Selective Cleaning Enhances Machine Learning Accuracy for Drug Repurposing: Multiscale Discovery of MDM2 Inhibitors
Abstract
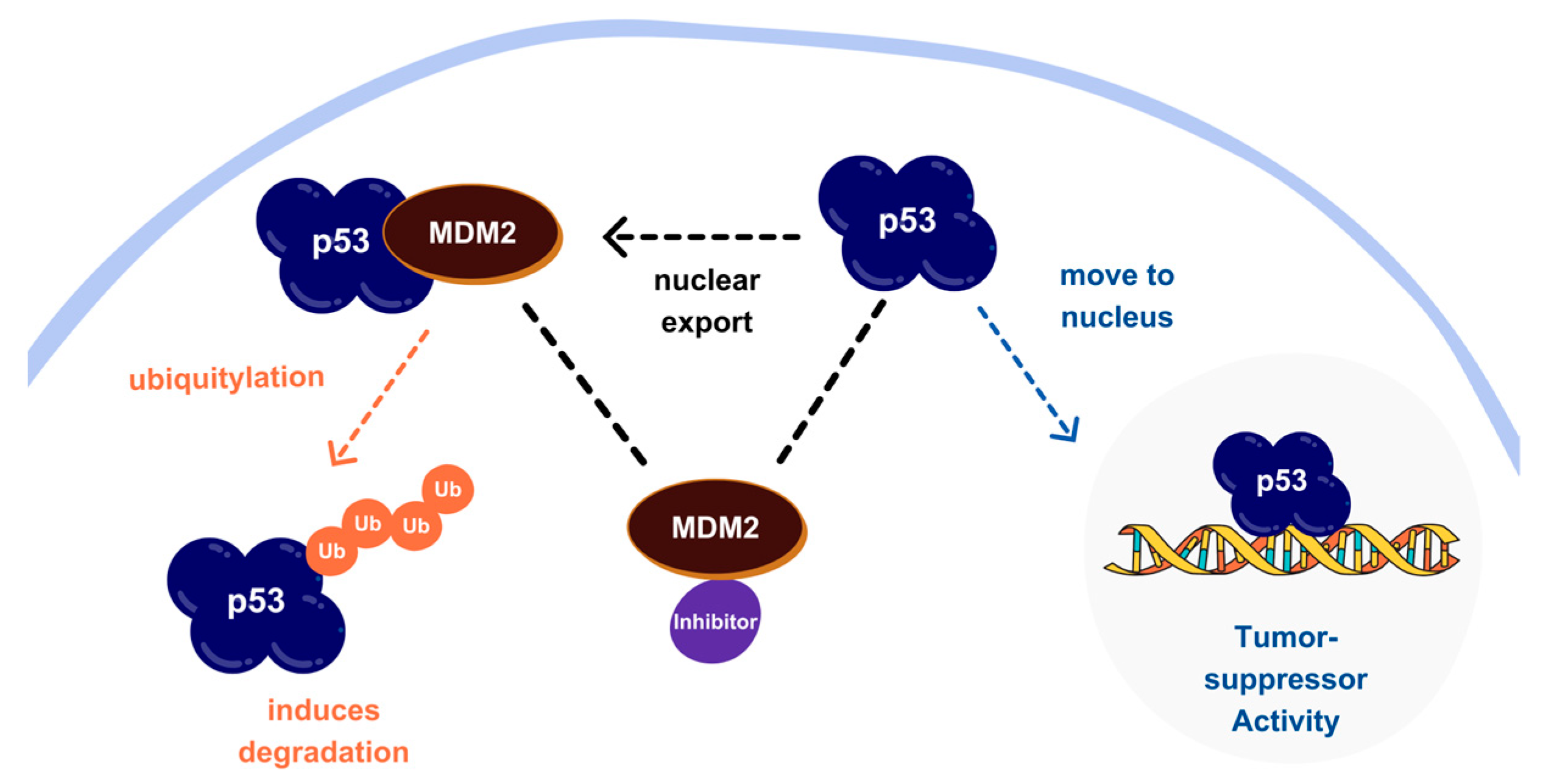
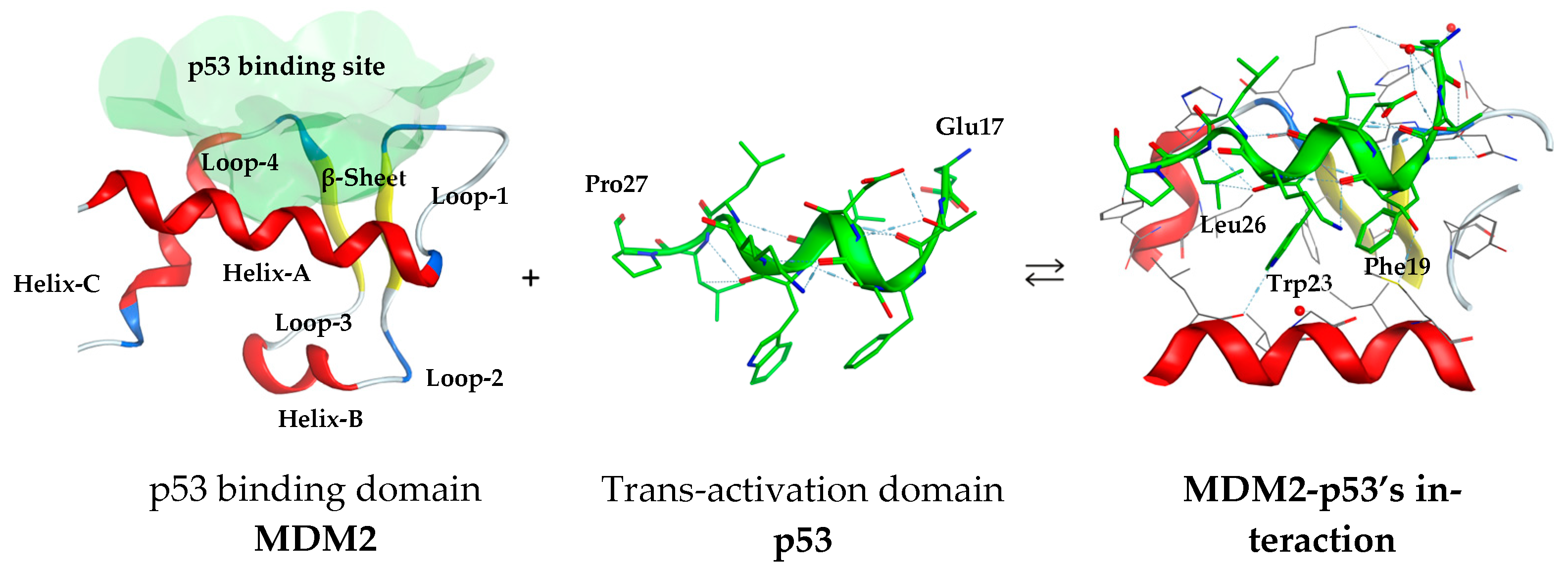
1. Introduction
2. Results and Discussion
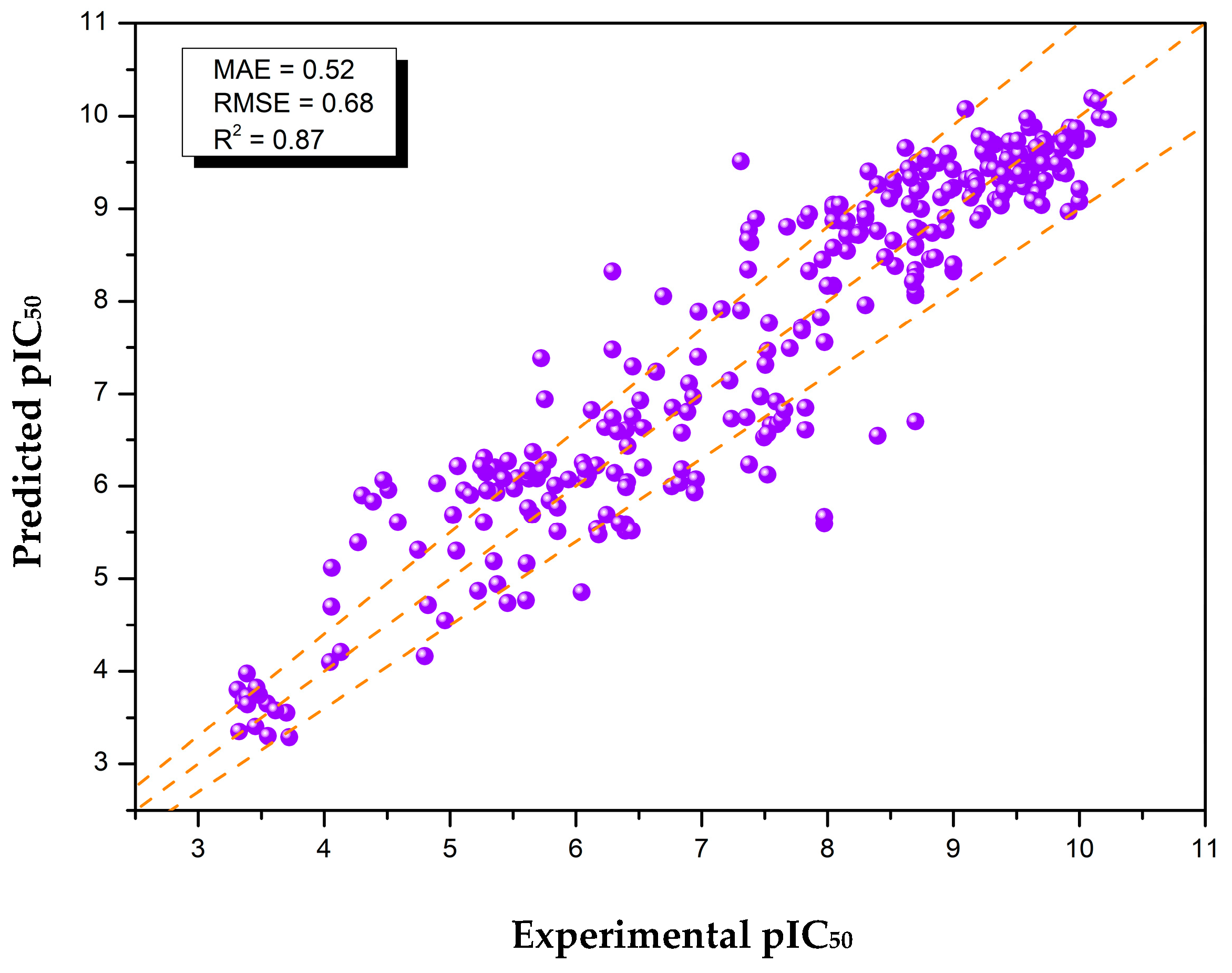
2.1. Optimized Data and ML Model for Virtual Screening
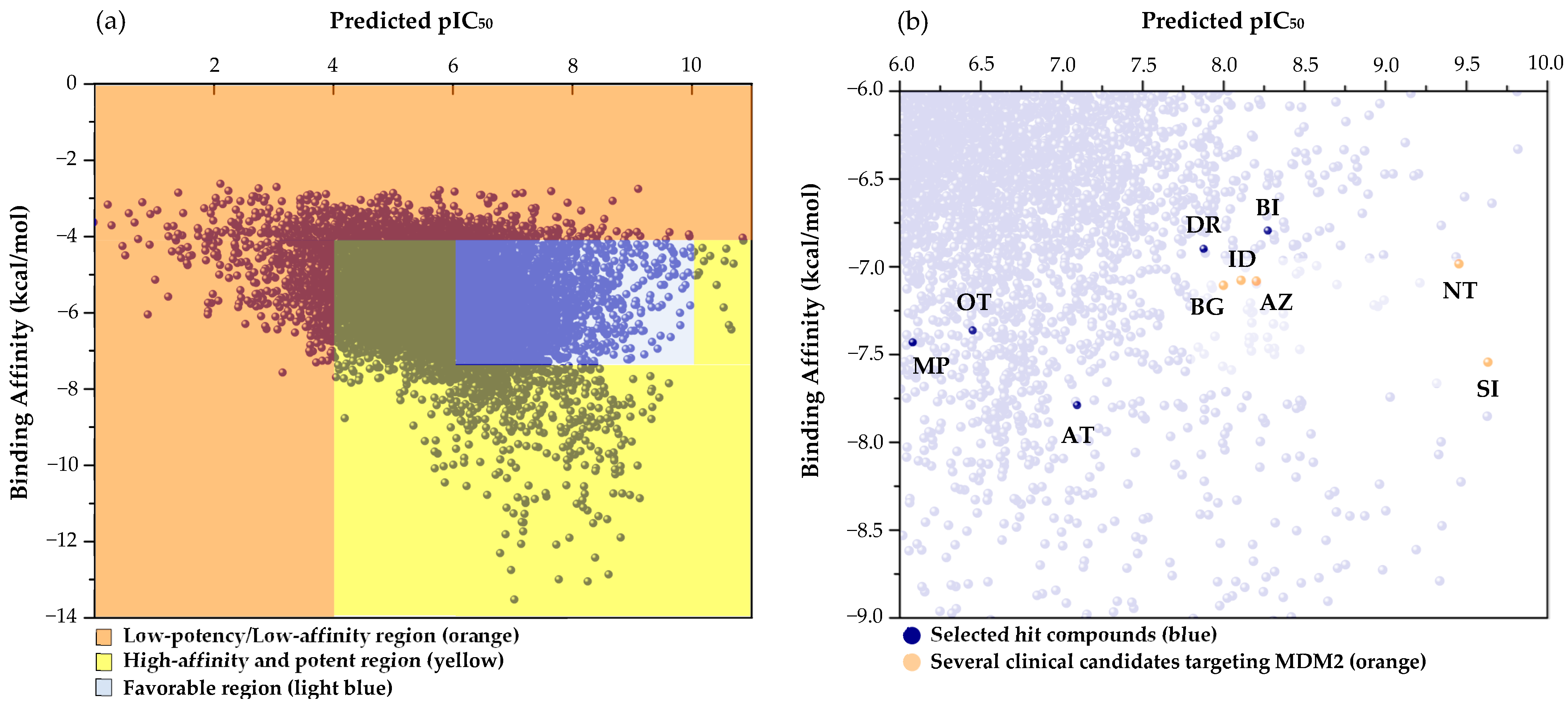
2.2. Integrated Virtual Screening Hits for Potential MDM2 Inhibitors
2.3. Affinity and Site-Selectivity Validation of the Top Hits Using Redocking Simulations
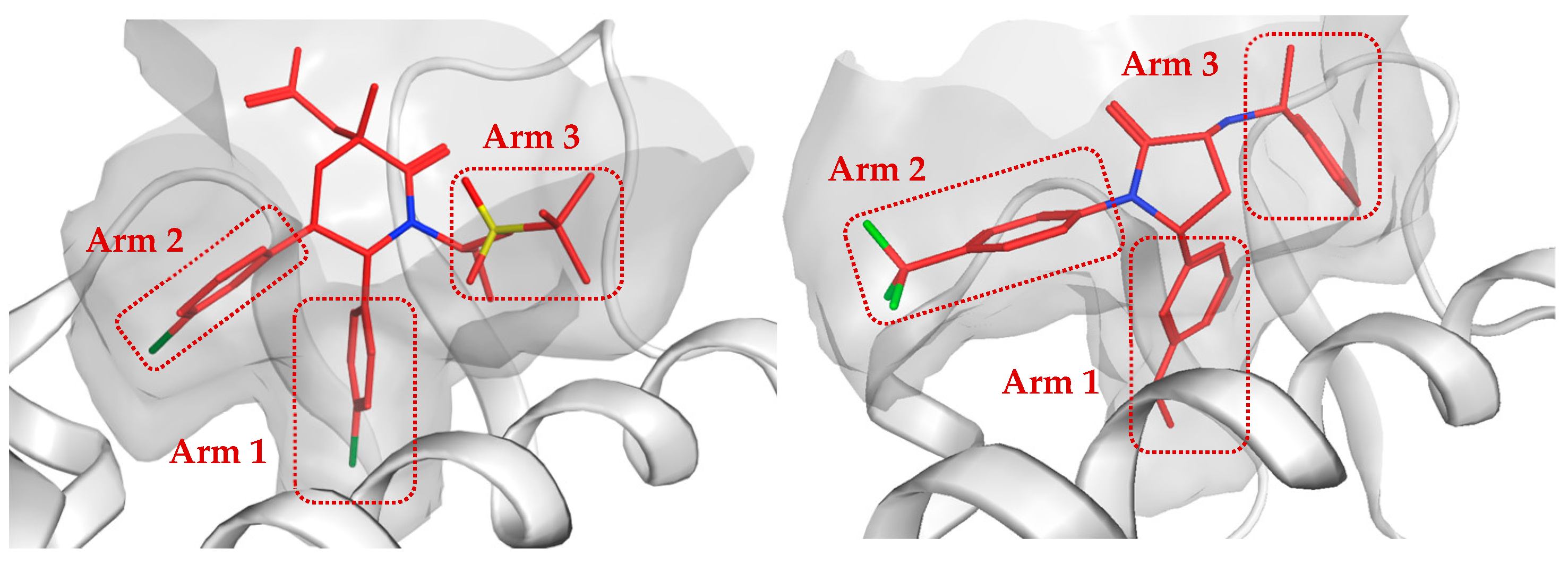
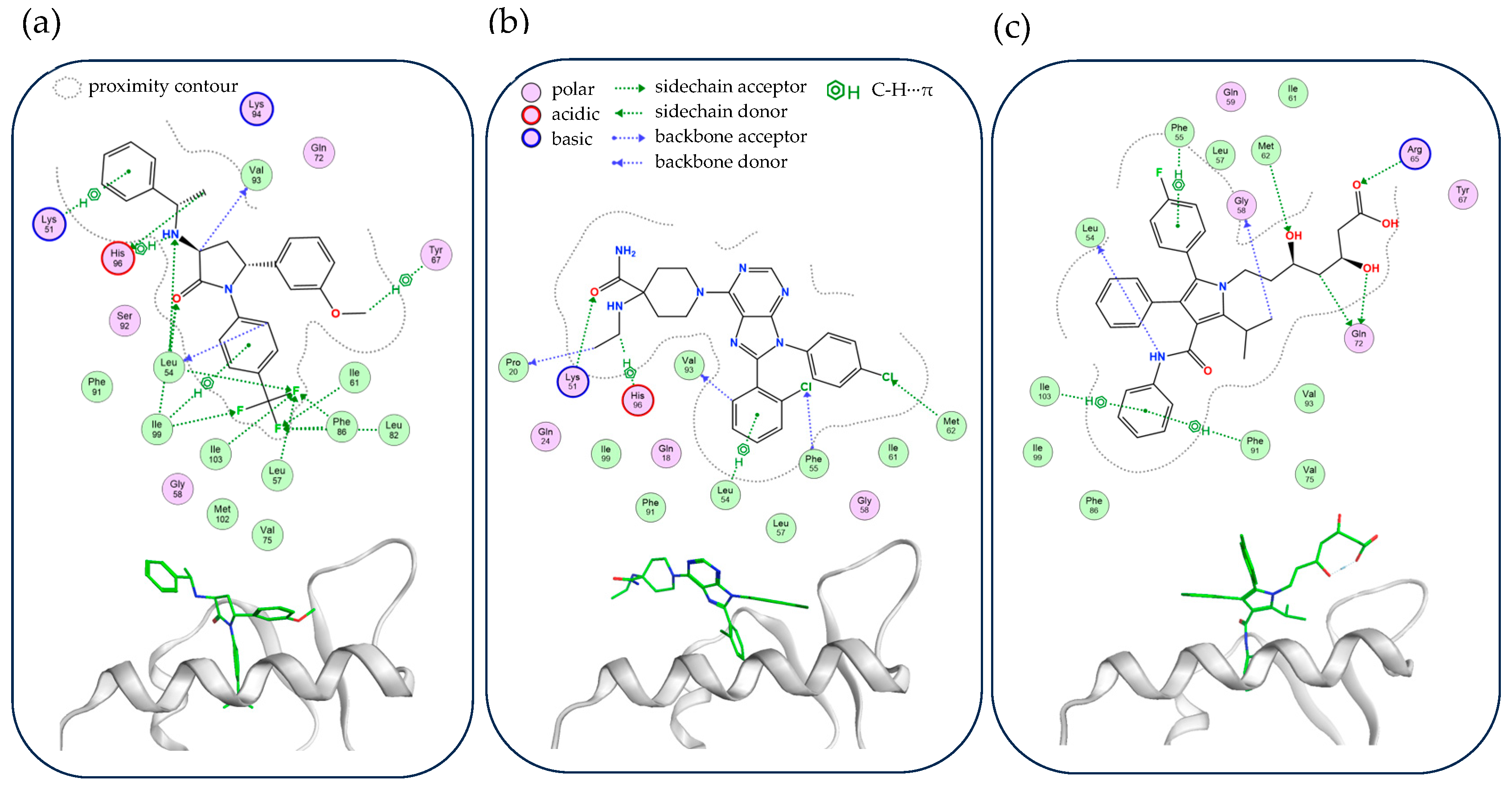
2.4. Analysis of Protein–Ligand Interactions in Optimized MDM2-H2 Compound Complexes
2.5. Binding Stability and Deep Pocket Insertion of MDM2-H2 Compound Complexes from MD Simulations
3. Methods
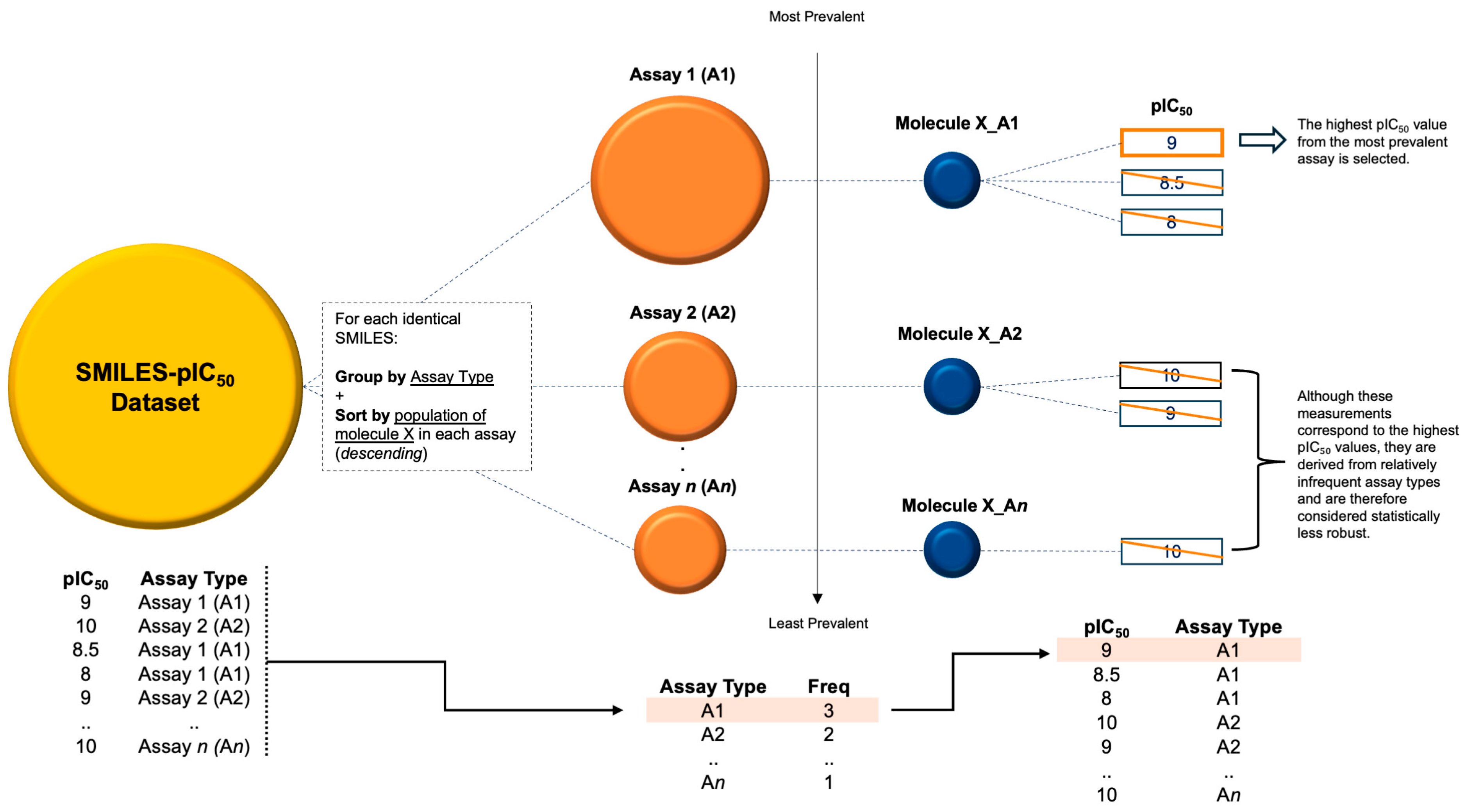
3.1. Training Dataset Preparation
3.2. ML Selection and Optimization
- Feature Extraction: We extracted a set of 300 two-dimensional molecular descriptors—computed via Chemprop’s integrated RDKit feature generator—for use as input features in our scikit-learn models.
- Data Splitting: The dataset was divided into training (D1_training) and validation sets, with 20% allocated for validation.
- Model Training: Various ML models were trained using the scikit-learn 1.4.0 package [53], including k-nearest neighbor, decision tree, random forest, AdaBoost, XGBoost, gradient boosting, histogram gradient boosting, stochastic gradient descent, and multi-layer perceptron. Additionally, deep learning models were trained using ChemProp 1.6.1 [41], leveraging ChemProp’s neural fingerprints and pIC50 target values.
- Hyperparameter Optimization: Each model underwent hyperparameter tuning to enhance performance, and different data optimization procedures were compared using the validation set.
- Final Evaluation: The best performing models were tested on the independent D1_testing dataset to assess their predictive accuracy.
3.3. Repurposing Dataset Preparation
3.4. ML-Based Virtual Screening
3.5. Structure-Based Virtual Screening (SBVS)
3.6. Redocking Analysis and Toxicity Prediction
3.7. ONIOM Simulation
3.8. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCL2 | B-Cell Lymphoma-2 Protein |
| COVID-19 | Coronavirus Disease 2019 |
| FDA | The United States Food and Drug Administration |
| IC50 | Half-Maximal Inhibitory Concentration |
| MDM2 | Mouse Double Minute 2 Protein |
| MDM4 | Mouse Double Minute 4 Protein |
| SMILES | Simplified Molecular Input Line Entry System |
References
- “Estimated Number of Deaths From 2022 to 2025, Both Sexes, Age [0-85+]”, Cancer Tomorrow. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/isotype?types=1&single_unit=500000&years=2025 (accessed on 19 November 2024).
- “Worldwide Cancer Data”, World Cancer Research Fund International. Available online: https://www.wcrf.org/cancer-trends/worldwide-cancer-data/ (accessed on 26 February 2024).
- Reifenberger, G.; Liu, L.; Ichimura, K.; Ee, S.; Collins, V. Amplification and Overexpression of the MDM2 Gene in a Subset of Human Malignant Gliomas without P53 Mutations. Cancer Res. 1993, 53, 2736–2739. [Google Scholar] [PubMed]
- Watanabe, T.; Hotta, T.; Ichikawa, A.; Kinoshita, T.; Nagai, H.; Uchida, T.; Murate, T.; Saito, H. The MDM2 Oncogene Overexpression in Chronic Lymphocytic Leukemia and Low-Grade Lymphoma of B-Cell Origin. Blood 1994, 84, 3158–3165. [Google Scholar] [CrossRef] [PubMed]
- Dembla, V.; Somaiah, N.; Barata, P.; Hess, K.; Fu, S.; Janku, F.; Karp, D.D.; Naing, A.; Piha-Paul, S.A.; Subbiah, V.; et al. Prevalence of MDM2 Amplification and Coalterations in 523 Advanced Cancer Patients in the MD Anderson Phase 1 Clinic. Oncotarget 2018, 9, 33232–33243. [Google Scholar] [CrossRef] [PubMed]
- Horie, S.; Endo, K.; Kawasaki, H.; Terada, T. Overexpression of MDM2 Protein in Intrahepatic Cholangiocarcinoma: Relationship with P53 Overexpression, Ki-67 Labeling, and Clinicopathological Features. Virchows Arch. 2000, 437, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Gluck, I.; Simon, A.J.; Catane, R.; Pfeffer, R.; Schachter, J.; Rechavi, G.; Bar, J. Germline Analysis of Thymidine/Guanidine Polymorphism at Position 309 of the Mdm2 Promoter in Malignant Melanoma Patients. Melanoma Res. 2009, 19, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Konopleva, M.; Martinelli, G.; Daver, N.; Papayannidis, C.; Wei, A.; Higgins, B.; Ott, M.; Mascarenhas, J.; Andreeff, M. MDM2 Inhibition: An Important Step Forward in Cancer Therapy. Leukemia 2020, 34, 2858–2874. [Google Scholar] [CrossRef] [PubMed]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Elenbaas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the MDM2 Oncoprotein Bound to the p53 Tumor Suppressor Transactivation Domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Lee, S.H.; Kim, D.H.; Ahn, M.J.; Kim, J.S.; Woo, J.Y.; Torizawa, T.; Kainosho, M.; Han, K.H. Structural Details on mdm2-p53 Interaction. J. Biol. Chem. 2005, 280, 38795–38802. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Aguilar, A.; Bernard, D.; Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 2015, 58, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Verstovsek, S.; Al-Ali, H.K.; Mascarenhas, J.; Perkins, A.; Vannucchi, A.M.; Mohan, S.R.; Scott, B.L.; Woszczyk, D.; Koschmieder, S.; García-Delgado, R.; et al. BOREAS: A global, phase III study of the MDM2 inhibitor navtemadlin (KRT-232) in relapsed/refractory myelofibrosis. Future Oncol. 2022, 18, 4059–4069. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, P.; Beckermann, B.M.; Catalani, O.; Esteve, J.; Gamel, K.; Konopleva, M.Y.; Martinelli, G.; Monnet, A.; Papayannidis, C.; Park, A.; et al. MIRROS: A randomized, placebo-controlled, Phase III trial of cytarabine ± idasanutlin in relapsed or refractory acute myeloid leukemia. Future Oncol. 2020, 16, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Schuetze, S.M.; Jayadeva, G.; Santoro, M. 140TiP Brightline-4: A phase III open-label, single-arm, multicentre study to assess the safety and efficacy of brigimadlin (BI 907828), an MDM2-p53 antagonist, in patients with treatment-naïve or pretreated advanced dedifferentiated liposarcoma. ESMO Open 2024, 9, 102565. [Google Scholar] [CrossRef]
- Daver, N.G.; Wei, A.H.; Stein, E.M.; DeAngelo, D.J.; Pathak, D.; Xu, Y.; Grzesiak, S.; Venditti, A. PB1849: Trial In Progress: Phase Ib/Ii Study Of Siremadlin In Combination With Venetoclax + Azacitidine In Patients With Acute Myeloid Leukemia (Aml) Who Are Ineligible For Intensive Chemotherapy. HemaSphere 2022, 6, 1729–1730. [Google Scholar] [CrossRef]
- Shaheen, M.F.; Segar, J.M.; Chmielowski, B.; Drabick, J.J.; McKean, M.; Reeves, J.A.; Karapetis, C.S.; Orloff, M.M.; Tolcher, A.W.; Beck, J.T.T.; et al. A Phase 2 Study of Alrizomadlin (APG-115) in Combination with Pembrolizumab in Patients with Unresectable or Metastatic Cutaneous Melanoma That Has Failed Immuno-Oncologic (IO) Drugs. J. Clin. Oncol. 2023, 41, 9559. [Google Scholar] [CrossRef]
- Bauer, S.; Demetri, G.; Jeay, S.; Dummer, R.; Guerreiro, N.; Tan, D.S.; Kumar, A.; Meille, C.; Van Bree, L.; Halilovic, E.; et al. A Phase I, Open-Label, Multi-Center, Dose Escalation Study of Oral NVP-CGM097, a P53/HDM2-Protein-Protein Interaction Inhibitor, in Adult Patients with Selected Advanced Solid Tumors. Ann. Oncol. 2016, 27, vi116. [Google Scholar] [CrossRef]
- Gounder, M.M.; Bauer, T.M.; Schwartz, G.K.; Weise, A.M.; Lorusso, P.; Kumar, P.; Tao, B.; Hong, Y.; Patel, P.; Lu, Y.; et al. A First-in-Human Phase I Study of Milademetan, an MDM2 Inhibitor, in Patients With Advanced Liposarcoma, Solid Tumors, or Lymphomas. J. Clin. Oncol. 2023, 41, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Andreeff, M.; Kelly, K.R.; Yee, K.; Assouline, S.; Strair, R.; Popplewell, L.; Bowen, D.; Martinelli, G.; Drummond, M.W.; Vyas, P.; et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 2016, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Guan, S.; Zhao, Y.; Yu, Y.; Wang, Y.; Shi, Y.; Mao, X.; Yang, K.L.; Sun, W.; Xu, X.; et al. Novel MDM2 Inhibitor SAR405838 (MI-773) Induces P53-Mediated Apoptosis in Neuroblastoma. Oncotarget 2016, 7, 82757–82769. [Google Scholar] [CrossRef] [PubMed]
- Walker, I.; Newell, H. Do Molecularly Targeted Agents in Oncology Have Reduced Attrition Rates? Nat. Rev. Drug Discov. 2009, 8, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.; Pearson, A.D.J. How Can Attrition Rates Be Reduced in Cancer Drug Discovery? Expert Opin. Drug Discov. 2013, 8, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, L.; Kirk, R. High Drug Attrition Rates—Where Are We Going Wrong? Nat. Rev. Clin. Oncol. 2011, 8, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Saikin, S.K.; Kreisbeck, C.; Sheberla, D.; Becker, J.S.; Aspuru-Guzik, A. Closed-Loop Discovery Platform Integration Is Needed for Artificial Intelligence to Make an Impact in Drug Discovery. Expert Opin. Drug Discov. 2019, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A Review of Computational Drug Repositioning: Strategies, Approaches, Opportunities, Challenges, and Directions. J. Cheminform. 2020, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Yoon, H.; Kim, G.; Lee, H.; Lee, Y. Revolutionizing Medicinal Chemistry: The Application of Artificial Intelligence (AI) in Early Drug Discovery. Pharmaceuticals 2023, 16, 1259. [Google Scholar] [CrossRef] [PubMed]
- Schuhmacher, A.; Hinder, M.; von Stegmann und Stein, A.; Hartl, D.; Gassmann, O. Analysis of pharma R&D productivity—A new perspective needed. Drug Discov. Today 2023, 28, 103726. [Google Scholar] [CrossRef]
- Kulkarni, V.S.; Alagarsamy, V.; Solomon, V.R.; Jose, P.A.; Murugesan, S. Drug Repurposing: An Effective Tool in Modern Drug Discovery. Russ. J. Bioorganic Chem. 2023, 49, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.; Mäser, P.; Mahmoud, A.B. Drug Repurposing in the Chemotherapy of Infectious Diseases. Molecules 2024, 29, 635. [Google Scholar] [CrossRef] [PubMed]
- Gil, C.; Martinez, A. Is Drug Repurposing Really the Future of Drug Discovery or Is New Innovation Truly the Way Forward? Expert Opin. Drug Discov. 2021, 16, 829–831. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Byrne, R.; Schneider, G.; Yang, S. Concepts of Artificial Intelligence for Computer-Assisted Drug Discovery. Chem. Rev. 2019, 119, 10520–10594. [Google Scholar] [CrossRef] [PubMed]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef] [PubMed]
- Warner, W.A.; Sanchez, R.; Dawoodian, A.; Li, E.; Momand, J. Identification of FDA-approved Drugs that Computationally Bind to MDM2. Chem. Biol. Drug Des. 2012, 80, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Zawacka-Pankau, J.E. The Undervalued Avenue to Reinstate Tumor Suppressor Functionality of the P53 Protein Family for Improved Cancer Therapy-Drug Repurposing. Cancers 2020, 12, 2717. [Google Scholar] [CrossRef] [PubMed]
- Munisamy, M.; Mukherjee, N.; Thomas, L.; Pham, A.T.; Shakeri, A.; Zhao, Y.; Kolesar, J.; Rao, P.P.N.; Rangnekar, V.M.; Rao, M. Therapeutic Opportunities in Cancer Therapy: Targeting the P53-MDM2/MDMX Interactions. Am. J. Cancer Res. 2021, 11, 5762–5781. [Google Scholar] [PubMed]
- Ghafoor, N.A.; Yildiz, A. Targeting MDM2—p53 Axis through Drug Repurposing for Cancer Therapy: A Multidisciplinary Approach. ACS Omega 2023, 8, 34583–34596. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Hu, W.; Wang, Y.; Chen, W.; Wen, H.; Liu, J.; Li, W.; Wang, B. Searching for Novel MDM2/MDMX Dual Inhibitors through a Drug Repurposing Approach. J. Enzyme Inhib. Med. Chem. 2023, 39, 2288810. [Google Scholar] [CrossRef] [PubMed]
- Blanco-González, A.; Cabezón, A.; Seco-González, A.; Conde-Torres, D.; Antelo-Riveiro, P.; Piñeiro, Á.; Garcia-Fandino, R. The Role of AI in Drug Discovery: Challenges, Opportunities, and Strategies. Pharmaceuticals 2023, 16, 891. [Google Scholar] [CrossRef] [PubMed]
- Aittokallio, T. What Are the Current Challenges for Machine Learning in Drug Discovery and Repurposing? Expert Opin. Drug Discov. 2022, 17, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Heid, E.; Greenman, K.P.; Chung, Y.; Li, S.; Graff, D.E.; Florence, H.; Wu, H.; Green, W.H.; Mcgill, C.J. Chemprop: A Machine Learning Package for Chemical Property Prediction. J. Chem. Inf. Model. 2024, 64, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the 5th International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar] [CrossRef]
- Hua, T.; Vemuri, K.; Pu, M.; Qu, L.; Han, G.W.; Wu, Y.; Zhao, S.; Shui, W.; Li, S.; Korde, A.; et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 2016, 167, 750–762.e14. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lozano, G. Molecular Pathways: Targeting Mdm2 and Mdm4 in Cancer Therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.C.; Francoz, S.; Maetens, M.; Wahl, G.; Toledo, F.; Lozano, G. Keeping P53 in Check: Essential and Synergistic Functions of Mdm2 and Mdm4. Cell Death Differ. 2006, 13, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Konopleva, M.; Samudio, I.J.; Schober, W.D.; Bornmann, W.G.; Andreeff, M. Concomitant Inhibition of MDM2 and Bcl-2 Protein Function Synergistically Induce Mitochondrial Apoptosis in AML. Cell Cycle 2006, 5, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Akmal, M.F.; Wahyuningrum, D.; Ivansyah, A.L. Theoretical Insight and Molecular Recognition of Oxatub[4]Arene-Based Organic Macrocycle as a Supramolecular Host for Antipsychotic Drug Risperidone. J. Mol. Liq. 2022, 366, 120195. [Google Scholar] [CrossRef]
- Zdrazil, B.; Felix, E.; Hunter, F.; Manners, E.J.; Blackshaw, J.; Corbett, S.; de Veij, M.; Ioannidis, H.; Lopez, D.M.; Mosquera, J.F.; et al. The ChEMBL Database in 2023: A Drug Discovery Platform Spanning Multiple Bioactivity Data Types and Time Periods. Nucleic Acids Res. 2024, 52, D1180–D1192. [Google Scholar] [CrossRef] [PubMed]
- The Pandas Development Team. pandas-dev/pandas: Pandas, v2.1.4; Zenodo: Geneva, Switzerland, 2023. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; ISBN 1441412697. [Google Scholar]
- “MolVS: Molecule Validation and Standardization”. GitHub. Available online: https://github.com/mcs07/MolVS?tab=readme-ov-file (accessed on 7 August 2024).
- “pandasql”. Python Package Index (PyPI). Available online: https://pypi.org/project/pandasql/#description (accessed on 7 August 2024).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Ursu, O.; Holmes, J.; Knockel, J.; Bologa, C.G.; Yang, J.J.; Mathias, S.L.; Nelson, S.J.; Oprea, T.I. DrugCentral: Online Drug Compendium. Nucleic Acids Res. 2017, 45, D932–D939. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef] [PubMed]
- Kallen, J.; Izaac, A.; Chau, S.; Wirth, E.; Schoepfer, J.; Mah, R.; Schlapbach, A.; Stutz, S.; Vaupel, A.; Guagnano, V.; et al. Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes. ChemMedChem 2019, 14, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group ULC. Molecular Operating Environment (MOE); Chemical Computing Group ULC: Montreal, QC, Canada, 2024. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Houston, D.R.; Walkinshaw, M.D. Consensus Docking: Improving the Reliability of Docking in a Virtual Screening Context. J. Chem. Inf. Model. 2013, 53, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Z.; Xue, H.; Hu, N.; Liu, Y.; Sun, H.; Yu, D.; Qin, L.; Shi, G.; Wang, F.; et al. Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2. J. Med. Chem. 2024, 67, 7836–7858. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Kemmler, E.; Dunkel, M.; Preissner, R. ProTox 3.0: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2024, 52, W513–W520. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.a.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.a.; Nakatsuji, H.; et al. Gaussian 16; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Chai, J.-D.; Head-Gordon, M. Long-Range Corrected Hybrid Density Functionals with Damped Atom–Atom Dispersion Corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [PubMed]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A Complete Basis Set Model Chemistry. I. The Total Energies of Closed-Shell Atoms and Hydrides of the First-Row Elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Vela, A.; Gázquez, J.L. Extended Hückel Parameters from Density Functional Theory. J. Phys. Chem. 1988, 92, 5688–5693. [Google Scholar] [CrossRef]
- Sturgeon, J.B.; Laird, B.B. Symplectic Algorithm for Constant-Pressure Molecular Dynamics Using a Nosé–Poincaré Thermostat. J. Chem. Phys. 2000, 112, 3474–3482. [Google Scholar] [CrossRef]
- Khelfaoui, H.; Harkati, D.; Saleh, B.A. Molecular Docking, Molecular Dynamics Simulations and Reactivity, Studies on Approved Drugs Library Targeting ACE2 and SARS-CoV-2 Binding with ACE2. J. Biomol. Struct. Dyn. 2020, 39, 7246–72621. [Google Scholar] [CrossRef] [PubMed]


| Name | Max. Clinical Phase | Type of Compounds | pIC50 | Ref |
|---|---|---|---|---|
| Navtemadlin (NV) | III | Piperidine/-one | 9.22 | [12] |
| Idasanutlin (ID) | III | Nutlins | 8.22 | [13] |
| Brigimadlin (BG) | III | Spiro-oxindoles | 7.92 | [14] |
| Siremadlin (SI) | II | Pyrroloimidazolone | 9.64 | [15] |
| Alrizomadlin (AZ) | II | Spiro-oxindoles | 8.42 | [16] |
| CGM097 | I | Isoquinoline | 8.77 | [17] |
| Milademetan | I | Spiro-oxindoles | 7.75 | [18] |
| RG7112 | I | Nutlins | 7.74 | [19] |
| MI-773 | I | Spiro-oxindoles | 7.00 | [20] |
| ML Model | RMSE | R2 |
|---|---|---|
| k-Nearest Neighbor | 0.84 | 0.77 |
| Decision Tree | 0.99 | 0.69 |
| Random Forest | 0.82 | 0.78 |
| AdaBoost | 0.99 | 0.69 |
| XGBoost | 0.86 | 0.76 |
| Gradient Boosting | 0.81 | 0.79 |
| Histogram Gradient Boosting | 0.81 | 0.79 |
| Stochastic Gradient Descent | 0.93 | 0.72 |
| Multi-Layer Perceptron | 0.79 | 0.80 |
| Graph Convolutional Neural Network | 0.73 | 0.84 |
| Data Optimization | Number of Data Points | RMSE | R2 |
|---|---|---|---|
| Standard Cleaning a | 1926 | 0.74 | 0.81 |
| Standard Cleaning b | 3419 | 0.73 | 0.84 |
| Standard Cleaning + Filtering | 2954 | 0.64 | 0.86 |
| Full Optimization (With Selective Cleaning) | 2954 | 0.58 | 0.87 |
| Molecule Name | Mechanism of Action | Max Clinical Phase | Two-Dimensional Structure | Binding Affinity (kcal/mol) | Predicted pIC50 |
|---|---|---|---|---|---|
| MePPEP (MP) | CB1 antagonist | II | 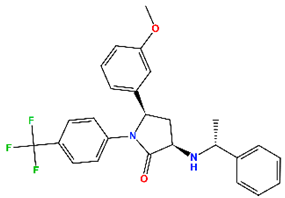 | −7.43 | 6.08 |
| Otenabant (OT) | CB1 antagonist | III | 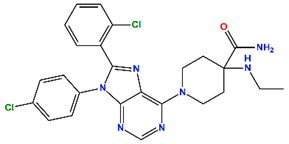 | −7.36 | 6.45 |
| Atorvastatin (AT) | HMG-CoA reductase inhibitor | Approved | 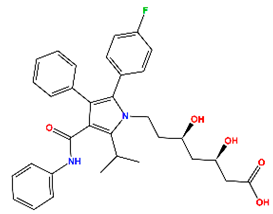 | −7.79 | 7.10 |
| BIRT-2584 (BI) | ITGAL and ITGB2 antagonist | II | 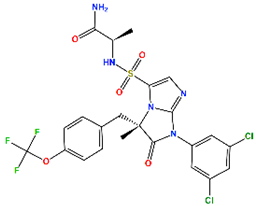 | −6.79 | 8.27 |
| Drinabant (DR) | CB1 antagonist | Preclinical | 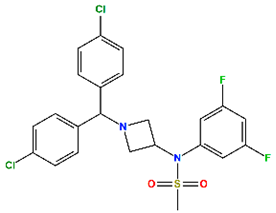 | −6.90 | 7.88 |
| Ligand | Docking Score | ||||||
|---|---|---|---|---|---|---|---|
| MOE (kcal/mol) | Autodock Vina (MDM2; kcal/mol) | GOLD (MDM2) | |||||
| MDM4 | BCL2 | MDM2_p1 | MDM2_p2 | p1−p2 | |||
| MP | −9.6 | −6.6 | −7.6 | −7.1 | 0.5 | −7.7 | 72.7 |
| OT | −9.4 | −7.4 | −7.5 | −6.9 | 0.5 | −8.1 | 64.0 |
| AT | −11.1 | −7.3 | −8.4 | −8.1 | 0.3 | −7.8 | 61.4 |
| BI | −10.1 | −7.1 | −7.4 | −7.5 | −0.1 | −8.0 | 65.4 |
| DR | −9.3 | −6.4 | −7.2 | −6.8 | 0.4 | −8.0 | 72.1 |
| Residue | MP | OT | AT | Type of Interaction |
|---|---|---|---|---|
| Arg65 | +++ | - | +++ | Hydrogen bond |
| Leu54 a | + | +++ | +++ | Hydrogen bond |
| Gln72 a | + | - | ++ | Hydrogen bond |
| Lys51 | - | +++ | - | Hydrogen bond |
| Met62 a | - | + | ++ | C-H···Cl/C-H···O interaction |
| Ile61 | +++ | ++ | +++ | C-H···F interaction |
| Ile99 b | + | ++ | ++ | C-H···π interaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akmal, M.F.; Wong, M.W. Selective Cleaning Enhances Machine Learning Accuracy for Drug Repurposing: Multiscale Discovery of MDM2 Inhibitors. Molecules 2025, 30, 2992. https://doi.org/10.3390/molecules30142992
Akmal MF, Wong MW. Selective Cleaning Enhances Machine Learning Accuracy for Drug Repurposing: Multiscale Discovery of MDM2 Inhibitors. Molecules. 2025; 30(14):2992. https://doi.org/10.3390/molecules30142992
Chicago/Turabian StyleAkmal, Mohammad Firdaus, and Ming Wah Wong. 2025. "Selective Cleaning Enhances Machine Learning Accuracy for Drug Repurposing: Multiscale Discovery of MDM2 Inhibitors" Molecules 30, no. 14: 2992. https://doi.org/10.3390/molecules30142992
APA StyleAkmal, M. F., & Wong, M. W. (2025). Selective Cleaning Enhances Machine Learning Accuracy for Drug Repurposing: Multiscale Discovery of MDM2 Inhibitors. Molecules, 30(14), 2992. https://doi.org/10.3390/molecules30142992








