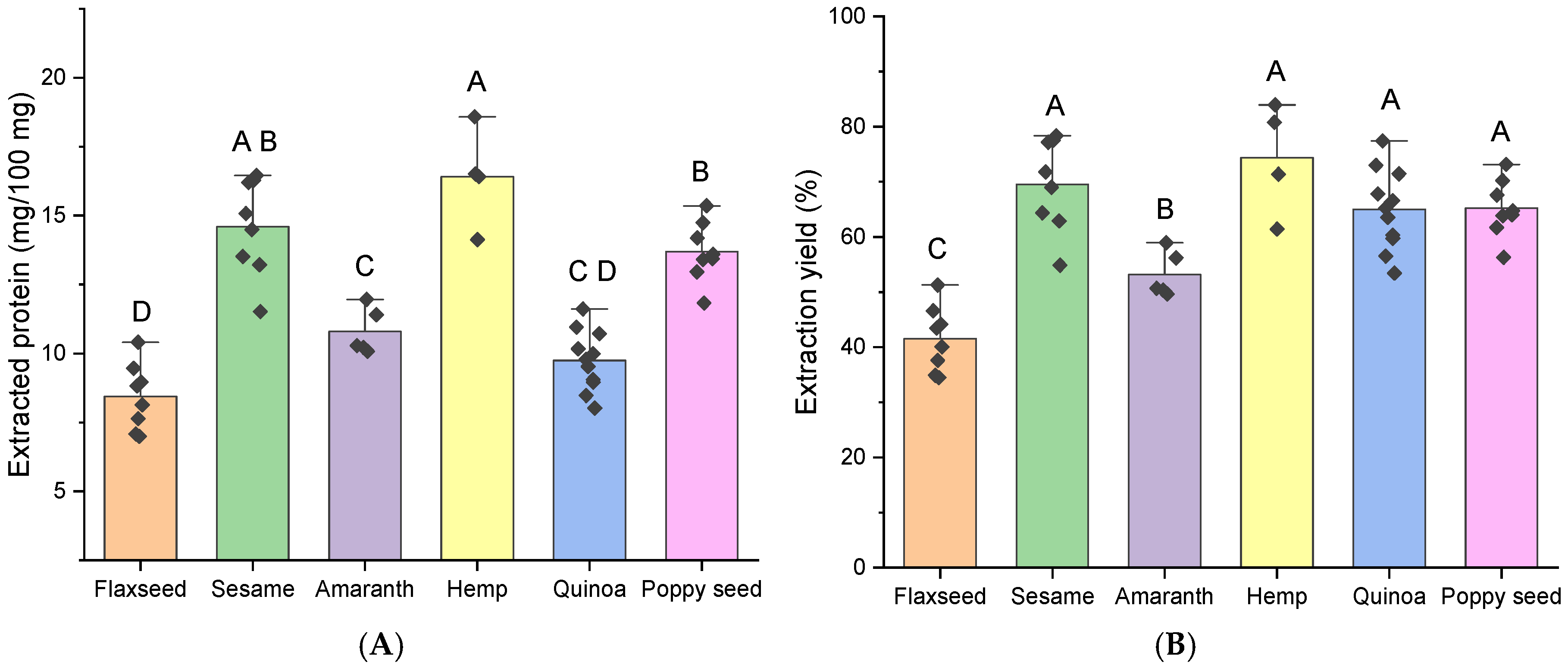
2.1.1. Crude Protein
2.1.2. Fat Content
2.2. Protein Extraction
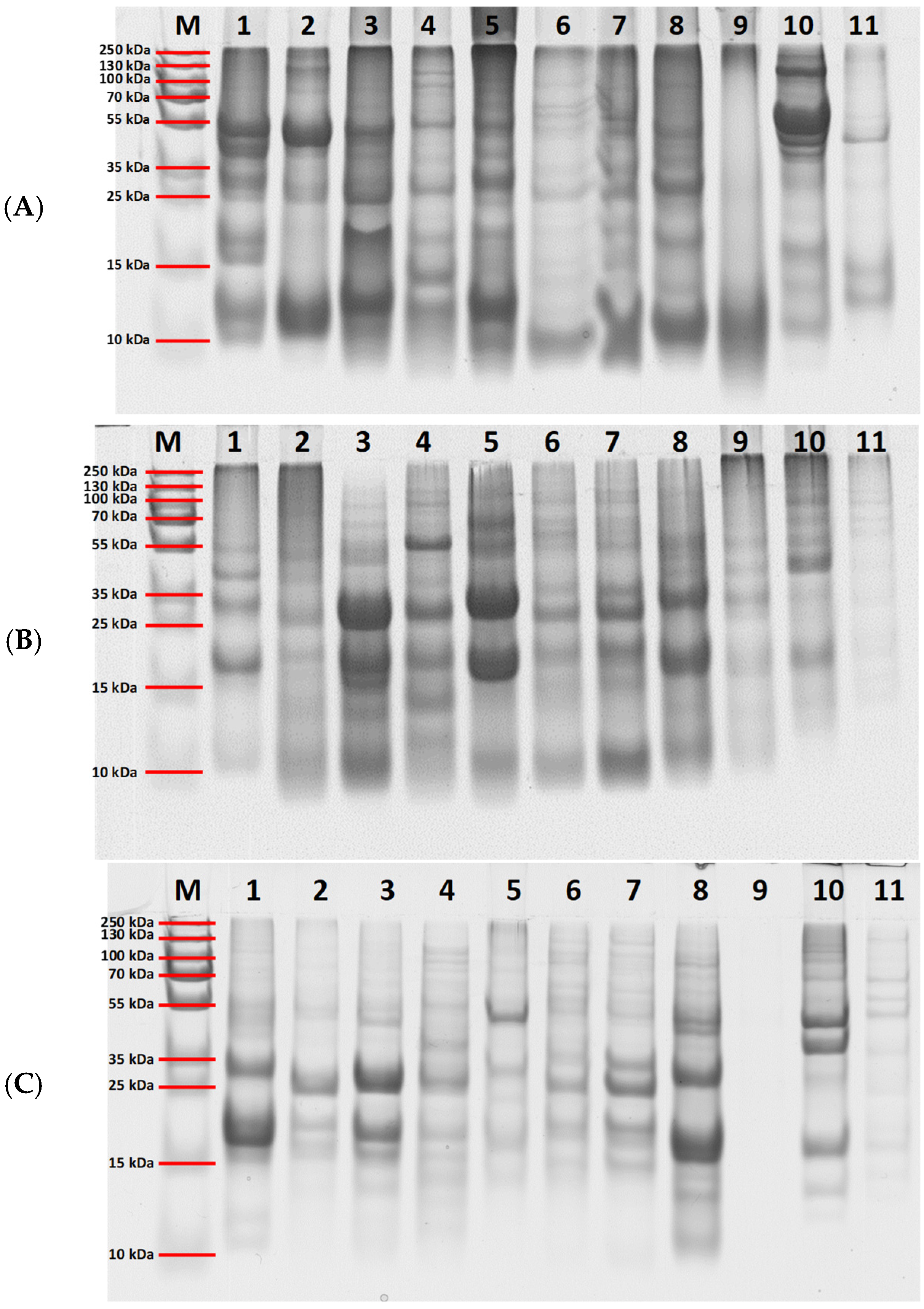
2.3. Profile of the Extracted Proteins
2.4. Targeted Discovery and Validation of Species-Specific Peptide Markers
2.4.1. Identification of Robust Peptide Markers for Superseed Authentication
2.4.2. Specificity and Stability of Selected Marker Peptides
2.4.3. Validation of the Targeted LC-MS/MS Method
3. Materials and Methods
3.1. Biological Samples
3.2. Chemicals
3.3. Sample Preparation and Protein Extraction
3.3.1. Protein Extraction Using the Ambi/Urea Method
3.3.2. SDS Buffer Extraction Method
3.3.3. TCA Extraction Method
3.4. Sample Characterization
3.4.1. Protein Content
3.4.2. Electrophoretic Characterization
3.5. Targeted Mass Spectrometry for Peptide Marker Search
3.5.1. Sample Preparation
3.5.2. Mass Spectrometric Analysis
3.5.3. Method Validation
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mintel. Super Growth for “Super” Foods: New Product Development Shoots up 202% Globally over the Past Five Years. Available online: https://www.mintel.com/press-centre/super-growth-for-super-foods-new-product-development-shoots-up-202-globally-over-the-past-five-years/ (accessed on 16 April 2025).
- Burton, R.A.; Andres, M.; Cole, M.; Cowley, J.M.; Augustin, M.A. Industrial hemp seed: From the field to value-added food ingredients. J. Cannabis Res. 2022, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Brigante, F.I.; Garcia, M.E.; Lopez Radcenco, A.; Moyna, G.; Wunderlin, D.A.; Baroni, M.V. Identification of chia, flax and sesame seeds authenticity markers by nmr-based untargeted metabolomics and their validation in bakery products containing them. Food Chem. 2022, 387, 132925. [Google Scholar] [CrossRef] [PubMed]
- Hannan, M.A.; Rahman, M.A.; Sohag, A.A.M.; Uddin, M.J.; Dash, R.; Sikder, M.H.; Rahman, M.S.; Timalsina, B.; Munni, Y.A.; Sarker, P.P.; et al. Black cumin (Nigella sativa L.): A comprehensive review on phytochemistry, health benefits, molecular pharmacology, and safety. Nutrients 2021, 13, 1784. [Google Scholar] [CrossRef] [PubMed]
- Calderon Bravo, H.; Vera Cespedes, N.; Zura-Bravo, L.; Munoz, L.A. Basil seeds as a novel food, source of nutrients and functional ingredients with beneficial properties: A review. Foods 2021, 10, 1467. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Khanna, K.R.; Shukla, S.; Dixit, B.S.; Banerji, R. Prospects of breeding opium poppies (Papaver somniferum L.) as a high-linoleic-acid crop. Plant Breed. 2006, 114, 89–91. [Google Scholar] [CrossRef]
- Tavakkoli, A.; Mahdian, V.; Razavi, B.M.; Hosseinzadeh, H. Review on clinical trials of black seed (Nigella sativa) and its active constituent, thymoquinone. J. Pharmacopunct. 2017, 20, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Bozan, B.; Temelli, F. Chemical composition and oxidative stability of flax, safflower and poppy seed and seed oils. Bioresour. Technol. 2008, 99, 6354–6359. [Google Scholar] [CrossRef] [PubMed]
- Rubilar, M.; Gutiérrez, C.; Verdugo, M.; Shene, C.; Sineiro, J. Flaxseed as a source of functional ingredients. J. Soil Sci. Plant Nutr. 2010, 10, 373–377. [Google Scholar] [CrossRef]
- Coşkuner, Y.; Karababa, E. Some physical properties of flaxseed (Linum usitatissimum L.). J. Food Eng. 2007, 78, 1067–1073. [Google Scholar] [CrossRef]
- Cerino, P.; Buonerba, C.; Cannazza, G.; D’Auria, J.; Ottoni, E.; Fulgione, A.; Di Stasio, A.; Pierri, B.; Gallo, A. A review of hemp as food and nutritional supplement. Cannabis Cannabinoid Res. 2021, 6, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Farinon, B.; Molinari, R.; Costantini, L.; Merendino, N. The seed of industrial hemp (Cannabis sativa L.): Nutritional quality and potential functionality for human health and nutrition. Nutrients 2020, 12, 1935. [Google Scholar] [CrossRef] [PubMed]
- Nissen, L.; Zatta, A.; Stefanini, I.; Grandi, S.; Sgorbati, B.; Biavati, B.; Monti, A. Characterization and antimicrobial activity of essential oils of industrial hemp varieties (Cannabis sativa L.). Fitoterapia 2010, 81, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Z.; Zheng, C.; Liu, C. A comprehensive review of bioactive compounds and processing technology of sesame seed. Oil Crop. Sci. 2022, 7, 88–94. [Google Scholar] [CrossRef]
- Pathak, N.; Bhaduri, A.; Rai, A.K. Sesame: Bioactive compounds and health benefits. In Bioactive Molecules in Food; Springer: Cham, Switzerland, 2019; pp. 181–200. [Google Scholar]
- Wei, P.; Zhao, F.; Wang, Z.; Wang, Q.; Chai, X.; Hou, G.; Meng, Q. Sesame (Sesamum indicum L.): A comprehensive review of nutritional value, phytochemical composition, health benefits, development of food, and industrial applications. Nutrients 2022, 14, 4079. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Mogra, R. Psyllium (Plantago ovata) husk: A wonder food for good health. Int. J. Sci. Res. (IJSR) 2015, 4, 5. [Google Scholar]
- de la Barca, A.M.; Rojas-Martinez, M.E.; Islas-Rubio, A.R.; Cabrera-Chavez, F. Gluten-free breads and cookies of raw and popped amaranth flours with attractive technological and nutritional qualities. Plant Foods Hum. Nutr. 2010, 65, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; Le Maux, S.; Dubrulle, C.; Barre, C.; FitzGerald, R.J. Quinoa (Chenopodium quinoa willd.) protein hydrolysates with in vitro dipeptidyl peptidase iv (dpp-iv) inhibitory and antioxidant properties. J. Cereal Sci. 2015, 65, 112–118. [Google Scholar] [CrossRef]
- Martínez, E.N.; Castellani, O.F.; Añón, M.C. Common molecular features among amaranth storage proteins. J. Agric. Food Chem. 1997, 45, 3832–3839. [Google Scholar] [CrossRef]
- Penarrieta, J.M.; Alvarado, J.A.; Akesson, B.; Bergenstahl, B. Total antioxidant capacity and content of flavonoids and other phenolic compounds in canihua (Chenopodium pallidicaule): An andean pseudocereal. Mol. Nutr. Food Res. 2008, 52, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Salo, H.M.; Nguyen, N.; Alakarppa, E.; Klavins, L.; Hykkerud, A.L.; Karppinen, K.; Jaakola, L.; Klavins, M.; Haggman, H. Authentication of berries and berry-based food products. Compr. Rev. Food Sci. Food Saf. 2021, 20, 5197–5225. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Drakopoulou, S.K.; Aalizadeh, R.; Thomaidis, N.S. Targeted and untargeted metabolomics as an enhanced tool for the detection of pomegranate juice adulteration. Foods 2019, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.; Frew, R.; Kebede, B. A review of conventional and rapid analytical techniques coupled with multivariate analysis for origin traceability of soybean. Crit. Rev. Food Sci. Nutr. 2024, 64, 6616–6635. [Google Scholar] [CrossRef] [PubMed]
- Mburu, M.; Komu, C.; Paquet-Durand, O.; Hitzmann, B.; Zettel, V. Chia oil adulteration detection based on spectroscopic measurements. Foods 2021, 10, 1798. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Zhang, L.; Yang, R.; Wang, X.; Yu, L.; Yue, X.; Ma, F.; Mao, J.; Wang, X.; Li, P. Adulteration detection of essence in sesame oil based on headspace gas chromatography-ion mobility spectrometry. Food Chem. 2022, 370, 131373. [Google Scholar] [CrossRef] [PubMed]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef]
- Li, X.; Wang, D.; Ma, F.; Yu, L.; Mao, J.; Zhang, W.; Jiang, J.; Zhang, L.; Li, P. Rapid detection of sesame oil multiple adulteration using a portable raman spectrometer. Food Chem. 2023, 405, 134884. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.; Minaei, S.; Beheshti, B. A dedicated electronic nose combined with chemometric methods for detection of adulteration in sesame oil. J. Food Sci. Technol. 2023, 60, 2681–2694. [Google Scholar] [CrossRef] [PubMed]
- Karppinen, K.; Avetisyan, A.; Hykkerud, A.L.; Jaakola, L. A dpcr method for quantitative authentication of wild lingonberry (Vaccinium vitis-idaea) versus cultivated american cranberry (V. Macrocarpon). Foods 2022, 11, 1476. [Google Scholar] [CrossRef] [PubMed]
- Erban, A.; Fehrle, I.; Martinez-Seidel, F.; Brigante, F.; Mas, A.L.; Baroni, V.; Wunderlin, D.; Kopka, J. Discovery of food identity markers by metabolomics and machine learning technology. Sci. Rep. 2019, 9, 9697. [Google Scholar] [CrossRef] [PubMed]
- Brigante, F.I.; Lucini Mas, A.; Pigni, N.B.; Wunderlin, D.A.; Baroni, M.V. Targeted metabolomics to assess the authenticity of bakery products containing chia, sesame and flax seeds. Food Chem. 2020, 312, 126059. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Kuster, B. Plant proteome dynamics. Annu. Rev. Plant Biol. 2022, 73, 67–92. [Google Scholar] [CrossRef] [PubMed]
- Miernyk, J.A. Seed proteomics. Methods Mol. Biol. 2014, 1072, 361–377. [Google Scholar] [PubMed]
- Kadereit, J.W.; Körner, C.; Nick, P.; Sonnewald, U. Strasburger—Lehrbuch der Pflanzenwissenschaften; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Heldt, H.W.; Piechulla, B. Pflanzenbiochemie; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Marambe, H.K.; Wanasundara, J.P.D. Protein from flaxseed (Linum usitatissimum L.). In Sustainable Protein Sources; Academic Press: Cambridge, MA, USA, 2017; pp. 133–144. [Google Scholar]
- Sá, A.G.A.; Pacheco, M.T.B.; Moreno, Y.M.F.; Carciofi, B.A.M. Cold-pressed sesame seed meal as a protein source: Effect of processing on the protein digestibility, amino acid profile, and functional properties. J. Food Compos. Anal. 2022, 111, 104634. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, J.; Fu, Y.; Zhang, T.; Jiang, L.; Sui, X. Structural, nutritional, and functional properties of amaranth protein and its application in the food industry: A review. Sustain. Food Proteins 2023, 1, 45–55. [Google Scholar] [CrossRef]
- Dakhili, S.; Abdolalizadeh, L.; Hosseini, S.M.; Shojaee-Aliabadi, S.; Mirmoghtadaie, L. Quinoa protein: Composition, structure and functional properties. Food Chem. 2019, 299, 125161. [Google Scholar] [CrossRef] [PubMed]
- Melo, D.; Alvarez-Orti, M.; Nunes, M.A.; Espirito Santo, L.; Machado, S.; Pardo, J.E.; Oliveira, M. Nutritional and chemical characterization of poppy seeds, cold-pressed oil, and cake: Poppy cake as a high-fibre and high-protein ingredient for novel food production. Foods 2022, 11, 3027. [Google Scholar] [CrossRef] [PubMed]
- Koza, J.; Jurgonski, A. Partially defatted rather than native poppy seeds beneficially alter lipid metabolism in rats fed a high-fat diet. Sci. Rep. 2023, 13, 14171. [Google Scholar] [CrossRef] [PubMed]
- Ixtaina, V.Y.; Nolasco, S.M.; Tomás, M.C. Physical properties of chia (Salvia hispanica L.) seeds. Ind. Crops Prod. 2008, 28, 286–293. [Google Scholar] [CrossRef]
- Iglesias-Puig, E.; Haros, M. Evaluation of performance of dough and bread incorporating chia (Salvia hispanica L.). Eur. Food Res. Technol. 2013, 237, 865–874. [Google Scholar] [CrossRef]
- Oomah, B.D.; Kenaschuk, E.O.; Cui, W.; Mazza, G. Variation in the composition of water-soluble polysaccharides in flaxseed. J. Agric. Food Chem. 2002, 43, 1484–1488. [Google Scholar] [CrossRef]
- Liu, J.; Shim, Y.Y.; Tse, T.J.; Wang, Y.; Reaney, M.J.T. Flaxseed gum a versatile natural hydrocolloid for food and non-food applications. Trends Food Sci. Technol. 2018, 75, 146–157. [Google Scholar] [CrossRef]
- Wang, W.; Tai, F.; Chen, S. Optimizing protein extraction from plant tissues for enhanced proteomics analysis. J. Sep. Sci. 2008, 31, 2032–2039. [Google Scholar] [CrossRef] [PubMed]
- Kučka, M.; Harenčár, Ľ.; Ražná, K.; Nôžková, J.; Kowalczewski, P.Ł.; Deyholos, M.; Dziedzic, K.; Rybicka, I.; Zembrzuska, J.; Kačániová, M.; et al. Great potential of flaxseed mucilage. Eur. Food Res. Technol. 2023, 250, 877–893. [Google Scholar] [CrossRef]
- Lorenc, F.; Jarosova, M.; Bedrnicek, J.; Smetana, P.; Barta, J. Structural characterization and functional properties of flaxseed hydrocolloids and their application. Foods 2022, 11, 2304. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.W.Y.; Lei, B.; Li-Chan, E.C.Y. Isolation and structural characterization of the major protein fraction from norman flaxseed (Linum usitatissimum L.). Food Chem. 2005, 90, 271–279. [Google Scholar] [CrossRef]
- Rivas, R.N.; Dench, J.E.; Caygill, J.C. Nitrogen extractability of sesame (Sesamum indicum L.) seed and the preparation of two protein isolates. J. Sci. Food Agric. 2006, 32, 565–571. [Google Scholar] [CrossRef]
- Bojorquez-Velazquez, E.; Barrera-Pacheco, A.; Espitia-Rangel, E.; Herrera-Estrella, A.; Barba de la Rosa, A.P. Protein analysis reveals differential accumulation of late embryogenesis abundant and storage proteins in seeds of wild and cultivated amaranth species. BMC Plant Biol. 2019, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sun, Y.; Li, Y.; Wu, Q.; Wang, L. Identification and characterization of the seed storage proteins and related genes of Cannabis sativa L. Front. Nutr. 2021, 8, 678421. [Google Scholar]
- Bensaddek, D.; Nicolas, A.; Lamond, A.I. Signal enhanced proteomics: A biological perspective on dissecting the functional organisation of cell proteomes. Curr. Opin. Chem. Biol. 2019, 48, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.M.; Jordan, B.T.; Mehlferber, M.M.; Jeffery, E.D.; Chatzipantsiou, C.; Kaur, S.; Millikin, R.J.; Dai, Y.; Tiberi, S.; Castaldi, P.J.; et al. Enhanced protein isoform characterization through long-read proteogenomics. Genome Biol. 2022, 23, 69. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Huschek, G.; Homann, T.; Rawel, H.M. Effect of sample preparation on the detection and quantification of selected nuts allergenic proteins by LC-MS/MS. Molecules 2021, 26, 4698. [Google Scholar] [CrossRef] [PubMed]
- Salo-väänänen, P.P.; Koivistoinen, P.E. Determination of protein in foods: Comparison of net protein and crude protein (n × 6.25) values. Food Chem. 1996, 57, 27–31. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Wiedemann, L.T.; Nichani, K.; Henze, A.; Rawel, H.M. The optimization of the osborne extraction method for the fractionation and characterization of oat proteins. Separations 2024, 11, 271. [Google Scholar] [CrossRef]
- Sagu, S.T.; Landgraber, E.; Rackiewicz, M.; Huschek, G.; Rawel, H. Relative abundance of alpha-amylase/trypsin inhibitors in selected sorghum cultivars. Molecules 2020, 25, 5982. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Zimmermann, L.; Landgraber, E.; Homann, T.; Huschek, G.; Ozpinar, H.; Schweigert, F.J.; Rawel, H.M. Comprehensive characterization and relative quantification of alpha-amylase/trypsin inhibitors from wheat cultivars by targeted HPLC-MS/MS. Foods 2020, 9, 1448. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Rawel, H.M.; Rohn, S. Targeted bottom-up mass spectrometry approach for the relative quantification of post-translational modification of bovine kappa-casein during milk fermentation. Molecules 2022, 27, 5834. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Ulbrich, N.; Morche, J.R.; Nichani, K.; Ozpinar, H.; Schwarz, S.; Henze, A.; Rohn, S.; Rawel, H.M. Formation of cysteine adducts with chlorogenic acid in coffee beans. Foods 2024, 13, 1660. [Google Scholar] [CrossRef] [PubMed]

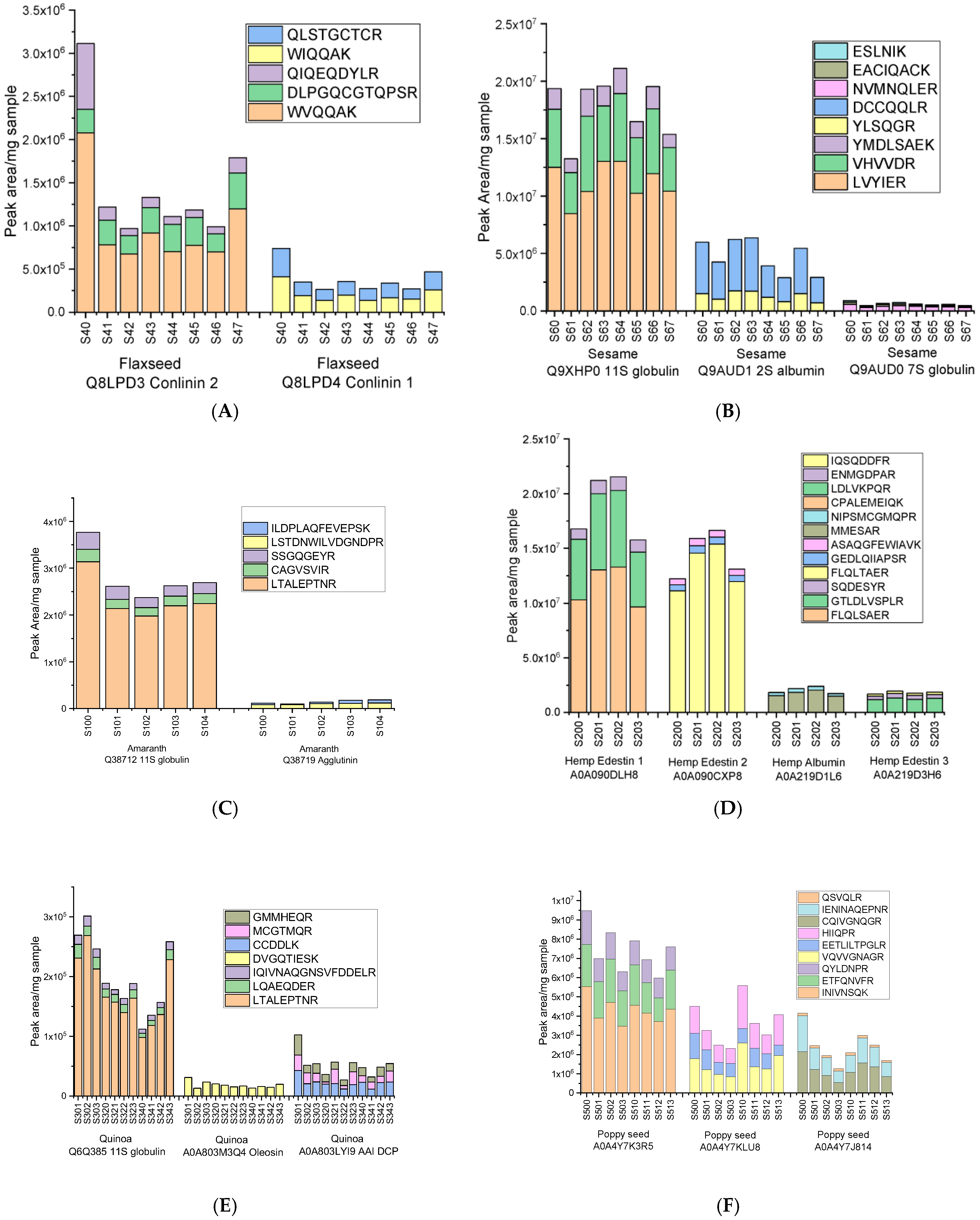
| Superseed Species | Protein Accession | Protein Name | Gene | Amino Acids | Mass (Da) | Marker Peptide | Intensity Range (Peak Area/mg) | Assigned Role |
|---|---|---|---|---|---|---|---|---|
| Flaxseed | Q8LPD3 | Conlinin 2 (2S albumin) | Cnl2 | 168 | 19,012 | WVQQAK | 675,000–2,100,000 | Quantifier |
| DLPGQCGTQPSR | 209,000–761,000 | Qualifier | ||||||
| QIQEQDYLR | 82,000–271,000 | Qualifier | ||||||
| Q8LPD4 | Conlinin 1 (2S albumin) | Cnl1 | 169 | 19,063 | WIQQAK | 137,000–410,000 | Quantifier | |
| QLSTGCTCR | 120,000–330,000 | Qualifier | ||||||
| Sesame | Q9XHP0 | 11S globulin seed storage | n.d. | 459 | 51,830 | LVYIER | 10,170,000–12,958,000 | Quantifier |
| VHVVDR | 3,819,000–6,576,000 | Qualifier | ||||||
| YMDLSAEK | 1,142,000–2,202,000 | Qualifier | ||||||
| Q9AUD1 | 2S albumin | LOC10 5174067 | 153 | 17,504 | YLSQGR | 620,000–1,655,000 | Quantifier | |
| DCCQQLR | 2,105,000–4,642,000 | Qualifier | ||||||
| Q9AUD0 | 7S globulin | n.d. | 585 | 67,069 | NVMNQLER | 216,000–486,000 | Quantifier | |
| EACIQACK | 46,000–179,000 | Qualifier | ||||||
| ESLNIK | 63,000–148,000 | Qualifier | ||||||
| Amaranth | Q38712 | 11S globulin | n.d. | 501 | 56,672 | LTALEPTNR | 2,139,000–3,134,000 | Qualifier |
| CAGVSVIR | 179,000–268,000 | Qualifier | ||||||
| SSGQGEYR | 216,000–362,000 | Quantifier | ||||||
| Q38719 | Agglutinin | AHA | 304 | 34,958 | LSTDNWILVDGNDPR | 80,000–107,000 | Quantifier | |
| ILDPLAQFEVEPSK | 18,000–69,000 | Qualifier | ||||||
| TYDGLVHIK | 23,000–58,000 | Qualifier | ||||||
| Hemp | A0A090DLH8 | Edestin 1 (11S globulin) | ede1A | 511 | 58,504 | FLQLSAER | 9,657,000–13,279,000 | Quantifier |
| GTLDLVSPLR | 4,987,000–7,015,000 | Qualifier | ||||||
| SQDESYR | 952,000–1,242,000 | Qualifier | ||||||
| A0A090CXP8 | Edestin 2 (11S globulin) | ede2C | 491 | 55,986 | FLQLTAER | 11,128,000–15,372,000 | Quantifier | |
| GEDLQIIAPSR | 535,000–676,000 | Qualifier | ||||||
| ASAQGFEWIAVK | 548,000–662,000 | Qualifier | ||||||
| A0A219D1L6 | Albumin (2S albumin) | Cs2S-1 | 142 | 16,742 | MMESAR | 1,487,000–2,039,000 | Quantifier | |
| NIPSMCGMQPR | 241,000–331,000 | Qualifier | ||||||
| CPALEMEIQK | 17,000–31,000 | Qualifier | ||||||
| A0A219D3H6 | Edestin 3 (11S globulin) | CsEde3B | 491 | 55,938 | LDLVKPQR | 1,325,000–1,190,000 | Quantifier | |
| ENMGDPAR | 331,000–416,000 | Qualifier | ||||||
| IQSQDDFR | 178,000–210,000 | Qualifier | ||||||
| Quinoa | Q6Q385 | 11S seed storage globulin | 11S | 480 | 53,641 | LTALEPTNR | 98,000–269,000 | Qualifier |
| LQAEQDER | 6000–23,000 | Qualifier | ||||||
| IQIVNAQGNSVFDDELR | 7000–17,000 | Quantifier | ||||||
| A0A803M3Q4 | Oleosin | n.d. | 185 | 19,083 | DVGQTIESK | 13,000–31,000 | Quantifier | |
| A0A803LYI9 | AAI domain-containing protein | n.d. | 129 | 15,358 | CCDDLK | 12,000–43,000 | Quantifier | |
| MCGTMQR | 3000–26,000 | Qualifier | ||||||
| GMMHEQR | 8000–34,000 | Qualifier | ||||||
| Poppy seed | A0A4Y7J814 | Protein-serine/threonine phosphatase | C5167_ 014922 | 853 | 95,547 | CQIVGNQGR | 540,000–2,148,000 | Qualifier |
| IENINAQEPNR | 602,000–1,870,000 | Quantifier | ||||||
| QSVQLR | 96,000–141,000 | Qualifier | ||||||
| A0A4Y7K3R5 | Cupin type-1 domain-containing protein | C5167_ 011661 | 468 | 53,356 | INIVNSQK | 3,473,000–5,527,000 | Quantifier | |
| ETFQNVFR | 1,232,000–2,253,000 | Qualifier | ||||||
| QYLDNPR | 996,000–1,748,000 | Qualifier | ||||||
| A0A4Y7KLU8 | Cupin type-1 domain-containing protein | C5167_ 048617 | 514 | 57,714 | VQVVGNAGR | 846,000–2,601,000 | Quantifier | |
| EETLILTPGLR | 546,000–1,332,000 | Qualifier | ||||||
| HIIQPR | 777,000–2,231,000 | Qualifier |
| Protein ID | Peptide | Validation Criterion | ||||
|---|---|---|---|---|---|---|
| IS Recovery (%) | Repeatability (% RSD) | Reproducibility (% RSD) | Linearity (R2) | |||
| Amaranth | Q38712 | SSGQGEYR | 89.4 ± 1.4 | 0.2 | 1.1 | 0.9984 |
| Q38719 | LSTDNWILVDGNDPR | 2.5 | 5.9 | 0.9989 | ||
| Flaxseed | Q8LPD3 | WVQQAK | 90.2 ± 1.7 | 0.1 | 3.3 | 0.9941 |
| Q8LPD4 | WIQQAK | 0.7 | 5.5 | 0.9982 | ||
| Hemp | A0A090CXP8 | FLQLTAER | 97.6 ± 0.9 | 0.4 | 2.4 | 0.9986 |
| A0A090DLH8 | FLQLSAER | 0.7 | 4.8 | 0.999 | ||
| A0A219D1L6 | MMESAR | 0.2 | 0.8 | 0.9922 | ||
| Poppy | A0A4Y7J814 | IENINAQEPNR | 93.7 ± 0.7 | 2.1 | 7.0 | 0.9993 |
| A0A4Y7K3R5 | INIVNSQK | 0.3 | 4.9 | 0.9985 | ||
| A0A4Y7KLU8 | VQVVGNAGR | 0.7 | 3.3 | 0.9967 | ||
| Quinoa | A0A803LYI9 | CCDDLK | 90.0 ± 4.3 | 0.4 | 5.5 | 0.9985 |
| A0A803M3Q4 | DVGQTIESK | 0.8 | 8.2 | 0.9977 | ||
| Q6Q385 | IQIVNAQGNSVFDDELR | 5.8 | 7.5 | 0.9988 | ||
| Sesame | Q9AUD1 | YLSQGR | 97.1 ± 1.1 | 8.5 | 6.4 | 0.9975 |
| Q9XHP0 | LVYIER | 1.2 | 1.2 | 0.9975 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagu, S.T.; Schnepf, B.; Stenzel, P.; Nichani, K.; Erban, A.; Kopka, J.; Rawel, H.M.; Henze, A. Discovery of Species-Specific Peptide Markers for Superseed Authentication Using Targeted LC-MS/MS Proteomics. Molecules 2025, 30, 2993. https://doi.org/10.3390/molecules30142993
Sagu ST, Schnepf B, Stenzel P, Nichani K, Erban A, Kopka J, Rawel HM, Henze A. Discovery of Species-Specific Peptide Markers for Superseed Authentication Using Targeted LC-MS/MS Proteomics. Molecules. 2025; 30(14):2993. https://doi.org/10.3390/molecules30142993
Chicago/Turabian StyleSagu, Sorel Tchewonpi, Beatrice Schnepf, Peter Stenzel, Kapil Nichani, Alexander Erban, Joachim Kopka, Harshadrai M. Rawel, and Andrea Henze. 2025. "Discovery of Species-Specific Peptide Markers for Superseed Authentication Using Targeted LC-MS/MS Proteomics" Molecules 30, no. 14: 2993. https://doi.org/10.3390/molecules30142993
APA StyleSagu, S. T., Schnepf, B., Stenzel, P., Nichani, K., Erban, A., Kopka, J., Rawel, H. M., & Henze, A. (2025). Discovery of Species-Specific Peptide Markers for Superseed Authentication Using Targeted LC-MS/MS Proteomics. Molecules, 30(14), 2993. https://doi.org/10.3390/molecules30142993








