Rapid Analysis of Compounds from Piperis Herba and Piperis Kadsurae Caulis and Their Differences Using High-Resolution Liquid–Mass Spectrometry and Molecular Network Binding Antioxidant Activity
Abstract
1. Introduction
2. Results
2.1. Optimization of Extraction and Mass Spectrometry Conditions
2.2. Molecular Network Visualization and Analysis
2.3. Identification of the Compounds of Piperis Herba and Piperis Kadsurae Caulis
2.4. Identification of the Amide Alkaloids
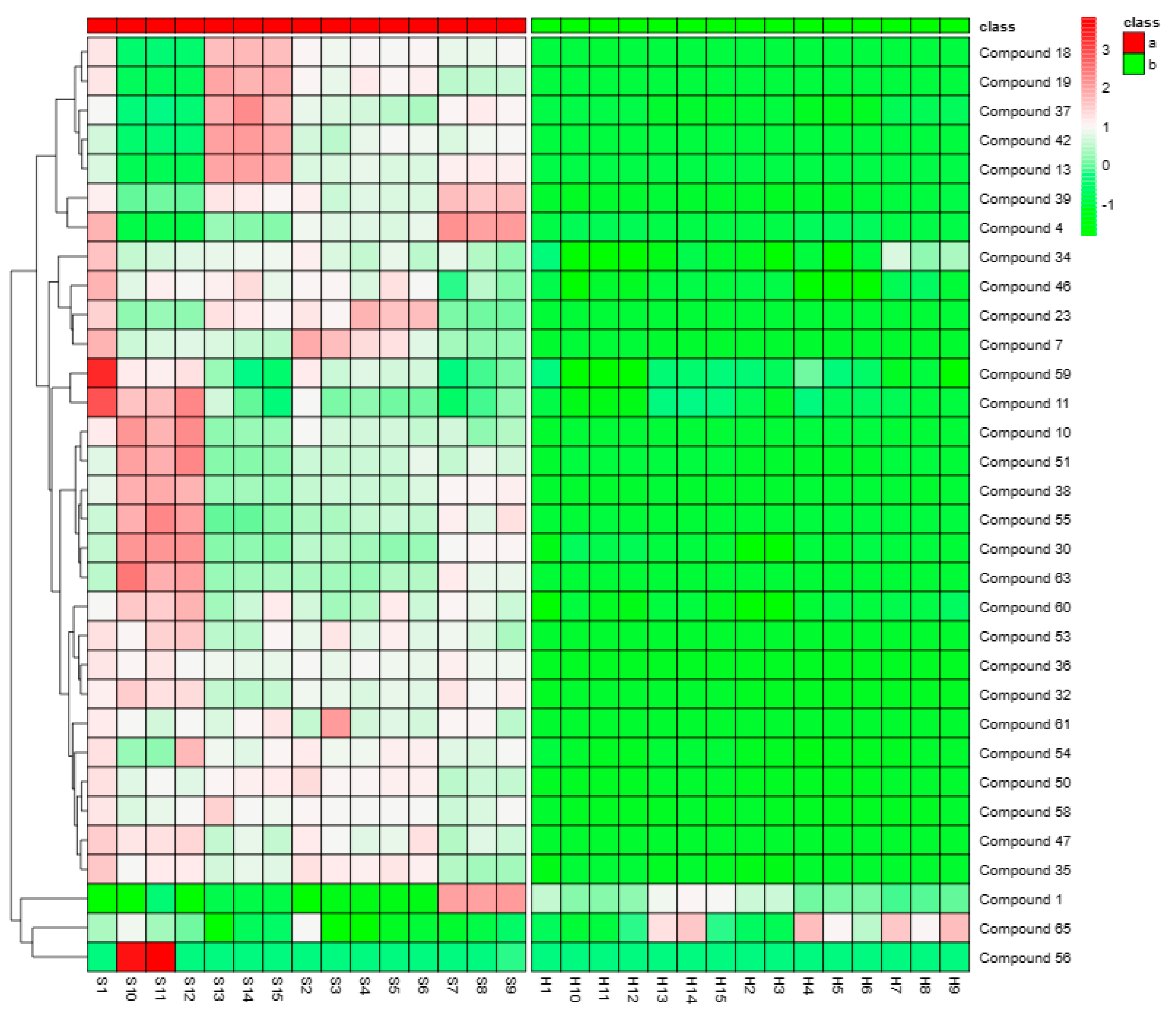
2.5. Analysis of the Differences in the Compounds between Piperis Herba and Piperis Kadsurae Caulis
2.6. Results of DPPH and ABTS Free Radical Scavenging Experiments in Piperis Herba and Piperis Kadsurae Caulis
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Collection and Preparation of Medicinal Herbs
4.3. UHPLC-MS Analysis
4.4. Molecular Network and Compound Identification Analysis
4.5. Analysis of the Differential Compounds in Piperis Herba and Piperis Kadsurae Caulis
4.6. Comparison of Antioxidant Activity between Piperis Herba and Piperis Kadsurae Caulis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.Q.; Geng, T.; Huang, W.Z.; Wang, Z.Z.; Xiao, W. Research progress of platelet activating factor(PAF) receptor antagonist. Zhongguo Zhong Yao Za Zhi 2018, 43, 1392–1403. [Google Scholar]
- Zhou, J.L.; Su, D.X.; Li, X.Z.; Xu, M.L.; Ma, X.X.; Zhuang, X.Y. Research Progress on Analgesic Effect of Piper Genus Native to Yunnan Province. Chin. J. Tradit. Chin. Med. 2022, 40, 255–258. [Google Scholar]
- Carsono, N.; Tumilaar, S.G.; Kurnia, D.; Latipudin, D.; Satari, M.H. A Review of Bioactive Compounds and Antioxidant Activity Properties of Piper Species. Molecules 2022, 27, 6774. [Google Scholar] [CrossRef] [PubMed]
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China. Part I; Chemical Industry Press: Beijing, China, 2020; p. 306. [Google Scholar]
- Nongmai, C.; Kanokmedhakul, K.; Promgool, T.; Paluka, J.; Suwanphakdee, C.; Kanokmedhakul, S. Chemical constituents and antibacterial activity from the stems and leaves of Piper wallichii. J. Asian Nat. Prod. Res. 2022, 24, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Hu, X.; Xu, R.; Ba, Y.; Chen, X.; Wang, X.; Cao, B.; Wu, X. Amide alkaloids characterization and neuroprotective properties of Piper nigrum L.: A comparative study with fruits, pericarp, stalks and leaves. Food Chem. 2022, 368, 130832. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhu, Y.; Zhang, Y.L.; Zeng, M.N.; Cao, Y.G.; Sun, P.T.; Cao, B.; Du, K.; Zhao, X.; Wang, X.W.; et al. Neolignans and amide alkaloids from the stems of Piper kadsura and their neuroprotective activity. Phytochemistry 2022, 203, 113336. [Google Scholar] [CrossRef] [PubMed]
- Wasilewicz, A.; Bojkova, D.; Beniddir, M.A.; Cinatl, J.; Rabenau, H.F.; Grienke, U.; Rollinger, J.M.; Kirchweger, B. Molecular networking unveils anti-SARS-CoV-2 constituents from traditionally used remedies. J. Ethnopharmacol. 2024, 30, 319. [Google Scholar] [CrossRef]
- Li, W.; Mei, S.; Zhou, H.; Salman, F.M.; Hu, T.; Wu, T. Metabolite fingerprinting of the ripening process in Pixian douban using a feature-based molecular network and metabolomics analysis. Food Chem. 2023, 418, 135940. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, J.; Zhou, C.; Yan, B.; Cai, Q.; He, H.; Duan, X.; Fan, H. Differential Analysis of Serum Principal Components Treated with Compound Sophora Decoction and Related Compounds Based on High-Resolution Mass Spectrometry (HRMS). Evid.-Based Complement. Altern. Med. eCAM 2020, 2020, 7518479. [Google Scholar] [CrossRef]
- Villanueva-Bermejo, D.; Siles-Sanchez, M.L.N.; Martin, H.D.; Rodríguez, G.M.; Jaime, L.; Santoyo, S.; Fornari, T. Theoretical framework to evaluate antioxidant synergistic effects from the coextraction of marjoram, rosemary and parsley. Food Chem. 2024, 437, 137919. [Google Scholar] [CrossRef]
- Iordanescu, O.A.; Bala, M.; Gligor, P.D.; Zippenfening, S.E.; Cugerean, M.I.; Petroman, M.I.; Hadaruga, D.I.; Hadaruga, N.G.; Riviş, M. A DPPH· Kinetic Approach on the Antioxidant Activity of Various Parts and Ripening Levels of Papaya (Carica papaya L.) Ethanolic Extracts. Plants 2021, 10, 1679. [Google Scholar] [CrossRef]
- GNPS. Available online: https://gnps.ucsd.edu/ (accessed on 2 November 2023).
- Hou, K.; Chen, F.; Zu, Y.; Yang, L. Ionic liquids-lithium salts pretreatment followed by ultrasound-assisted extraction of vitexin-4″-O-glucoside, vitexin-2″-O-rhamnoside and vitexin from Phyllostachys edulis leaves. J. Chromatogr. A 2016, 1431, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; He, F.; Wang, C.J.; Xie, Y.; Zhang, Y.Y.; Sang, Z.; Qiu, P.; Luo, P.; Xiao, S.Y.; Li, J.; et al. Discovery of chemical markers for improving the quality and safety control of Sinomenium acutum stem by the simultaneous determination of multiple alkaloids using UHPLC-QQQ-MS/MS. Sci. Rep. 2020, 10, 14182. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, M.; Yu, C.; Zhang, G.; Tang, X. Simultaneous determination of vitexin-4″-O-glucoside, vitex-in-2″-O-rhamnoside, rutin and vitexin from hawthorn leaves flavonoids in rat plasma by UPLC-ESI-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010, 878, 1837–1844. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.S.; Hung, C.J.; Lin, C.L.; Chang, T.H.; Chen, C.L.; Sung, P.J.; Cheng, M.J.; Huang, H.Y.; Chen, J.J. New Norneolignan and Bioactive Constituents of Clitoria ternatea. Chem. Nat. Compd. 2020, 56, 1000–1004. [Google Scholar] [CrossRef]
- Ma, R.H.; Yang, J.; Qi, L.W.; Xin, G.Z.; Wang, C.Z.; Yuan, C.S.; Wen, X.D.; Li, P. In vivo microdialysis with LC-MS for analysis of spinosin and its interaction with cyclosporin A in rat brain, blood and bile. J. Pharm. Biomed. Anal. 2012, 61, 22–29. [Google Scholar] [CrossRef]
- Ee, G.C.; Lim, C.M.; Lim, C.K.; Rahmani, M.; Shaari, K.; Bong, C.F. Alkaloids from Piper sarmentosum and Piper nigrum. Nat. Prod. Res. 2009, 23, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Bhattacharya, P.; Basak, S.; Gayen, S.; Nandy, A.; Saha, A. Pharmacoinformatics study of Piperolactam A from Piper betle root as new lead for non steroidal anti fertility drug development. Comput. Biol. Chem. 2017, 67, 213–224. [Google Scholar] [CrossRef]
- Macedo, A.L.; Dos, S.T.; Valverde, A.L.; Moreira, D.L.; Vasconcelos, T.R.A. An Overview of Neolignans of the Genus Piper L.: Isolation Methods and Biological Activities. Mini Rev. Med. Chem. 2017, 17, 693–720. [Google Scholar] [CrossRef]
- Luca, S.V.; Minceva, M.; Gertsch, J.; Skalicka-Woźniak, K. LC-HRMS/MS-based phytochemical profiling of Piper spices: Global association of piperamides with endocannabinoid system modulation. Food Res. Int. 2021, 141, 110123. [Google Scholar] [CrossRef]
- Cao, J.; Tang, T.T.; Li, Q.T.; Fang, Y.T.; Zheng, Y.F.; Ji, J.; Cheng, J.M. HPLC characterization study and determination of amide components in Piper wallichii (Miq.) Hand.-Mazz. J. Chin. Med. Mater. 2022, 45, 922–926. [Google Scholar]
- Li, X.; Ren, J.N.; Fan, G.; Yang, S.Z.; Zhang, L.L.; Pan, S.Y. Separation and purification of nootkatone from fermentation broth of Yarrowia lipolytica with high-speed counter-current chromatography. J. Food Sci. Technol. 2022, 59, 4487–4498. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Goto, M.; Wang, L.T.; Hsieh, K.Y.; Morris-Natschke, S.L.; Tang, G.H.; Long, C.L.; Lee, K.H. Multidrug resistance-selective antiproliferative activity of Piper amide alkaloids and synthetic analogues. Bioorg. Med. Chem. Lett. 2014, 24, 4818–4821. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.W.; Kim, Y.K.; Kim, K.; Lee, H.S.; Choi, J.H.; Lee, W.S.; Jun, C.D.; Park, J.H.; Lee, J.M.; Rho, M.C. Alkamides from the fruits of Piper longum and Piper nigrum displaying potent cell adhesion inhibition. Bioorg. Med. Chem. Lett. 2008, 18, 4544–4546. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Su, Y.; Luo, J.F.; Gu, W.; Niu, H.M.; Li, Y.; Wang, Y.H.; Long, C.L. New amide alkaloids from Piper longum fruits. Nat. Prod. Bioprospect. 2013, 3, 277–281. [Google Scholar] [CrossRef]
- Dabur, R.; Mittal, A. Detection and qualitative analysis of fatty acid amides in the urine of alcoholics using HPLC-QTOF-MS. Alcohol 2016, 52, 71–78. [Google Scholar] [CrossRef]
- Munigunti, R.; Nelson, N.; Mulabagal, V.; Gupta, M.P.; Brun, R.; Calderon, A.I. Identification of oleamide in Guatteria recurvisepala by LC/MS-based Plasmodium falciparum thioredoxin reductase ligand binding method. Planta Med. 2011, 77, 1749–1753. [Google Scholar] [CrossRef]
- Meng, L.Z.; Gong, S.L.; He, Y.B.; Liu, Y. Organic Spectral Analysis, 4th ed.; Wuhan University Press: Wuhan, China, 2016; p. 63. [Google Scholar]
- Zhang, Y.; Xu, D.; Xing, X.; Yang, H.; Gao, W.; Li, P. The chemistry and activity-oriented characterization of isoflavones difference between roots of Pueraria lobata and P. thomsonii guided by feature-based molecular networking. Food Chem. 2023, 422, 136198. [Google Scholar] [CrossRef]
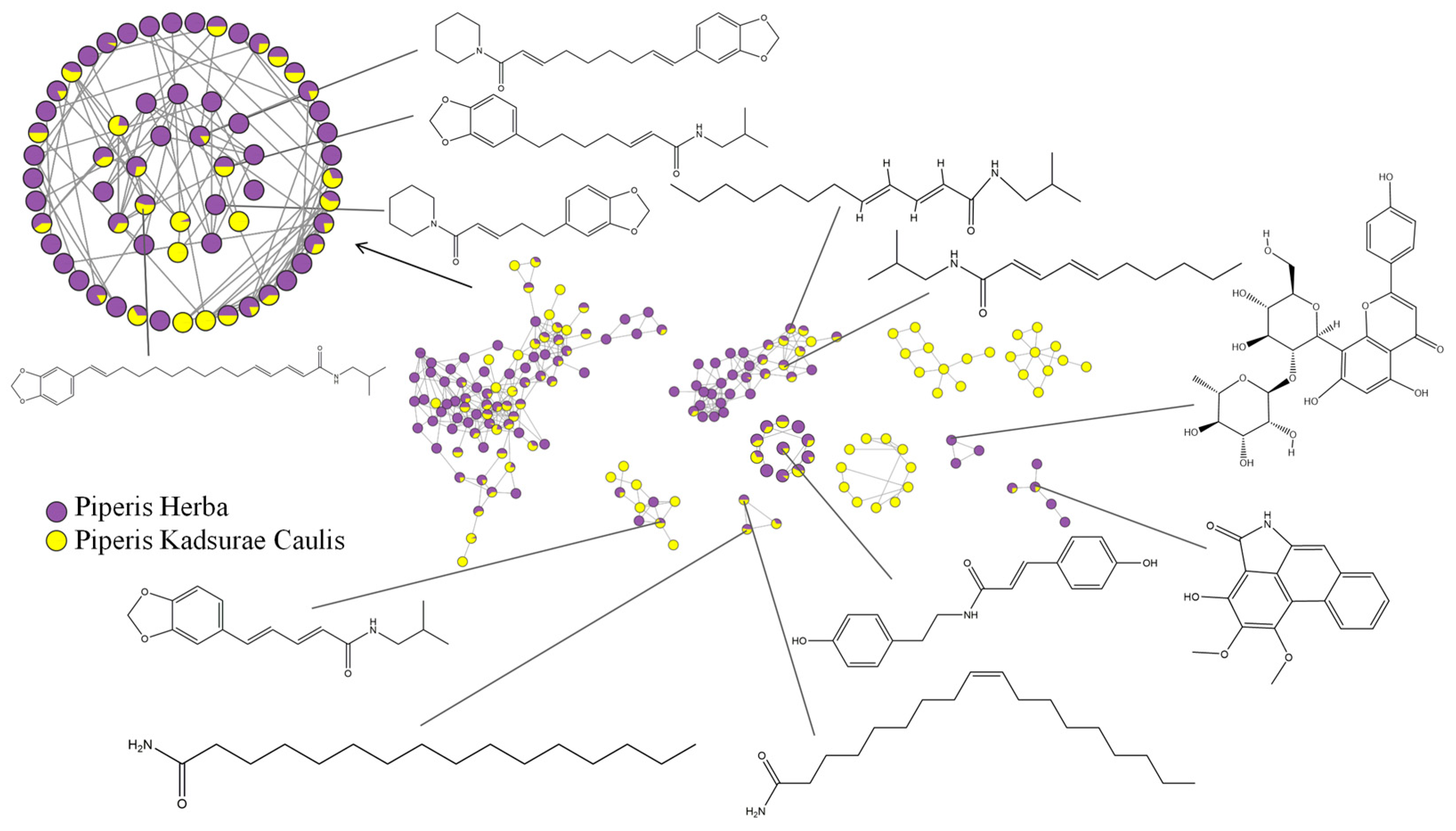

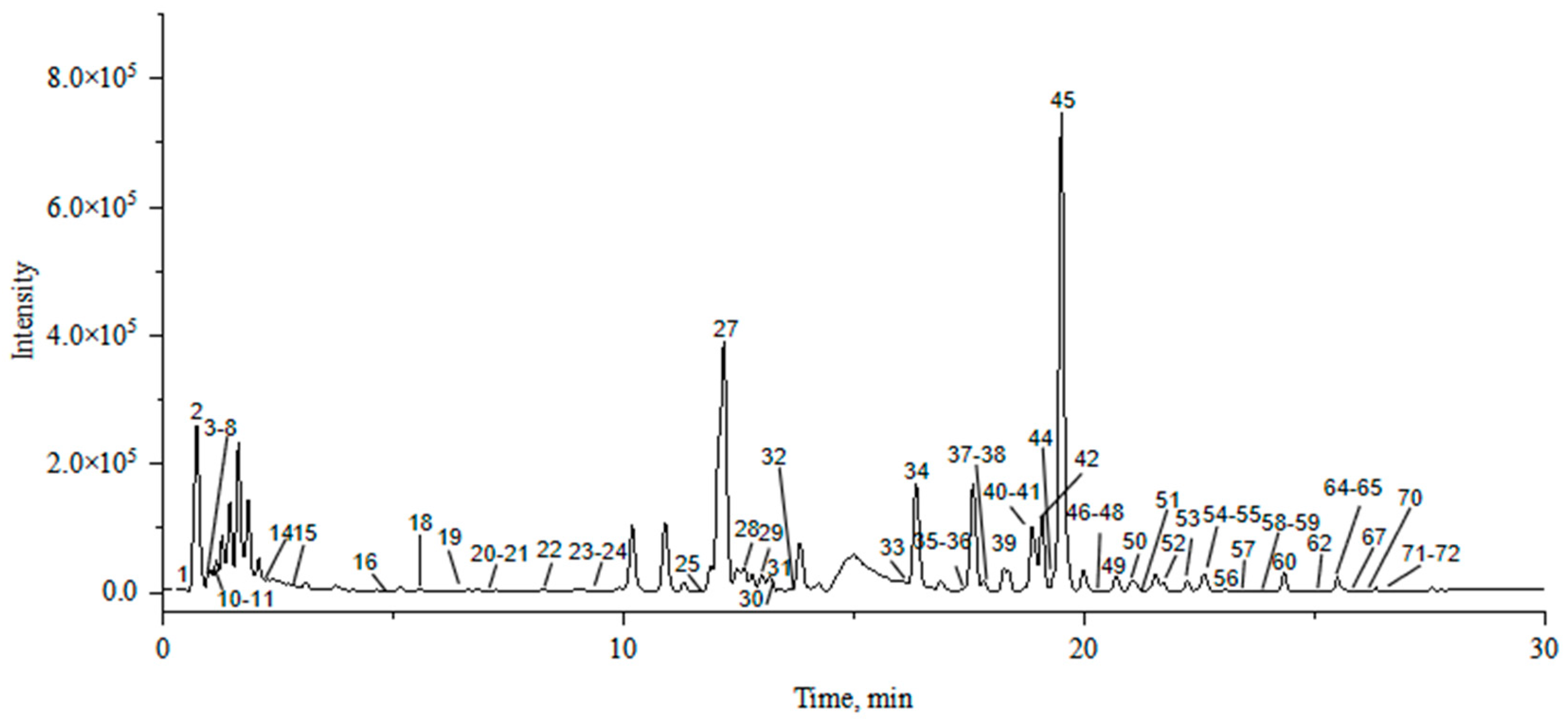





| NO. | tR min | Molecular Formula | [M+H]+ | Error (ppm) | MS2 | Compound | S1 | H1 | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.66 | C10H19NO7 | 266.1236 | 0.8 | 248.1128, 230.1024, 182.0824 | D-1-[(3-Carboxypropyl)amino]-1-deoxyfructose | + | + | [13] |
| 2 | 0.73 | C10H13NO2 | 180.1017 | −1 | 163.0751, 145.0651, 115.0550 | 1-methyl-1,2,3,4-tetrahydroisoquinoline-6,7-diol | + | + | [13] |
| 3 | 0.95 | C19H21NO2 | 296.1643 | −0.9 | 265.1218 | (-)-Nuciferine | − | + | [13] |
| 4 | 0.96 | C27H30O15 | 595.1653 | −0.7 | 433.1128, 415.1017, 313.0700 | Vitexia-Glucoside | + | + | [14] |
| 5 | 0.96 | C10H10O3 | 179.0696 | −3.7 | 147.0441, 119.0491 | Coniferyl aldehyde | + | + | [13] |
| 6 | 0.97 | C25H27NO10 | 502.1699 | −1.7 | 337.1072, 305.0804, 201.0549 | 2-[[(E)-3-[2-(4-hydroxy-3-methoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydro-1-benzofuran-5-yl]prop-2-enoyl]amino]pentanedioic acid | + | + | [13] |
| 7 | 0.98 | C17H19NO3 | 286.1434 | −1.2 | 237.0907, 107.0500 | Coclaurine | + | + | [15] |
| 8 | 0.98 | C21H24O5 | 221.1898 | −0.8 | 219.1062, 151.0781, 135.0441 | Isodihydrofutoquinol B | − | + | [13] |
| 9 | 1 | C27H30O14 | 579.1702 | −1.2 | 433.1124 | Vitexin-2-O-rhamnoside | + | − | [16] |
| 10 | 1.06 | C19H23NO4 | 330.1695 | −1.5 | 192.1016, 137.0592 | Reticuline | + | + | [17] |
| 11 | 1.1 | C20H23NO4 | 342.1697 | −0.8 | 311.1264, 265.0864 | Isocorydine | + | + | [13] |
| 12 | 1.12 | C28H32O15 | 609.1813 | −0.1 | 447.1292, 429.1188, 327.0854 | Spinosin | + | − | [18] |
| 13 | 1.16 | C21H20O10 | 433.1127 | −0.5 | 313.0710, 283.0604 | Vitexin | + | − | [14] |
| 14 | 2.43 | C11H16O3 | 197.1171 | −0.8 | 179.1061, 133.1008, 105.0696 | Loliolid | + | + | [13] |
| 15 | 2.85 | C17H17NO3 | 284.1281 | −0.2 | 284.1298, 147.0485, 121.0656 | Paprazine | + | + | [19] |
| 16 | 4.73 | C38H40N2O10 | 685.2752 | −0.6 | 548.1907, 520.1955, 351.0863 | (1R,2S)-7-hydroxy-1-(4-hydroxy-3,5-dimethoxyphenyl)-2-N,3-N-bis[2-(4-hydroxyphenyl)ethyl]-6,8-dimethoxy-1,2-dihydronaphthalene-2,3-dicarboxamide | + | + | [13] |
| 17 | 5.15 | C18H20N2O2 | 297.1596 | −0.5 | 176.1158, 175.1251, 105.0421 | N-(4-benzamidobutyl)benzamide | + | − | [13] |
| 18 | 5.51 | C14H15NO3 | 246.1125 | 0 | 175.0384, 145.0281, 98.0597 | 1-(3-(1,3-Benzodioxol-5-yl)-1-oxo-2-propenyl)pyrrolidine | + | + | [13] |
| 19 | 6.21 | C14H15NO3 | 246.1125 | 0 | 175.0380, 145.0276, 117.0337 | (2E,4E)-5-(1,3-benzodioxol-5-yl)-N,N-dimethylpenta-2,4-dienamide | + | + | [13] |
| 20 | 7.22 | C16H11NO3 | 266.0807 | −1.6 | 266.0811, 251.0577, 195.0687 | Piperolactam A | + | + | [20] |
| 21 | 7.29 | C14H17NO2 | 232.1331 | −0.5 | 232.1361, 161.0624, 133.0646 | Piperlotine A | + | + | [21] |
| 22 | 8.38 | C17H13NO4 | 296.0916 | −0.4 | 281.0681, 263.0570, 207.0673 | Piperolactam D | + | + | [19] |
| 23 | 9.5 | C15H17NO3 | 260.1281 | 0 | 175.0482, 145.0375, 86.0974 | Ilepcimide | + | + | [13] |
| 24 | 9.53 | C16H17NO3 | 272.1276 | −0.9 | 201.0542, 135.0448, 86.0947 | Piperyline | + | + | [6] |
| 25 | 11.78 | C19H25NO4 | 332.1855 | −0.5 | 135.0436, 86.0962 | (2E,4E)-1-piperidin-1-yl-5-(2,3,4-trimethoxyphenyl)penta-2,4-dien-1-one | + | + | [13] |
| 26 | 12.14 | C23H31NO4 | 386.2323 | −0.8 | 225.1273, 135.0440, 86.0990 | (2,3-dimethoxyphenyl)-[1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl]methanol | + | − | [13] |
| 27 | 12.18 | C16H19NO3 | 274.1436 | −0.7 | 274.1435, 201.0544, | Piperlonguminine | + | + | [6] |
| 28 | 12.62 | C16H35NO2 | 274.2738 | −0.8 | 256.2624, 106.0862 | Lauryldiethanolamine | + | + | [13] |
| 29 | 13.03 | C17H21NO3 | 288.1592 | −0.8 | 135.0437 | Piperanine | + | + | [6] |
| 30 | 13.16 | C17H19NO3 | 286.1434 | −0.9 | 201.057, 86.0963 | Piperine | + | + | [6] |
| 31 | 13.27 | C18H39NO3 | 318.3001 | −0.7 | 318.3051, 300.2885 | Phytosphingosine | + | + | [13] |
| 32 | 13.76 | C18H21NO3 | 300.1595 | 0.1 | 161.0594, 131.0493, 103.0557 | (2E,4E)-7-(1,3-benzodioxol-5-yl)-1-pyrrolidin-1-ylhepta-2,4-dien-1-one | + | + | [13] |
| 33 | 16.18 | C16H14O4 | 271.0962 | −0.9 | 121.0232 | (E)-1-(4-hydroxy-2-methoxyphenyl)-3-(4-hydroxyphenyl)prop-2-en-1-one | − | + | [13] |
| 34 | 16.45 | C18H23NO3 | 302.1750 | −0.4 | 161.0682, 131.0579, 103.0621 | Futoamide | + | + | [22] |
| 35 | 17.12 | C19H21NO3 | 312.1592 | −0.7 | 169.0643, 131.0495, 86.0985 | Piperettine | + | + | [13] |
| 36 | 17.33 | C19H23NO3 | 314.1749 | −0.6 | 161.0676, 131.0575, 86.1027 | Pipersintenamide | + | + | [23] |
| 37 | 17.83 | C18H25NO3 | 304.1906 | −0.3 | 203.1064, 135.0449, 123.0438 | Pipercallosidine | + | + | [13] |
| 38 | 17.93 | C14H23NO | 222.1851 | −0.9 | 151.1115, 81.0343 | Spilanthol | + | + | [23] |
| 39 | 18.65 | C19H25NO3 | 316.1907 | −0.1 | 135.0469, 86.0971 | Piperolein A | + | + | [6] |
| 40 | 18.85 | C20H25NO3 | 328.1907 | −0.1 | 229.1249, 135.0484 | Retrofractamide A | + | + | [6] |
| 41 | 18.87 | C22H28O5 | 373.2010 | 0.1 | 151.0843, 139.0774 | Galgravin | − | + | [13] |
| 42 | 19.08 | C15H22O | 219.1742 | −0.8 | 163.1135, 93.0702, 81.0714 | Nootkatone | + | + | [24] |
| 43 | 19.12 | C18H32O2 | 281.2472 | −1 | 133.1006, 105.0690, 91.0538 | Linoleic acid | + | − | [13] |
| 44 | 19.39 | C18H39NO2 | 302.3051 | −0.8 | 284.2922, 106.0855, 88.0759 | Tetradecyldiethanolamine | + | + | [13] |
| 45 | 19.55 | C14H25NO | 224.2007 | −0.9 | 151.1123, 133.1007, 81.0534 | Pellitorine | + | + | [6] |
| 46 | 20.13 | C20H27NO3 | 330.2064 | 0.1 | 229.1235, 135.0450 | Retrofractamide C | + | + | [22] |
| 47 | 20.17 | C21H25NO3 | 340.1905 | −0.6 | 227.1070, 131.0490, 86.0962 | Dehydropipernonaline | + | + | [22] |
| 48 | 20.55 | C20H29NO3 | 332.2219 | −0.4 | 135.0443 | (E)-9-(1,3-benzodioxol-5-yl)-N-(2-methylpropyl)non-8-enamide | + | + | [13] |
| 49 | 20.7 | C32H30N2O4 | 507.2275 | −0.6 | 256.1321, 238.1221, 105.0331 | Asperphenamate | + | + | [13] |
| 50 | 21.09 | C21H27NO3 | 342.2062 | −0.4 | 229.1221, 135.0459, 86.0969 | Pipernonaline | + | + | [22] |
| 51 | 21.31 | C22H27NO3 | 354.2061 | −0.7 | 135.0438, 131.0485 | (2E,4E,10E)-11-(1,3-benzodioxol-5-yl)-1-pyrrolidin-1-ylundeca-2,4,10-trien-1-one | + | + | [13] |
| 52 | 21.69 | C15H27NO | 238.2164 | −0.8 | 168.1384, 81.0697 | (2E,4E)-N-Isobutylundeca-2,4-dienamide | + | + | [13] |
| 53 | 22.34 | C21H29NO3 | 344.2220 | −0.1 | 135.0468, 112.0754, 86.0969 | Piperolein B | + | + | [6] |
| 54 | 22.66 | C22H29NO3 | 356.2220 | −0.2 | 135.0430, 131.0481 | Retrofractamide B | + | + | [6] |
| 55 | 22.72 | C16H27NO | 250.2164 | −0.7 | 124.0754, 98.0599 | (2E,4E)-1-(1-Pyrrolidinyl)-2,4-dodecadien-1-one | + | + | [25] |
| 56 | 23.14 | C22H29NO3 | 356.2220 | −0.2 | 135.0450, 131.0489, 98.0606 | (4E,10E)-11-(1,3-benzodioxol-5-yl)-1-pyrrolidin-1-ylundeca-4,10-dien-1-one | + | + | [13] |
| 57 | 23.62 | C29H39N3O2 | 462.3117 | 0.4 | 406.2469, 338.1844, 198.1271 | Echinulin | + | + | [13] |
| 58 | 24.18 | C22H31NO3 | 358.2377 | 0.2 | 161.0597, 135.0449 | Piperchabamide D | + | + | [26] |
| 59 | 24.27 | C23H29NO3 | 368.2216 | −1.2 | 225.1358, 135.0444, 86.0968 | Piperundecalidine | + | + | [22] |
| 60 | 24.38 | C16H29NO | 252.2321 | -0.3 | 196.1694, 179.1435, 95.0851 | (2E,4E)-N-(2-methylpropyl)dodeca-2,4-dienamide | + | + | [6] |
| 61 | 24.98 | C23H31NO3 | 370.2376 | −0.3 | 135.0437, 131.0494, 86.0962 | Piperchabamide B | + | − | [6] |
| 62 | 25.02 | C20H42O5 | 363.3104 | −0.2 | 195.1213, 133.0850 | 3,6,9,12-Tetraoxatetracosan-1-ol | + | + | [13] |
| 63 | 25.26 | C17H29NO | 264.2320 | −0.8 | 112.0754, 86.0963 | (2E,4E)-N-dodecadienoylpiperidine | + | − | [27] |
| 64 | 25.44 | C17H31NO | 266.2476 | −1 | 112.0749, 95.0849 | (2E,4E)-N-ethyl-3,7,11-trimethyldodeca-2,4-dienamide | + | + | [13] |
| 65 | 25.5 | C24H33NO3 | 384.2532 | −0.3 | 283.1693, 135.0469, 86.0987 | Guineensine | + | + | [6] |
| 66 | 25.5 | C23H33NO3 | 372.2530 | −0.9 | 135.0438, 86.0964 | (2E,11E)-12-(1,3-benzodioxol-5-yl)-N-(2-methylpropyl)dodeca-2,11-dienamide | + | − | [6] |
| 67 | 25.79 | C19H32O2 | 293.2474 | −0.5 | 243.2090, 137.1314 | Methyl alpha-eleostearate | + | + | [6] |
| 68 | 25.83 | C24H35NO3 | 386.2686 | −0.9 | 313.1802, 135.0439 | (2E,4E)-5-(1,3-benzodioxol-5-yl)-N,N-dihexylpenta-2,4-dienamide | + | − | [6] |
| 69 | 25.93 | C25H33NO3 | 396.2530 | −0.9 | 135.0438, 131.0485, 86.0960 | Piperchabamide C | + | − | [6] |
| 70 | 25.99 | C16H33NO | 256.2632 | −1.3 | 102.0910, 88.0756 | Palmitamide | + | + | [28] |
| 71 | 26.33 | C26H37NO3 | 412.2845 | −0.3 | 339.1948, 135.0435, 86.0960 | Brachystamide B | + | + | [6] |
| 72 | 26.33 | C18H35NO | 282.2789 | −1 | 265.2513, 247.2419, 149.1321 | Oleamide | + | + | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.; Liao, N.; Liu, H.; Gao, W.; Zhong, S.; Zheng, C.; Chen, H.; Xiao, L.; Zhu, Y.; Huang, S.; et al. Rapid Analysis of Compounds from Piperis Herba and Piperis Kadsurae Caulis and Their Differences Using High-Resolution Liquid–Mass Spectrometry and Molecular Network Binding Antioxidant Activity. Molecules 2024, 29, 439. https://doi.org/10.3390/molecules29020439
Shi D, Liao N, Liu H, Gao W, Zhong S, Zheng C, Chen H, Xiao L, Zhu Y, Huang S, et al. Rapid Analysis of Compounds from Piperis Herba and Piperis Kadsurae Caulis and Their Differences Using High-Resolution Liquid–Mass Spectrometry and Molecular Network Binding Antioxidant Activity. Molecules. 2024; 29(2):439. https://doi.org/10.3390/molecules29020439
Chicago/Turabian StyleShi, Dezhi, Nanxi Liao, Hualan Liu, Wufeng Gao, Shaohui Zhong, Chao Zheng, Haijie Chen, Lianlian Xiao, Yubo Zhu, Shiwen Huang, and et al. 2024. "Rapid Analysis of Compounds from Piperis Herba and Piperis Kadsurae Caulis and Their Differences Using High-Resolution Liquid–Mass Spectrometry and Molecular Network Binding Antioxidant Activity" Molecules 29, no. 2: 439. https://doi.org/10.3390/molecules29020439
APA StyleShi, D., Liao, N., Liu, H., Gao, W., Zhong, S., Zheng, C., Chen, H., Xiao, L., Zhu, Y., Huang, S., Zhang, Y., Hu, Y., Zheng, Y., Ji, J., & Cheng, J. (2024). Rapid Analysis of Compounds from Piperis Herba and Piperis Kadsurae Caulis and Their Differences Using High-Resolution Liquid–Mass Spectrometry and Molecular Network Binding Antioxidant Activity. Molecules, 29(2), 439. https://doi.org/10.3390/molecules29020439







