Experimental and Computational Analysis of Synthesis Conditions of Hybrid Nanoflowers for Lipase Immobilization
Abstract
1. Introduction
2. Results and Discussion
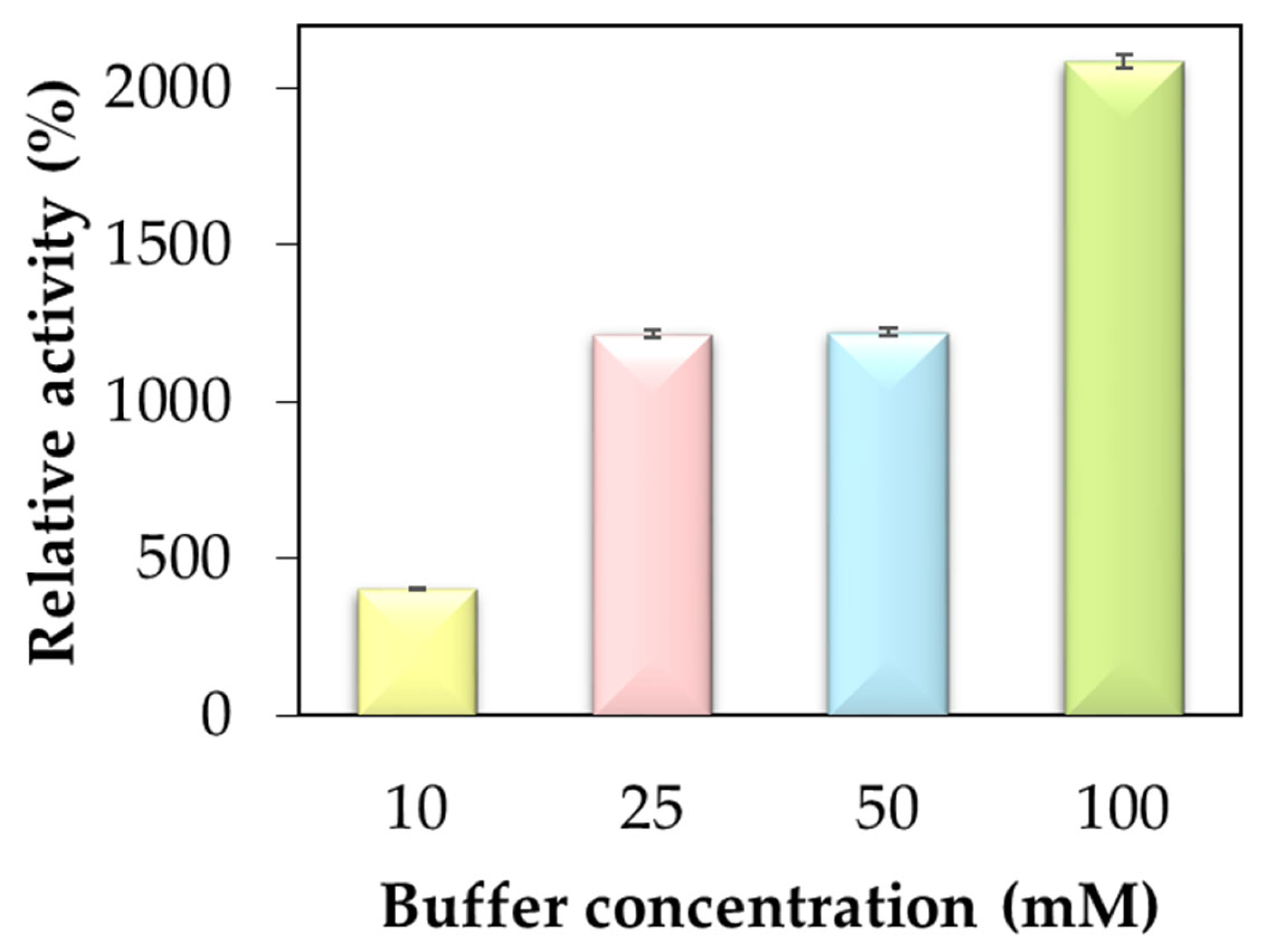
2.1. Activity of hNF-Lipase
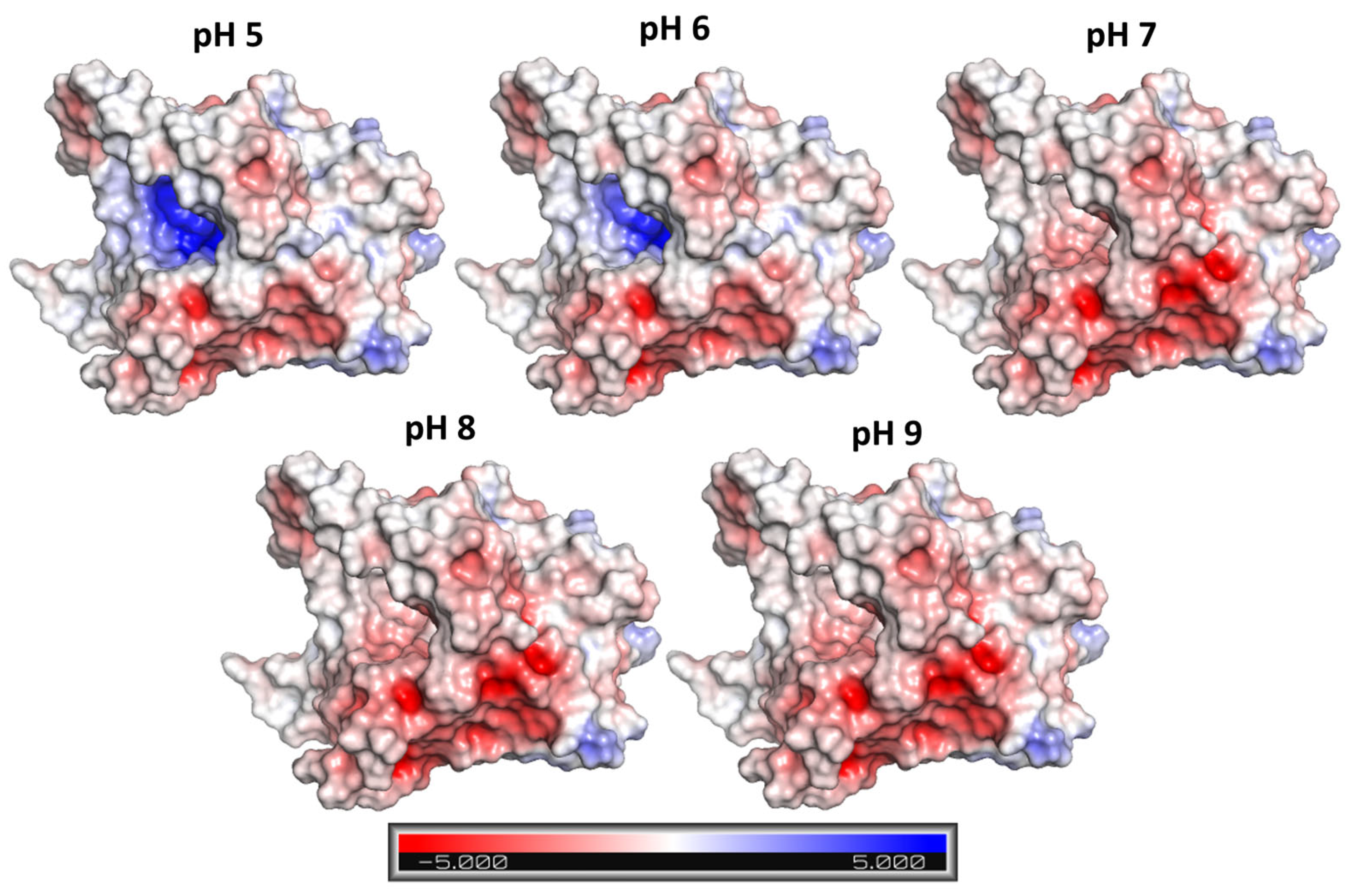
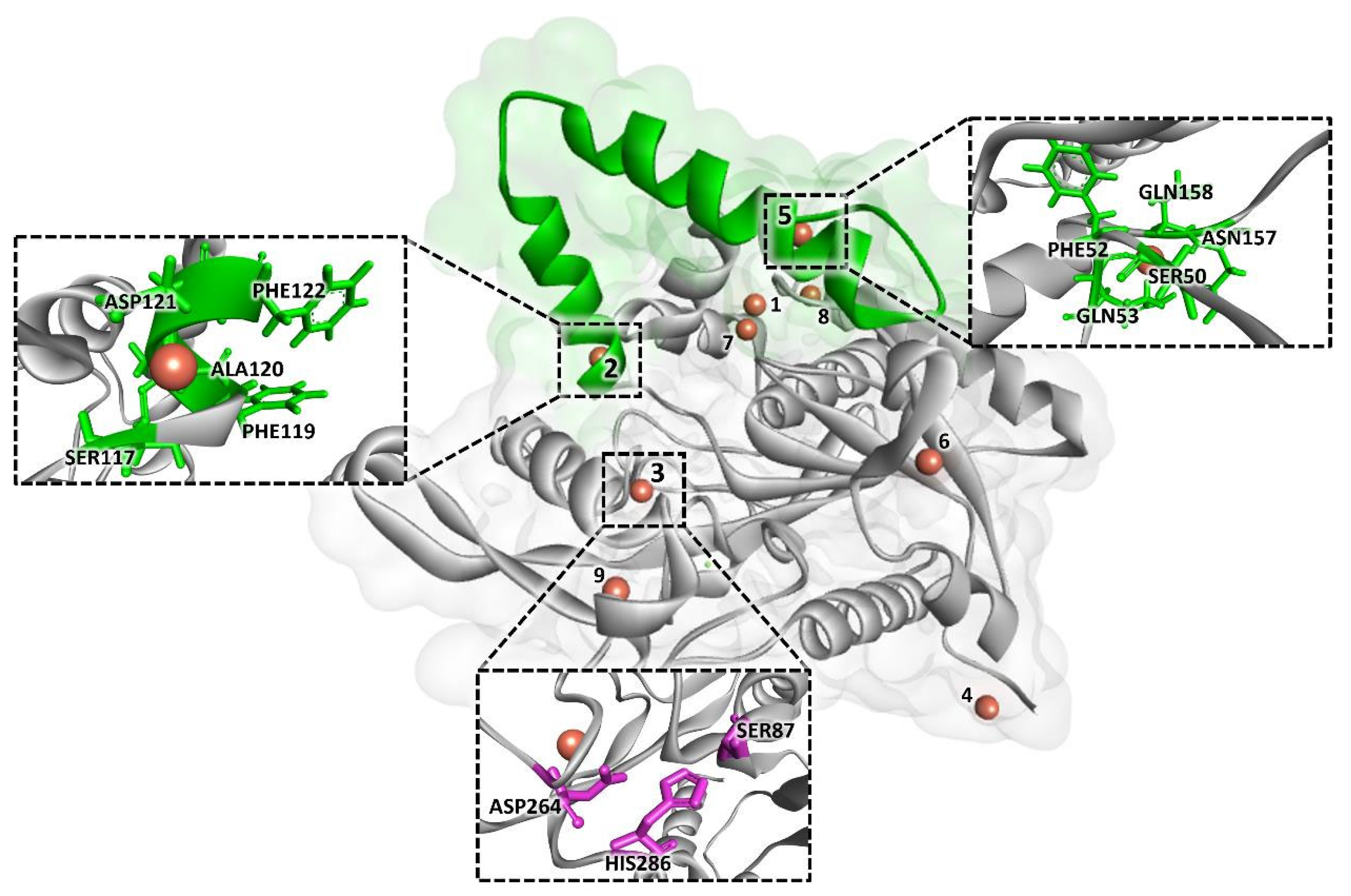
2.2. Computational Analysis
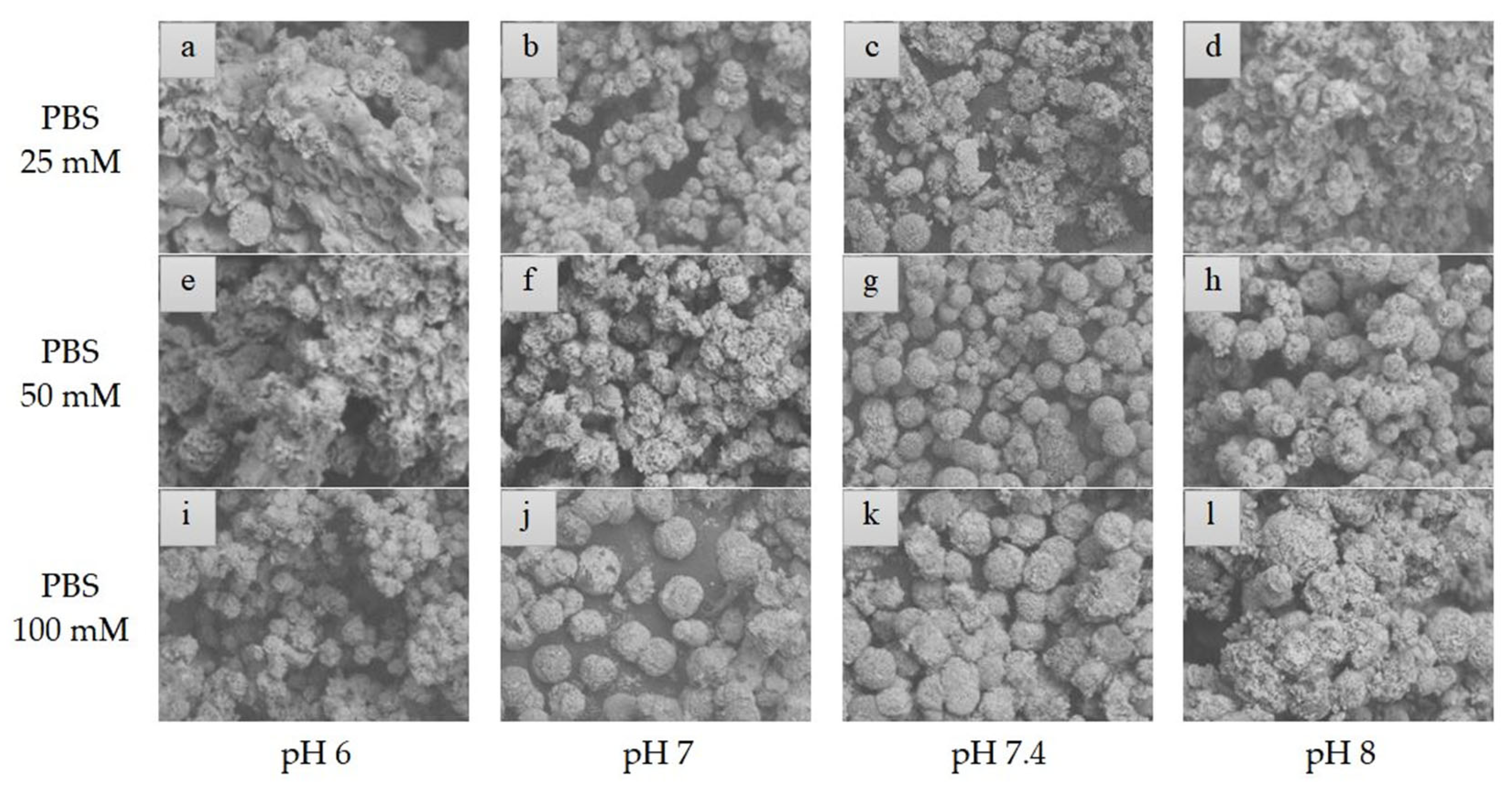
2.3. Morphology of hNF-Lipase
2.4. FTIR of hNF-Lipase
3. Material and Methods
3.1. Materials
3.2. Synthesis of Hybrid Nanoflowers Lipase
3.3. Quantification of Total Proteins
3.4. Lipase Activity
3.5. Computational Analysis
3.6. Characterization
3.6.1. Morphological Characterization of hNF-Lipase
3.6.2. Physicochemical Characterization of hNF-Lipase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Feng, Y.; Wang, G.; Wang, Z.; Bilal, M.; Lv, H.; Jia, S.; Cui, J. Production and use of immobilized lipases in/on nanomaterials: A review from the waste to biodiesel production. Int. J. Biol. Macromol. 2020, 152, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, H.; Su, Z. Enzyme-based hybrid nanoflowers with high performances for biocatalytic, biomedical, and environmental applications. Coord. Chem. Rev. 2020, 416, 213342. [Google Scholar] [CrossRef]
- Yin, Y.; Xiao, Y.; Lin, G.; Xiao, Q.; Lin, Z.; Cai, Z. An enzyme-inorganic hybrid nanoflower based immobilized enzyme reactor with enhanced enzymatic activity. J. Mater. Chem. B 2015, 3, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Yang, C.; Ma, C.; Tang, J. Enzyme-inorganic hybrid nanoflowers: Classification, synthesis, functionalization and potential applications. Chem. Eng. J. 2021, 415, 129075. [Google Scholar] [CrossRef]
- Cui, J.; Zhao, Y.; Liu, R.; Zhong, C.; Jia, S. Surfactant-activated lipase hybrid nanoflowers with enhanced enzymatic performance. Sci. Rep. 2016, 6, 27928. [Google Scholar] [CrossRef]
- de Souza, R.L.; de Faria, E.L.P.; Figueiredo, R.T.; Freitas, L.D.S.; Iglesias, M.; Mattedi, S.; Zanin, G.M.; Santos, O.A.A.D.; Coutinho, J.A.P.; Lima, Á.S.; et al. Protic ionic liquid as additive on lipase immobilization using silica sol-gel. Enzym. Microb. Technol. 2013, 52, 141–150. [Google Scholar] [CrossRef]
- Li, C.; Zhao, J.; Zhang, Z.; Jiang, Y.; Bilal, M.; Jiang, Y.; Jia, S.; Cui, J. Self-assembly of activated lipase hybrid nanoflowers with superior activity and enhanced stability. Biochem. Eng. J. 2020, 158, 107582. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Shah, S.Z.H.; Iqbal, H.M.N. Engineering enzyme-coupled hybrid nanoflowers: The quest for optimum performance to meet biocatalytic challenges and opportunities. Int. J. Biol. Macromol. 2019, 135, 677–690. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, W.; Elfeky, N.M.; Wang, Y.; Zhao, D.; Zhou, H.; Wang, J.; Bao, Y. Self-assembly of lipase hybrid nanoflowers with bifunctional Ca2+ for improved activity and stability. Enzym. Microb. Technol. 2020, 132, 109408. [Google Scholar] [CrossRef]
- Sharma, N.; Parhizkar, M.; Cong, W.; Mateti, S.; Kirkland, M.; Puri, M.; Sutti, A. Metal ion type significantly affects the morphology but not the activity of lipase–metal–phosphate nanoflowers. RSC Adv. 2017, 7, 25437. [Google Scholar] [CrossRef]
- Altinkaynak, C.; Gulmez, C.; Atakisi, O.; Özdemir, N. Evaluation of organic-inorganic hybrid nanoflower’s enzymatic activity in the presence of different metal ions and organic solvents. Int. J. Biol. Macromol. 2020, 164, 162–171. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, X.; Kong, D.; Li, G.; Li, Q. Immobilization of thermophilic lipase in inorganic hybrid nanoflower through biomimetic mineralization. Colloids Surf. B Biointerfaces 2021, 197, 111450. [Google Scholar] [CrossRef]
- Fotiadou, R.; Patila, M.; Hammami, M.A.; Enotiadis, A.; Moschovas, D.; Tsirka, K.; Spyrou, K.; Giannelis, E.P.; Avgeropoulos, A.; Paipetis, A.; et al. Development of effective lipase-hybrid nanoflowers enriched with carbon and magnetic nanomaterials for biocatalytic transformations. Nanomaterials 2019, 9, 808. [Google Scholar] [CrossRef]
- Li, K.; Wang, J.; He, Y.; Abdulrazaq, M.A.; Yan, Y. Carbon nanotube-lipase hybrid nanoflowers with enhanced enzyme activity and enantioselectivity. J. Biotechnol. 2018, 281, 87–98. [Google Scholar] [CrossRef]
- Almeida, L.C.; Barbosa, M.S.; de Jesus, F.A.; Santos, R.M.; Fricks, A.T.; Freitas, L.S.; Pereira, M.M.; Lima, Á.S.; Soares, C.M.F. Enzymatic transesterification of coconut oil by using immobilized lipase on biochar: An experimental and molecular docking study. Biotechnol. Appl. Biochem. 2021, 68, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.S.; Freire, C.C.C.; Souza, R.L.; Cabrera-Padilla, R.Y.; Pereira, M.M.; Freire, M.G.; Lima, Á.S.; Soares, C.M.F. Effects of phosphonium-based ionic liquids on the lipase activity evaluated by experimental results and molecular docking. Biotechnol. Prog. 2019, 35, e2816. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, X.; Yang, J.; Han, H.; Li, Q.; Tang, J. Lipase-inorganic hybrid nanoflower constructed through biomimetic mineralization: A new support for biodiesel synthesis. J. Colloid Interface Sci. 2018, 514, 102–107. [Google Scholar] [CrossRef]
- Zhong, L.; Jiao, X.; Hu, H.; Shen, X.; Zhao, J.; Feng, Y.; Li, C.; Du, Y.; Cui, J.; Jia, S. Activated magnetic lipase-inorganic hybrid nanoflowers: A highly active and recyclable nanobiocatalyst for biodiesel production. Renew. Energy 2021, 171, 825–832. [Google Scholar] [CrossRef]
- Ren, W.; Li, Y.; Wang, J.; Li, L.; Xu, L.; Wu, Y.; Wang, Y.; Fei, X.; Tian, J. Synthesis of magnetic nanoflower immobilized lipase and its continuous catalytic application. New J. Chem. 2019, 43, 11082–11090. [Google Scholar] [CrossRef]
- Escobar, S.; Velasco-Lozano, S.; Lu, C.H.; Lin, Y.F.; Mesa, M.; Bernal, C.; López-Gallego, F. Understanding the functional properties of bio-inorganic nanoflowers as biocatalysts by deciphering the metal-binding sites of enzymes. J. Mater. Chem. B 2017, 5, 4478–4486. [Google Scholar] [CrossRef]
- Ke, C.; Fan, Y.; Chen, Y.; Xu, L.; Yan, Y. A new lipase-inorganic hybrid nanoflower with enhanced enzyme activity. RSC Adv. 2016, 6, 19413–19416. [Google Scholar] [CrossRef]
- Aydemir, D.; Gecili, F.; Özdemir, N.; Ulusu, N.N. Synthesis and characterization of a triple enzyme-inorganic hybrid nanoflower (TrpE@ihNF) as a combination of three pancreatic digestive enzymes amylase, protease and lipase. J. Biosci. Bioeng. 2020, 129, 679–686. [Google Scholar] [CrossRef]
- Noma, S.A.A.; Yılmaz, B.S.; Ulu, A.; Özdemir, N.; Ateş, B. Development of l-asparaginase@hybrid Nanoflowers (ASNase@HNFs) Reactor System with Enhanced Enzymatic Reusability and Stability. Catal. Lett. 2021, 151, 1191–1201. [Google Scholar] [CrossRef]
- Zheng, L.; Sun, Y.; Wang, J.; Huang, H.; Geng, X.; Tong, Y.; Wang, Z. Preparation of a flower-like immobilized D-psicose 3-epimerase with enhanced catalytic performance. Catalysts 2018, 8, 468. [Google Scholar] [CrossRef]
- Li, Y.; Fei, X.; Liang, L.; Tian, J.; Xu, L.; Wang, X.; Wang, Y. The influence of synthesis conditions on enzymatic activity of enzyme-inorganic hybrid nanoflowers. J. Mol. Catal. B Enzym. 2016, 133, 92–97. [Google Scholar] [CrossRef]
- Guimarães, J.R.; Carballares, D.; Tardioli, P.W.; Rocha-Martin, J.; Fernandez-Lafuente, R. Tuning Immobilized Commercial Lipase Preparations Features by Simple Treatment with Metallic Phosphate Salts. Molecules 2022, 27, 4486. [Google Scholar] [CrossRef]
- Lisboa, M.C.; Rodrigues, C.A.; Barbosa, A.S.; Mattedi, S.; Freitas, L.S.; Mendes, A.A.; Dariva, C.; Franceschi, E.; Lima, Á.S.; Soares, C.M.F. New perspectives on the modification of silica aerogel particles with ionic liquid used in lipase immobilization with platform in ethyl esters production. Process Biochem. 2018, 75, 157–165. [Google Scholar] [CrossRef]
- Carvalho, N.B.; Vidal, B.T.; Barbosa, A.S.; Pereira, M.M.; Mattedi, S.; Freitas, L.D.S.; Lima, Á.S.; Soares, C.M.F. Lipase immobilization on silica xerogel treated with protic ionic liquid and its application in biodiesel production from different oils. Int. J. Mol. Sci. 2018, 19, 1829. [Google Scholar] [CrossRef]
- Yu, J.; Wang, C.; Wang, A.; Li, N.; Chen, X.; Pei, X.; Zhang, P.; Wu, S.G. Dual-cycle immobilization to reuse both enzyme and support by reblossoming enzyme-inorganic hybrid nanoflowers. RSC Adv. 2018, 8, 16088–16094. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Kwang, Y.H.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Show, P.L.; Tan, C.P.; Anuar, M.S.; Ariff, A.; Yusof, Y.A.; Chen, S.K.; Ling, T.C. Extractive fermentation for improved production and recovery of lipase derived from Burkholderia cepacia using a thermoseparating polymer in aqueous two-phase systems. Bioresour. Technol. 2012, 116, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Altinkaynak, C.; Yilmaz, I.; Koksal, Z.; Özdemir, H.; Ocsoy, I.; Özdemir, N. Preparation of lactoperoxidase incorporated hybrid nanoflower and its excellent activity and stability. Int. J. Biol. Macromol. 2016, 84, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Barbe, S.; Lafaquiere, V.; Guieysse, D.; Monsan, P.; Remaud-Siméon, M.; Andre, I. Insights into lid movements of Burkholderia cepacia lipase inferred from molecular dynamics simulations. Proteins Struct. Funct. Bioinform. 2009, 77, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Godoy, L.C.; Duquesne, S.; Bordes, F.; Sandoval, G.; Marty, A. Lipases and phospholipases: Methods and protocols. In Methods in Molecular Biology; Springer Science and Business Media: New York, NY, USA, 2012; Volume 861, pp. 3–30. [Google Scholar]
- Cui, J.; Jia, S. Organic–inorganic hybrid nanoflowers: A novel host platform for immobilizing biomolecules. Coord. Chem. Rev. 2017, 352, 249–263. [Google Scholar] [CrossRef]
- Altinkaynak, C.; Kocazorbaz, E.; Özdemir, N.; Zihnioglu, F. Egg white hybrid nanoflower (EW-hNF) with biomimetic polyphenol oxidase reactivity: Synthesis, characterization and potential use in decolorization of synthetic dyes. Int. J. Biol. Macromol. 2018, 109, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Alhayali, N.I.; Ozpozan, N.K.; Dayan, S.; Ozdemir, N.; Yilmaz, B.S. Catalase Fe3O4@Cu2+ hybrid biocatalytic nanoflowers fabrication and efficiency in the reduction of organic pollutants. Polyhedron 2021, 194, 114888. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Martinez-Rosell, G.; Giorgino, T.; de Fabritiis, G.; Martínez, G. PlayMolecule ProteinPrepare: A Web Application for Protein Preparation for Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 24, 1511–1516. [Google Scholar] [CrossRef]
- Lin, Y.F.; Cheng, C.W.; Shih, C.S.; Hwang, J.K.; Yu, C.S.; Lu, C.H. MIB: Metal Ion-Binding Site Prediction and Docking Server. J. Chem. Inf. Model. 2016, 56, 2287–2291. [Google Scholar] [CrossRef] [PubMed]
- Brandao, L.M.D.S.; Barbosa, M.S.; Souza, R.L.; Pereira, M.M.; Lima, Á.S.; Soares, C.M.F. Lipase activation by molecular bioimprinting: The role of interactions between fatty acids and enzyme active site. Biotechnol. Prog. 2021, 37, e3064. [Google Scholar] [CrossRef] [PubMed]


| Biocatalysts | Hydrolytic Activity (U g−1) | Specific Activity (U mg−1) | Relative Activity (%) | |
|---|---|---|---|---|
| BCL-free | 42.62 | - | 100 | |
| HNF-BCL pH 7.4 | 10 mM | 171.81 | 219.91 | 403.11 |
| HNF-BCL pH 6 | 25 mM | 217.34 | 257.33 | 509.96 |
| HNF-BCL pH 7 | 25 mM | 415.38 | 432.00 | 974.62 |
| HNF-BCL pH 7.4 | 25 mM | 518.00 | 538.07 | 1215.38 |
| HNF-BCL pH 8 | 25 mM | 182.32 | 188.01 | 427.77 |
| HNF-BCL pH 6 | 50 mM | 281.67 | 301.47 | 660.88 |
| HNF-BCL pH 7 | 50 mM | 398.42 | 432.39 | 934.81 |
| HNF-BCL pH 7.4 | 50 mM | 521.35 | 541.88 | 1223.26 |
| HNF-BCL pH 8 | 50 mM | 263.64 | 278.17 | 618.59 |
| HNF-BCL pH 6 | 100 mM | 362.19 | 410.30 | 849.82 |
| HNF-BCL pH 7 | 100 mM | 540.11 | 623.85 | 1267.27 |
| HNF-BCL pH 7.4 | 100 mM | 888.43 | 884.96 | 2084.96 |
| HNF-BCL pH 8 | 100 mM | 178.63 | 330.46 | 419.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, D.E.S.; Santos, L.M.F.; Freitas, J.P.A.; Almeida, L.C.d.; Santos, J.C.B.; Souza, R.L.d.; Pereira, M.M.; Lima, Á.S.; Soares, C.M.F. Experimental and Computational Analysis of Synthesis Conditions of Hybrid Nanoflowers for Lipase Immobilization. Molecules 2024, 29, 628. https://doi.org/10.3390/molecules29030628
Souza DES, Santos LMF, Freitas JPA, Almeida LCd, Santos JCB, Souza RLd, Pereira MM, Lima ÁS, Soares CMF. Experimental and Computational Analysis of Synthesis Conditions of Hybrid Nanoflowers for Lipase Immobilization. Molecules. 2024; 29(3):628. https://doi.org/10.3390/molecules29030628
Chicago/Turabian StyleSouza, Danivia Endi S., Lucas M. F. Santos, João P. A. Freitas, Lays C. de Almeida, Jefferson C. B. Santos, Ranyere Lucena de Souza, Matheus M. Pereira, Álvaro S. Lima, and Cleide M. F. Soares. 2024. "Experimental and Computational Analysis of Synthesis Conditions of Hybrid Nanoflowers for Lipase Immobilization" Molecules 29, no. 3: 628. https://doi.org/10.3390/molecules29030628
APA StyleSouza, D. E. S., Santos, L. M. F., Freitas, J. P. A., Almeida, L. C. d., Santos, J. C. B., Souza, R. L. d., Pereira, M. M., Lima, Á. S., & Soares, C. M. F. (2024). Experimental and Computational Analysis of Synthesis Conditions of Hybrid Nanoflowers for Lipase Immobilization. Molecules, 29(3), 628. https://doi.org/10.3390/molecules29030628








