Abstract
Natural compounds, including diterpenoids, play a critical role in various biological processes and are recognized as valuable components in cancer treatment. Isocyanides multicomponent reactions (IsMCRs) are one of the effective methods to obtain adducts at the carboxyl group with a peptide-like substituent. In this study, dehydroabietic acid and levopimaric acid diene adducts as the starting scaffolds were modified by the multicomponent Passerini (P-3CR) and Ugi (U-4CR) reactions to afford α-acyloxycarboxamides and α-acylaminocarboxamides. A group of twenty novel diterpene hybrids was subjected to NCI in vitro assessment, and a consistent structure–activity relationship was established. Eleven of the synthesized derivatives inhibited the growth of cancer cells of 4 to 39 cell lines in one dose assay, and the most active were derivatives 3d, 9d, and 10d holding a fragment of 1a,4a-dehydroquinopimaric acid. They were selected for a five-dose analysis and demonstrated a significant antiproliferative effect towards human cancer cell lines. The outstanding cytotoxic activity was observed for the P-3CR product 3d with growth inhibitory at submicromolar and micromolar concentrations (GI50 = 0.42–3 μM) against the most sensitive cell lines. The U-4CR products 9d and 10d showed selective activity against all leukemia cell lines with GI50 in the range of 1–17 µM and selectivity indexes of 5.49 and 4.72, respectively. Matrix COMPARE analysis using the GI50 vector showed a moderate positive correlation of compound 3d with standard anticancer agents that can influence kinase receptors and epidermal growth factor receptors (EGFRs). The ADMET analysis acknowledges the favorable prognosis using compounds as potential anticancer agents. The obtained results indicate that these new hybrids could be useful for the further development of anticancer drugs, and 1a,4a-dehydroquinopimaric acid derivatives could be recommended for in-depth studies and the synthesis of new antitumor analogs on their basis.
1. Introduction
Cancer remains a global health problem and a leading cause of death worldwide despite significant advances in medicinal chemistry research that have led to the development of various anticancer treatment options [1]. Since a large number of mutagenic processes allow cancer cells to acquire the properties of unlimited proliferation potential and resistance, the synthetic development of potential anticancer agents with low side effects is highly desirable to combat this deadly disease [2,3]. In recent years, modern clinical oncology has increasingly used effective cytostatics based on natural compounds, the action of which is aimed at new cellular targets to solve the problem of multidrug tumor resistance [4,5,6]. A targeted search for new cytostatic agents involves, first of all, the development of molecules that can actively influence the pathological cells of the body and their growth. One approach in the development process is to enhance the therapeutic potential of natural compounds through structural modifications [7,8,9].
Multicomponent reactions (MCRs) have become necessary effective methods for the formation of new structural bonds in the synthesis of a wide range of pharmacologically active derivatives of organic and natural compounds without the separation and purification of intermediates. The most well-known isocyanide-based MCRs are the three-component Passerini reaction (P-3CR) and the four-component Ugi reaction (U-4CR), which are driven by the conversion of a high-energy formal divalent isocyano-carbon into a tetravalent amide carbonyl carbon [10,11]. As a result of the interaction of isocyanides, aldehydes/ketones and carboxylic acids (P-3CR) or isocyanides, aldehydes/ketones, carboxylic acids and amines (U-4CR), the corresponding α-acyloxycarboxamides and α-acylaminocarboxamides are formed. Moreover, the combination of natural compounds and their derivatives via isocyanide-based multicomponent reactions is attracting widespread interest due to the rapid access to the large libraries of diversity-oriented products with promising pharmacological potential [12,13,14]. Thus, there are several examples of the synthesis of U-4CR и P-3CR products based on triterpenoids, in particular glycyrrhizin, glycyrrhetinic, and masticadienonic acids [15,16]. A set of 34 new α-acylamino- and α-acyloxy-oleanolic and maslinic derivatives was successfully synthesized, and the cytotoxic activity evaluation revealed their significant activity against A2780 ovarian carcinoma cells in the low micromolar concentration range and the ability to induce programmed cell death partially through the apoptosis pathway [17]. Using the U-4CR approach, a diverse library of betulinic acid, fusidic acid, and cholic acid conjugates with TEMPO (nitroxide) was obtained, with cytotoxic effects against PC3 prostate cancer cell lines (IC50 6.0–7.4 μM) and HT29 colon cancer cell lines (IC50 7.0–9.0 µM). Studies of the mechanism of action have shown that they are targeted to mitochondria, and the induction of apoptosis occurs due to the activation of the caspase pathway [18].
There are even fewer examples in the literature of the use of diterpenoids, including the abietane series, in U-4CR and P-3CR (Figure 1). Based on the labdane diterpenoid coronarin D, as a result of the U-4CR with L-phenylalanine, 2-aminopyridine, D-glucosamine and tert-butyl isocyanide, non-toxic derivatives I and II were synthesized which can reduce LPS-stimulated NO production in RAW cells on a par with the standard anti-inflammatory drug dexamethasone [19]. From dehydroabiethylamine as the amine component in U-4CR, new α-acylaminocarboxamides III–V were synthesized and in vitro cytotoxicity was studied against several human tumor cell lines. Rhodamine B conjugate V has been found to exhibit cytotoxic activity characterized by EC50 values in the low nanomolar range [20]. Dehydroabiethylamine was also used in the pseudo-seven-component Ugi reaction for the synthesis of bis-tetrazoles VI, which demonstrated significant inhibition of the enzymes acetylcholinesterase and butyrylcholinesterase in Ellman assays [21]. In our laboratory, based on the abietic acid and diene adducts of levopimaric acid using U-4CR, a number of diterpene dipeptides VII–IX have been synthesized that effectively inhibit the influenza A (H1N1) virus (IC50 2–32 μM, CC50 > 300 μM, SI 200) and are active against the pseudovirus SARS-CoV-2 [22]. Moreover, we recently reported that two diterpene U-4CR products obtained from the reaction of maleopimaric and dihydroquinopimaric acids with paraformaldehyde, benzylamine and ethyl 2-isocyanoacetate effectively inhibit leukemia, colon cancer, ovarian cancer, breast cancer and melanoma cell lines [23]. In addition, it was found that the azido-Ugi reaction of methyl maleopimarate amido-imide with isocyanides, paraformaldehyde and trimethylsilyl azide in one step leads to bis-1,5-disubstituted diterpene tetrazoles X with a selective cytotoxicity against NCI-60 cancer cell panels [24].
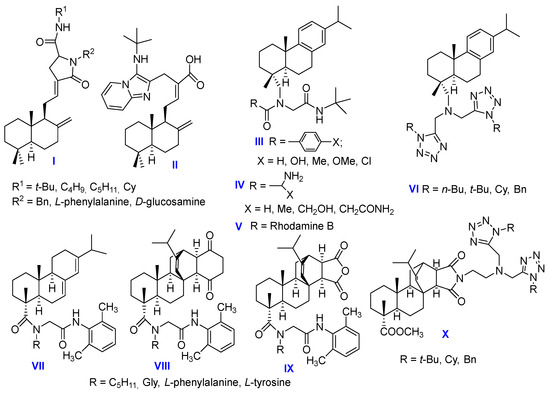
Figure 1.
The known U-4CR products of diterpene acids.
Taking into account the results obtained, here we have synthesized a library of twenty new resin acid derivatives using multicomponent Ugi and Passerini reactions and studied the cytotoxic potential of the synthesized products by high-throughput screening on 60 cell lines of nine different types of human cancer. Based on the screening results, new pharmacologically active derivatives were identified, and some patterns of pharmacological activity dependence on the substituent’s nature and the compound’s chemical structure were established.
2. Results and Discussion
2.1. Chemistry
We employed native dehydroabietic acid 1a and semi-synthetic derivatives of levopimaric diene adducts 1b,c,d as acid components in U-4CR and P-3CR. The most well-known and practically valuable products of the diene synthesis reaction are the adducts of levopimaric acid with maleic anhydride (maleopimaric acid 1b) and quinones. Quinopimaric acid, an adduct of levopimaric acid with p-benzoquinone, due to its easy ability for photocyclization, is most often used for further modifications in the form of its derivatives obtained as a result of reduction or oxidation reactions (2,3-dihydroquinopimaric acid 1c and 1a,4a -dehydroquinopimaric acid 1d) (Figure 2) [25].
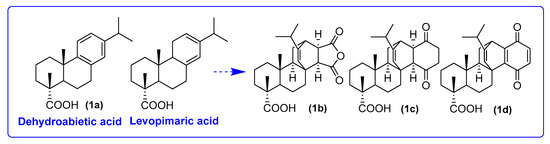
Figure 2.
Diterpene carboxylic acids for MCRs in this research.
For the synthesis of the first series of compounds, we exploited the P-3CR, which allows for the rapid and versatile generation of new diterpene derivatives displaying different α-acyloxyamides in the side chain. As it is known, this approach consists of the condensation of a carbonyl compound (aldehyde or ketone), isocyanides and carboxylic acid. With carboxylic acids 1a,b,c,d in our hands, we performed P-3CR with ethyl 2-isocyanoacetate and 2-morpholinoethyl isocyanate as the isocyanide component and paraformaldehyde as the highly reactive carbonyl partner in methanol for 3–4 days by monitoring the progress of reactions with thin-layer chromatography. It should be mentioned that paraformaldehyde was employed as the oxo-component to avoid stereoisomer formation during the conjugation process. As a result, a series of acyloxycarboxamides of dehydroabietic acid 2a,3a, maleopimaric acid 2b,3b, 2,3-dihydroquinopimaric acid 3c and 1a,4a-dehydroquinopimaric acid 2d,3d were obtained in 75–82% yields after purification by column chromatography (Scheme 1).
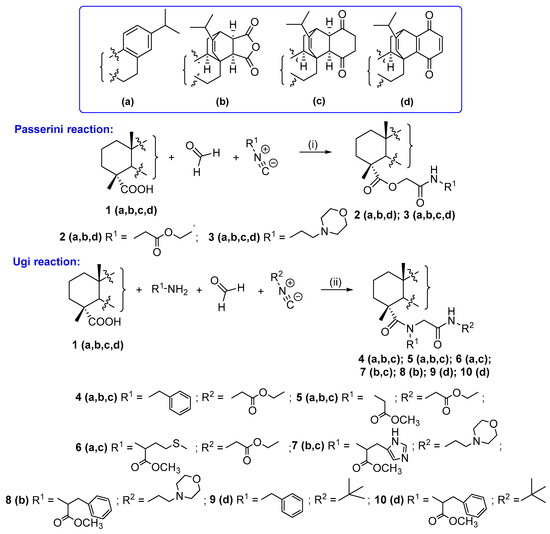
Scheme 1.
Conditions and reagents: P-3CR: ethyl 2-isocyanoacetate for 2a,b,d; 2-morpholinoethyl isocyanate for 3a,b,c,d; MeOH, rt, 3–4 days. Ugi 4CR: benzylamine and ethyl 2-isocyanoacetate for 4a,b,c; glycine methyl ester hydrochloride and ethyl 2-isocyanoacetate for 5a,b,c; L-methionine methyl ester hydrochloride and ethyl 2-isocyanoacetate for 6a,c; L-histidine methyl ester hydrochloride and 2-morpholinoethyl isocyanate for 7b,c; L-phenylalanine methyl ester hydrochloride and 2-morpholinoethyl isocyanate for 8b; benzylamine and tert-butyl isocyanide for 9d; and L-phenylalanine methyl ester hydrochloride and tert-butyl isocyanide for 10d, MeOH, rt, 5–7 days.
The second set of products was obtained by U-4CR, which included the conversion of a mixture of four components (amine, isocyanide, aldehyde/ketone and carboxylic acid) to α-acetoamidocarboxamides or a peptide-like scaffold. Benzylamine and methyl esters of L-amino acids (glycine, L-methionine, L-histidine and L-phenylalanine) were used as the amine equivalent, and ethyl 2-isocyanoacetate, 2-morpholinoethyl isocyanate or tert-butyl isocyanide was the source of isocyanide. Ugi condensation was carried out in methanol at room temperature for 5–7 days using equimolar concentrations of the components. The yields of the products varied in the range of 66–85% depending on the nature of the substituents at the nitrogen atom. As a result, a group of derivatives of dehydroabietic acid 4a–6a, maleopimaric acid 4b, 5b, 7b, 8b, 2,3-dihydroquinopimaric acid 4c–7c and 1a,4a-dehydroquinopimaric acid 9d, 10d was obtained (Scheme 1).
The complete identification of the structure of compounds 2–10 was made by means of NMR spectroscopy and mass spectra. The mass spectra of 2–10 exhibited molecular ion peaks corresponding to the molecular masses of the compounds. The 1H NMR spectra of the P-3CR and U-4CR products NH- atom signals as broadened signals at δ 5.55–9.15 ppm were contained. In the 13C NMR spectra, the CONH- carbon atom signals appeared at the δ 169.6–178.4 ppm, and for U-4CR products 4–10, CON- signals were also detected in the range of δ 172.8–183.9 ppm. The carbon signals of the dipeptide chain terminal amino acid keto groups in compounds like 5–8, 10 at δ 167.4–171.1 ppm were observed while the aromatic proton δ signals of the benzylamine substituent and L-phenylalanine proton atom signals resonated at δ 7.10–7.45 ppm. The characteristic signals of the amino acid methyl group protons and tert-butyl methyl atom protons appeared as singlets at δ 3.65–3.85 and δ 1.25 ppm, respectively (Supplemental Materials, Figures S39–S74).
Finally, in collaboration with the US National Cancer Institute, all the newly synthesized compounds were evaluated for antitumor activity [26,27,28,29,30,31,32].
2.2. One-Dose Assay
Among 20 synthesized compounds, 11 inhibited the growth of cancer cells. New derivatives 2a, 2d, 3a, 3b, 5a, 5b, 5c, 6a, 6c and 7b were inactive whereas compound 8b showed weak antitumor activity against one leukemia cell line. U-4CR и P-3CR products 2b, 3c, 4a, 4b, 4c and 7c and 3d, 9d and 10d demonstrate favorable cytotoxic activity in vitro, inhibiting the growth of 4 to 39 cell lines. Based on the results of the primary NCI-60 analysis, various structural relationships of activity could be observed. In particular, in a series of diterpene P-3CR products, the part of an acid component significantly affects the cytotoxic activity. α-Acyloxycarboxamides obtained from dehydroabietic acid are found to be non-cytotoxic. Among maleopimaric acid derivatives, activity against 5 cell lines was observed only for compound 2b, while the most active compounds were adducts of 2,3-dihydroquinopimaric acid 3c and 1a,4a-dehydroquinopimaric acid 3d, containing a morpholinoethyl isocyanate residue and showing activity against 10 and 26 cell lines, respectively (Table 1).

Table 1.
Cytotoxicity of compounds 2a, 2b, 2d, 3a, 3b, 3c, 3d, 4a, 4b, 4c, 5a, 5b, 5c, 6a, 6c, 7b, 7c, 8b, 9d and 10d at a concentration of 10−5 M on NCI-60 cancer cell lines.
In the context of evaluating the cytotoxic activity of Ugi products 4–10, the following structure–activity relationships were observed. In particular, the part of the amine component significantly affects the cytotoxic activity. The products obtained as a result of an interaction with glycine or L-methionine methyl esters do not exhibit cytotoxic activity in vitro. U-4CR products 7b and 7c, obtained from the reaction with L-histidine methyl ester, exhibit the least cytotoxic activity among this set of derivatives. The decoration of functionalizing diterpene acids with a benzylamine component had the most dramatic effect on cytotoxicity. In this set of derivatives, compounds 4a, 4b, and 4c are characterized by activity against 9–11 cell lines, and leukemia cell lines turned out to be the most sensitive to them. In addition, as in the case of the P-3CR product of 1a,4a-dehydroquinopimaric acid, a noticeable increase in cytotoxicity for a set of U-4CR-1a,4a-dehydroquinopimaric acid derivatives 9d and 10d was observed. The details of the utilized cell lines and results are provided in Table 1 and Supplementary Materials, Figures S1–S20.
2.3. Five-Dose Assay
The 1a,4a-dehydroquinopimaric acid derivatives 3d, 9d and 10d with the highest potency in the one-dose assay were selected for the five-dose study (Figure 3).
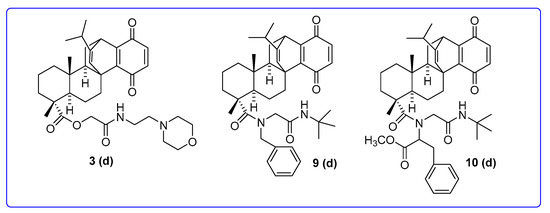
Figure 3.
Chemical structures of NSC845565 (3d), NSC850906 (9d) and NSC850907 (10d).
Protocols for NCI60 cell five-dose screening involved seeding of about 5000~40,000 cells/well (depending on the doubling time of individual cell lines) in 96-well plates, followed by treatment with studied compounds at concentrations of 0.01, 0.1, 1.0, 10 and 100 μM and incubation at 37 °C in 5% humidified CO2 for 48 h. Cells were fixed with a sulforhodamine B solution followed by a series of washing and staining to determine their viability. Growth inhibition was calculated relative to cells without drug treatment and time-zero control [27,33]. The results were presented as cell growth relative to the untreated cell control and to the time-zero number of cells. Growth inhibitions were indicated by values between 0 and 100, while lethality (cytotoxic effect) was indicated by values less than 0. The outcomes were used to create log concentration vs. % growth inhibition curves, and three response parameters (GI50, TGI and LC50) were calculated for each cell line. The GI50 value (growth inhibitory activity) corresponds to the concentration of the compound causing a 50% decrease in net cell growth, the TGI value (cytostatic activity) is the concentration of the compound resulting in total growth inhibition and the LC50 value (cytotoxic activity) is the concentration of the compound causing net 50% loss of initial cells at the end of the incubation period [33]. The results of the five-dose analysis are given in Table 2. The GI50 values for compounds 3d, 9d and 10d are shown in comparison with the standard anticancer agents doxorubicin (DRB) and 5-fluorouracil (5-FU), used by the NCI as a control [30].

Table 2.
In vitro cytotoxic effects of compounds 3d, 9d and 10d and standard drugs doxorubicin and 5-fluorouracil against NCI’s human tumor cell line screen in a five-dose analysis.
Thus, all compounds exhibited significant antiproliferative effects towards human cancer cell lines, and among them, the highest cytotoxic activity was observed for the P-3CR product 3d with growth inhibitory (GI50) at submicromolar (0.42 μM, CCRF-CEM) and micromolar concentrations (1–3 μM) and the U-4CR product 10d with a GI50 at micromolar concentrations (1–5 μM) against the most sensitive cell lines, respectively. A comparison of GI50 values for derivatives 3d, 9d, and 10d and standard drugs [30] shows that activity of studied compounds is generally lower than the activity of DRB (with the exception of colon cancer cell lines HCT-15 and ovarian cancer NCI/ADR-RES), but they have a higher activity than 5-FU. The Ugi adduct with benzylamine substituent 9d showed a selective activity against all leukemia cell lines with GI50 in the range of 1.21–17.1 μM (Table 2 and Figure 4, Figure 5 and Figure 6).

Figure 4.
Dose–response curves for all cell lines for compound 3d.
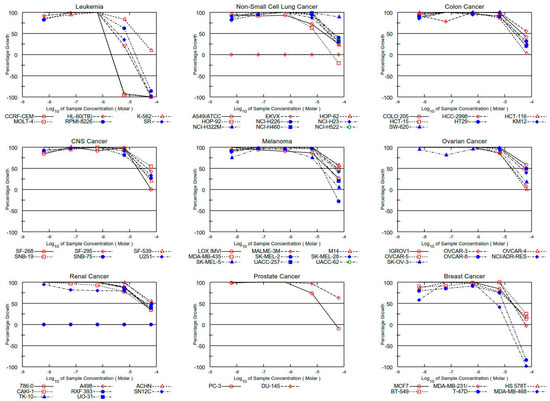
Figure 5.
Dose–response curves for all cell lines for compound 9d.
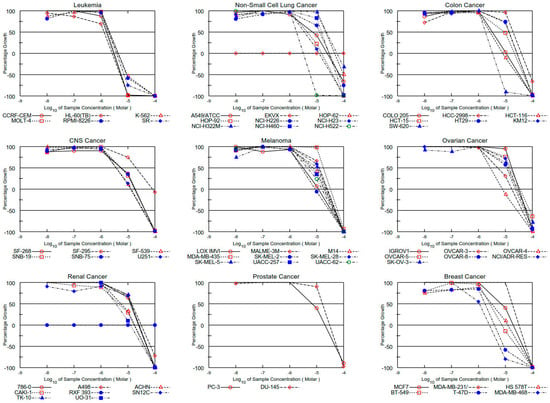
Figure 6.
Dose–response curves for all cell lines for compound 10d.
The mean values of the GI50 parameter for each subpanel are presented in Table 2. The data obtained showed that the panel of GI50 values of compounds 3d and 10d was significantly higher compared to that of compound 9d (6.26 and 8.49 vs. 30.17 μM). By reducing the average antiproliferative activity against individual subpanels, compound 3d forms the following cancer series of reduced activity: leukemia > melanoma > breast cancer > colon cancer > renal cancer > prostate cancer > ovarian cancer > NSLC > CNS cancer, whereas for compound 10d, this sequence is as follows: leukemia > breast cancer > CNS cancer > melanoma > colon cancer > renal cancer > ovarian cancer > prostate cancer > NSLC. For compound 9d, there is sensitivity only to the leukemia panel, while the difference in sensitivity between the other panels is not significant (Supplementary Materials, Figure S21–S38).
The selectivity index (SI) was calculated by dividing the average GI50 value of the full cell lines panel of compounds 3d, 9d and 10d by their average GI50 value of the individual subpanel (Table 3). Ratios between 3 and 6 mean moderate selectivity, and ratios greater than 6 indicate high selectivity towards the corresponding cell line, while compounds not meeting either of these criteria are rated nonselective [32]. In this context, compound 3d was found to be nonselective at all GI50 levels (SI 0.67–2.77), and Ugi-4CR products 9d and 10d were moderately selective at the GI50 levels towards leukemia (SI 5.49 and 4.72, respectively).

Table 3.
Average values of GI50 (µM) and selectivity ratios of compounds 3d, 9d and 10d against NCI subpanels in five-dose analysis.
2.4. COMPARE Correlations
Standard COMPARE analyses allow for the comparison of the selectivity patterns of tested compounds with standard antineoplastic agents of the known mode of action and NCI active synthetic and natural compounds, which are present in publicly available databases [34] (Table 4). The application of this algorithm may provide preliminary information about the mechanism of cell growth inhibition and cell death. A quantitative evaluation of the obtained results was carried out according to the Chaddock scale using the following interpretation of pair correlation coefficients: insignificant (0.00–0.30), weak (0.30–0.50), moderate (0.50–0.70), high (0.70–0.90) and very high (0.9–1.0) [35].

Table 4.
Standard agent COMPARE correlations for compounds 3d, 9d and 10d according to five-dose assays.
As follows from Table 4, a GI50 COMPARE analysis of compound 3d revealed moderate positive correlations with eprenatapopt, 3-bromopyruvic acid and ixazomib. TGI analysis also showed a moderate positive correlation with eprenetapopt, as well as with olmutinib, tepotinib and sunitinib, respectively. The cytotoxicity vector of compound 3d had a weak correlation coefficient with tepotinib. The anticarcinogenic effect of most of the identified drugs, the average graph of which correlates with the P-3CR product 3d, is associated with their apoptotic effect or the ability to inhibit enzymes, which increases the effectiveness of anticancer drugs and even leads to the death of cancer cells. Thus, the quinuclidinone derivative eprenetapopt exhibits antitumor activity through apoptosis of the mutant TP53 cancer cells and also exhibits synergy with other antitumor agents [36]. However, the mechanism of action of eprenetapopt as an anticancer agent still remains unresolved, although some studies suggest its ability to reduce the levels of the cellular antioxidant glutathione by increasing its turnover, causing an iron-dependent form of cell death known as ferroptosis [37]. 3-Bromopyruvic acid is an effective inhibitor of hexokinase II, causing cell death through its dissociation and thereby exerting an antitumor effect [38]. In addition, it reduces the expression of glycolytic enzymes PFKP, BPGM and GPI, depletes cells of glutathione and causes oxidative stress [39]. Ixazomib, a proteasome inhibitor, preferentially binds to and inhibits the chymotrypsin-like activity of the 20S proteasome subunit beta-5 [40], while olmutinib acts by covalently bonding to a cysteine residue near the kinase domain of the epidermal growth factor receptor (EGFR) [41], and the mechanism of action of tepotinib and sunitinib is associated with the selective inhibition of tyrosine kinase receptors involved in the processes of tumor growth, pathological angiogenesis and the formation of metastases [42,43].
According to the Chaddock scale [35], U-4CR analogs 9d and 10d had low positive correlations of the GI50 and TGI vectors with 6-mercaptopurine and regorafenib, as well as minor correlations with bendamustine and lapatinib at the LC50 level, and therefore, the resulting coefficients do not allow for high probability to explain the cytotoxicity mechanism of the tested compounds.
Taken together, the data obtained suggest that the most likely molecular targets for the derivative 3d may be proteins that carry out mitogenic signals from receptor molecules and epidermal growth factor receptors (EGFRs), and its action is associated with the ability to effectively reduce the activity of oncogenes that affect uncontrolled division tumor cells. However, the absence of high positive correlations with the above compounds requires further experimental studies to correctly interpret the results obtained. The obtained data on the antiproliferative activity of diterpene peptide-like derivatives and their molecular targets will make an important contribution to establishing the mechanisms of the putative action of abietane diterpenoids since such studies in the literature are limited to [20]. The design of such adducts is inspired by our previously described dipeptide derivatives of resin acids, which showed pronounced antiviral activity against the influenza A/Puerto Rico/8/34 (H1N1) virus and the SARS-CoV-2 pseudovirus [22]. Given that the parent 1a,4a-dehydroquinopimaric acid is not active in the NCI60 assay (Supplementary Materials, Figure S39) [44], it is also noteworthy that a rapid and cost-effective one-step protocol based on a multicomponent isocyanide method has yielded a collection of important diterpene derivatives as promising antitumor scaffolds, and the decoration of abietane acids by peptide-like substituents leads to submicromolar cytotoxicity.
2.5. ADMET Studies
The main ADMET properties of the tested compounds and the reference drug doxorubicin were determined and analyzed using AdmetLab 3.0 and OSIRIS DataWarrior 5.5.0 [45,46]. The calculated properties are summarized in Table 5. Compounds 3d and 9d have a high probability of bioavailability greater than 50%, while compound 10d has a probability of bioavailability greater than 50% of only 0.52. The bioavailability of doxorubicin is predicted to be less than 20%. Compounds 3d and 9d have a high drug similarity score. None of the compounds presented are predicted to be BCRP inhibitors. In contrast to doxorubicin, the investigated compounds are highly lipophilic and BBB-positive. Derivatives 3d and 9d do not affect the major cytochromes, whereas 10d is an inhibitor of CYP2C9 and CYP3A4 and may therefore increase the plasma concentration of drugs whose metabolism is mediated by these isoenzymes. Compound 10d has been shown to inhibit P-glycoprotein, while 3d and 9d have been identified as non-substrates/non-inhibitors, which may impede their transport in the body. A toxicity analysis showed that the compounds were highly unlikely to be cardiotoxic, cytotoxic or mutagenic, in contrast to the reference drug. The toxicity classes were also lower than those of doxorubicin. It is expected that 3d will be class 6 and 9d and 10d will be class 5. All compounds are expected to have respiratory toxicity and immunotoxicity, while 3d is also expected to have neuro- and nephrotoxicity. The probability of nephrotoxicity for compound 10d would be 0.54.

Table 5.
In silico ADMET-selected characteristics of compounds 3d, 9d and 10d, including probability [a].
Thus, these results render the most active compounds as suitable drug candidates, exceeding doxorubicin in terms of toxicity, absorption and distribution. Specific untoward reactions, such as organ toxicity, are to be monitored closely in preclinical studies.
3. Materials and Methods
3.1. General
The spectra were recorded at the Center for the Collective Use “Chemistry” of the UIC UFRC RAS and RCCU “Agidel” of the UFRC RAS. 1H and 13C-NMR spectra were recorded on a “Bruker AM-500” (Bruker, Billerica, MA, USA, 500 and 125.5 MHz, respectively, δ, ppm, Hz) in CDCl3, internal standard tetramethylsilane. Mass spectra were obtained on a liquid chromatograph–mass spectrometer LCMS-2010 EV (Shimadzu, Kyoto, Japan). Melting points were detected on a micro table “Rapido PHMK05“ (Nagema, Dresden, Germany). Optical rotations were measured on a polarimeter “Perkin-Elmer 241 MC” (Perkin Elmer, Waltham, MA, USA) in a tube length of 1 dm. Elemental analysis was performed on a Euro EA-3000 CHNS analyzer (Eurovector, Milan, Italy); the main standard is acetanilide. Thin-layer chromatography analyses were performed on Sorbfil plates (Sorbpolimer, Krasnodar, Russian Federation), using the solvent system chloroform–ethyl acetate, 40:1. Substances were detected by 10% H2SO4 with subsequent heating to 100–120 °C for 2–3 min. All the reagents and solvents were purchased from standard commercial vendors and were used without any further purification. For the synthesis of dehydroabietic acid 1a [47], maleopimaric acid 1b [48], 2,3-dihydroquinopimaric acid 1c [49] and 1a,4a-dehydroquinopimaric acid 1d [50], pine resin Pinus silvestris (containing about 25% levopimaric acid) was used. Compounds 4b and 4c were obtained according to the method described previously [23].
3.2. General Procedure for Passerini Reaction
Paraformaldehyde (1 mmol) was suspended in 10–20 mL dry methanol. The suspension was stirred for 2 h at room temperature. The diterpenic acid (1 mmol) and the corresponding isocyanide (1 mmol) (ethyl 2-isocyanoacetate for 2a,b,d or 2-morpholinoethyl isocyanate for 3a–d) were added, and the solution was stirred for an additional 3–4 days. The reaction mixture was poured into aqueous HCl (2M), and the precipitate formed was filtered off, washed until neutral, and dried in air. The residue was purified by column chromatography using petroleum ether/ethyl acetate as the eluent.
3.2.1. 2-((2-Ethoxy-2-oxoethyl)amino)-2-oxoethyl-7-isopropyl-1,4a-dimethyl-1,2,3,4,4a,9,10,10a-octahydrophenanthrene-1-carboxylate (2a)
Yield 75%, mp: 62 °C, [α]20D +51 (c 0.05, CHCl3), Rf 0.63. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.98–1.20 (4H, m, CH, CH2), 1.18 (3H, s, H-20), 1.20 (3H, s, H-16), 1.22 (3H, s, H-15), 1.24 (3H, s, H-19), 1.40 (3H, s, CH3), 1.45–2.37 (4H, m, CH, CH2), 2.75–3.00 (4H, m, H-6, H-7), 4.53–5.10 (6H, m, CH2), 5.55 (1H, br.s., NH), 6.85 (1H, s, H-17), 7.00 (1H, d, J = 7.5 Hz, H-12), 7.18 (1H, d, J = 7.5 Hz, H-11); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 179.7 (C-18), 169.6 (CO), 169.4 (CO), 146.9 (C-9), 145.6 (C-13), 136.3 (C-8), 135.1 (C-17), 124.0 (C-11), 123.7 (C-12), 61.5, 61.4, 53.2, 51.4, 47.3, 45.4, 41.2, 37.7, 36.2, 33.6, 31.9, 25.4, 24.6, 22.1, 19.5, 18.8, 14.2. Analysis calculated for C26H37NO5: C, 70.40; H, 8.41; N, 3.16. Found: C, 70.35; H, 8.39; N, 3.20. MS(APCI) m/z [M ]+ 444.3, calculated for C26H38NO5: 444.58.
3.2.2. 2-((2-Ethoxy-2-oxoethyl)amino)-2-oxoethyl-12-isopropyl-6,9a-dimethyl-1,3-dioxo-3,3a,4,5,5a,6,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-3b,11-ethenophenanthro[1,2-c]furan-6-carboxylate (2b)
Yield 81%, mp: 55 °C, [α]20D +21 (c 0.05, CHCl3), Rf 0.58. 1H NMR (δ, ppm, CDCl3+MeOD, 500 MHz): 0.55 (3H, s, CH3), 0.81–0.99 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.10 (3H, s, CH3), 1.21 (3H, s, CH3), 1.24–1.95 (12H, m, CH, CH2), 2.65 (d, 1H, J = 6.8 Hz, H-1a), 2.95–3.05 (2H, m, CH2,), 3.15 (1H, s, H-12), 3.50–4.10 (4H, m, CH2), 4.50–4.65 (2H, m, CH2), 5.50 (1H, br. s, H-14), 7.95 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3+MeOD, 125.5 MHz): 181.9 (C-22), 178.7 (CO), 173.8 (C-24), 173.0 (C-23), 171.2 (CO), 148.0 (C-13), 125.2 (C-14), 63.5, 61.0, 53.4, 53.0, 49.7, 46.6, 45.7, 39.1, 38.0, 37.6, 35.8, 35.7, 34.9, 32.7, 29.6, 27.1, 22.0, 21.9, 20.6, 19.9, 18.6, 17.2, 15.8. Analysis calculated for C30H41NO8: C, 66.28; H, 7.60; N, 2.58. Found: C, 66.32; H, 7.63; N, 2.54. MS(APCI) m/z [M + H]+ 544.4 calculated for C30H42NO8: 544.66.
3.2.3. 2-((2-Ethoxy-2-oxoethyl)amino)-2-oxoethyl-13-isopropyl-7,10a-dimethyl-1,4-dioxo-4,5,6,6a,7,8,9,10,10a,10b,11,12-dodecahydro-1H-4b,12-ethenochrysene-7-carboxylate (2d)
Yield 80%, mp: 75 °C, [α]20D +14 (c 0.05, CHCl3), Rf 0.55. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.66 (3H, s, H-18), 0.68–0.73 (3H, m, CH, CH2), 0.90 (3H, d, J = 6.9 Hz, H-16/H-17), 0.94 (3H, d, J = 6.9 Hz, H-16/H-17), 1.15 (3H, s, H-19), 1.20 (3H, s, CH3), 1.18–1.32 (5H, m, CH2), 1.40–1.68 (6H, m, CH, CH2), 2.35–2.99 (2H, m, CH), 3.23 (2H, s, CH2), 3.80 (2H, s, CH2), 4.10 (2H, s, CH2), 5.55 (1H, br.s., NH), 5.60 (1H, s, H-14), 6.50 (2H, dd, J = 6.8 Hz, 2H, H-2, H-3); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 185.3 (C-1), 184.7 (C-4), 179.1 (C-20), 170.8 (CO), 169.9 (CO), 152.8 (C-1a), 150.7 (C-4a), 149.6 (C-13), 137.6 (C-3), 133.6 (C-2), 125.4 (C-14), 60.7, 60.6, 56.0, 54.9, 49.3, 49.1, 47.1, 39.4, 39.0, 38.5, 37.8, 36.4, 32.3, 27.0, 21.7, 20.5, 20.2, 17.1, 16.7, 16.5, 16.3. Analysis calculated for C32H41NO7: C, 69.67; H, 7.49; N, 2.54. Found: C, 69.62; H, 7.48; N, 2.58. MS(APCI) m/z 552.9 [M + H]+, calculated for C32H42NO7: 552.68.
3.2.4. 2-((2-Morpholinoethyl)amino)-2-oxoethyl-7-isopropyl-1,4a-dimethyl-1,2,3,4,4a,9,10,10a-octahydrophenanthrene-1-carboxylate (3a)
Yield 77%, mp: 53 °C, [α]20D +8 (c 0.05, CHCl3), Rf 0.45. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.98–1.20 (4H, m, CH, CH2), 1.18 (3H, s, H-20), 1.20 (3H, s, H-16), 1.22 (3H, s, H-15), 1.24 (3H, s, H-19), 1.45–2.07 (8H, m, CH, CH2), 2.35–3.00 (4H, m, H-6, H-7), 3.80–4.80 (10H, m, CH2), 7.15 (1H, s, H-17), 7.18 (1H, d, J = 7.5 Hz, H-12), 7.28 (1H, d, J = 7.5 Hz, H-11), 7.98 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 179.7 (C-18), 169.6 (CO), 153.0 (C-9), 146.9 (C-13), 132.7 (C-8), 130.6 (C-17), 125.1 (C-11), 123.5 (C-12), 61.5, 61.4, 61.2, 61.0, 60.8, 60.2, 43.6, 37.7, 37.2, 37.1, 36.5, 33.6, 32.5, 30.2, 25.4, 24.6, 23.7, 19.5, 18.8, 16.1. Analysis calculated for C28H42N2O4: C, 71.46; H, 9.00; N, 5.95. Found: C, 71.50; H, 8.99; N, 6.00. MS(APCI) m/z 471.3 [M]+, calculated for C28H42N2O4: 470.65.
3.2.5. 2-((2-Morpholinoethyl)amino)-2-oxoethyl-12-isopropyl-6,9a-dimethyl-1,3-dioxo-3,3a,4,5,5a,6,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-3b,11-ethenophenanthro[1,2-c]furan-6-carboxylate (3b)
Yield 82%, mp: 48 °C, [α]20D +18 (c 0.05, CHCl3), Rf 0.49. 1H NMR (δ, ppm, CDCl3+MeOD, 500 MHz): 0.55 (3H, s, CH3), 0.81–0.99 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.10 (3H, s, CH3), 1.24–1.95 (16H, m, CH, CH2), 2.65 (d, 1H, J = 6.8 Hz, H-1a), 2.95–3.05 (2H, m, CH2,), 3.15 (1H, s, H-12), 3.50–4.10 (8H, m, CH2), 5.49 (2H, br.s., CH2), 5.65 (1H, br. s, H-14), 7.95 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3+MeOD, 125.5 MHz): 178.7 (C-22), 173.7 (C-24), 173.0 (C-23), 171.2 (CO), 148.0 (C-13), 125.2 (C-14), 66.7, 66.7, 63.2, 53.4, 53.2, 53.0, 52.7, 49.7, 46.6, 45.7, 39.1, 38.0, 37.6, 35.8, 35.7, 34.9, 32.7, 29.6, 27.1, 22.0, 21.9, 20.6, 19.9, 18.6, 17.2, 15.8. Analysis calculated for C32H46N2O7: C, 67.34; H, 8.12; N, 4.91. Found: C, 67.32; H, 8.15; N, 4.94. MS(APCI) m/z 571.4 [M + H]+, calculated for C32H47N2O7: 571.73.
3.2.6. 2-((2-Morpholinoethyl)amino)-2-oxoethyl-13-isopropyl-7,10a-dimethyl-1,4-dioxo-2,3,4,4a,5,6,6a,7,8,9,10,10a,10b,11,12,12a-hexadecahydro-1H-4b,12-ethenochrysene-7-carboxylate (3c)
Yield 79%, mp: 92 °C, [α]20D +11 (c 0.05, CHCl3), Rf 0.40.1H NMR (δ, ppm, CDCl3, 500 MHz): 0.59 (3H, s, CH3), 0.61–0.90 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.18 (3H, s, CH3), 1.28–1.92 (6H, m, CH, CH2), 2.14–2.25 (11H, m, CH, CH2), 2.62–2.88 (9H, m, CH, CH2), 3.25 (2H, s, CH2), 3.60–3.80 (4H, m, CH2, CH3), 5.40 (1H, br. s, H-14), 5.60–5.75 (2H, m, CH2), 6.25 (1H, br.s, NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 210.1 (C-4), 208.9 (C-1), 184.8 (C-20), 179.1 (CO), 147.4 (C-13), 125.4 (C-14), 60.6, 60.5, 58.7, 56.0, 55.9, 54.9, 52.0, 50.6, 49.3, 49.0, 41.5, 39.2, 38.9, 37.7, 37.0, 36.7, 35.1, 34.7, 32.9, 27.7, 25.4, 21.8, 20.8, 19.9, 16.9, 16.7, 16.4, 15.9. Analysis calculated for C34H50N2O6: C, 70.07; H, 8.65; N, 4.81. Found: C, 70.00; H, 8.60; N, 4.89. MS(APCI) m/z 583.9 [M + H]+, calculated for C34H51N2O6: 583.78.
3.2.7. 2-((2-Morpholinoethyl)amino)-2-oxoethyl-13-isopropyl-7,10a-dimethyl-1,4-dioxo-4,5,6,6a,7,8,9,10,10a,10b,11,12-dodecahydro-1H-4b,12-ethenochrysene-7-carboxylate (3d)
Yield 75%, mp: 110 °C, [α]20D +20 (c 0.05, CHCl3), Rf 0.39. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.64 (3H, s, H-18), 0.68–0.73 (3H, m, CH, CH2), 0.90 (3H, d, J = 6.9 Hz, H-16/H-17), 0.94 (3H, d, J = 6.9 Hz, H-16/H-17), 1.15 (3H, s, H-19), 1.20 (3H, s, CH3), 1.18–1.33 (5H, m, CH2), 1.40–1.68 (9H, m, CH, CH2), 2.35–2.99 (2H, m, CH), 2.90–2.93 (2H, м, CH2), 4.05–4.10 (2H, м, CH2), 4.20–4.30 (4H, м, CH2), 5.55 (1H, s, H-14), 5.62 (1H, br.s., NH), 6.50 (2H, dd, J = 6.8 Hz, 2H, H-2, H-3); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 200.4 (C-1), 195.9 (C-4), 184.1 (C-20), 167.8 (CO), 151.2 (C-1a), 144.7 (C-13), 142.8 (C-4a), 137.6 (C-3), 135.6 (C-2), 124.9 (C-14), 68.2, 67.6, 59.2, 59.0, 54.9, 49.3, 49.1, 47.1, 39.4, 39.0, 38.2, 38.5, 37.8, 36.4, 32.3, 27.0, 25.3, 22.0, 21.7, 20.5, 20.2, 17.1, 16.8, 16.6. Analysis calculated for C34H46N2O6: C, 70.56; H, 8.01; N, 4.84. Found: C, 70.60; H, 8.08; N, 4.90. MS(APCI) m/z 579.9 [M + H]+, calculated for C34H47N2O6: 579.75.
3.3. General Procedure for Ugi Reaction
Paraformaldehyde (1 mmol) was suspended in 10–20 mL dry methanol, followed by the addition of an amine or methyl ester of the corresponding amino acid (1.2 mmol) (benzylamine for 4a–c, 9d, methyl esters of glycine for 5a–c, methyl esters of L-methionine for 6a,c, methyl esters of L-histidine for 7b,c and methyl esters of L-phenylalanine for 8b, 10d). The suspension was stirred for 2 h at room temperature. The diterpenic acid (1 mmol) and the corresponding isocyanide (1 mmol) (ethyl 2-isocyanoacetate for 4a–c, 5a–c, 6a,c, 2-morpholinoethyl isocyanate for 7b,c or tert-butyl isocyanide for 9d, 10d) were added, and the solution was stirred for an additional 5–7 days. The reaction mixture was poured into aqueous HCl (2M), and the precipitate formed was filtered off, washed until neutral, and dried in air. The residue was purified by column chromatography using petroleum ether/ethyl acetate as the eluent.
3.3.1. Ethyl N-benzyl-N-(7-isopropyl-1,4a-dimethyl-1,2,3,4,4a,9,10,10a-octahydrophenanthrene-1-carbonyl)glycylglycinate (4a)
Yield 85%, mp: 61 °C, [α]20D -16 (c 0.05, CHCl3), Rf 0.50. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.98–1.20 (4H, m, CH, CH2), 1.18 (3H, s, H-20), 1.20 (3H, s, H-16), 1.22 (3H, s, H-15), 1.24 (3H, s, H-19), 1.42 (3H, s, CH3), 1.45–2.37 (4H, m, CH, CH2), 2.75–3.10 (4H, m, H-6, H-7), 3.73 (2H, d, J = 16.4 Hz, CH2), 4.00 (2H, d, J = 16.4 Hz, CH2), 4.15–4.20 (2H, m, CH2), 4.65 (1H, d, J = 16.4 Hz, CH2), 5.10 (1H, d, J = 16.4 Hz, CH2), 6.65 (1H, br.s., NH), 6.85 (1H, s, H-17), 7.00 (1H, d, J = 7.5 Hz, H-12), 7.18 (1H, d, J = 7.5 Hz, H-11), 7.20–7.45 (5H, m, H-Ar); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 179.7 (C-18), 169.6 (CO), 169.4 (CO), 146.9 (C-9), 145.6 (C-13), 136.3 (C-8), 135.1, 128.9, 128.8, 127.7, 127.6, 127.0, 126.9 (C-17), 124.0 (C-11), 123.7 (C-12), 61.5, 53.2, 51.4, 47.3, 45.4, 41.2, 37.7, 36.2, 33.6, 32.5, 31.9, 30.2, 25.4, 24.6, 22.1, 19.5, 18.8, 14.2. Analysis calculated for C33H44N2O4: C, 74.40; H, 8.33; N, 5.26. Found: C, 74.50; H, 8.29; N, 5.30. MS(APCI) m/z 532.3 [M]+, calculated for C33H45N2O4: 532.73.
3.3.2. Ethyl N-(7-isopropyl-1,4a-dimethyl-1,2,3,4,4a,9,10,10a-octahydrophenanthrene-1-carbonyl)-N-(2-methoxy-2-oxoethyl)glycylglycinate (5a)
Yield 72%, mp: 51 °C, [α]20D -17 (c 0.05, CHCl3), Rf 0.42. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.78–1.10 (4H, m, CH, CH2), 1.18 (3H, s, H-20), 1.20 (3H, s, H-16), 1.22 (3H, s, H-15), 1.24 (3H, s, H-19), 1.42 (3H, s, CH3), 1.45–2.37 (4H, m, CH, CH2), 2.75–3.00 (4H, m, H-6, H-7), 3.65–3.73 (5H, m, CH2, CH3), 3.85–4.40 (6H, m, CH2), 6.85 (1H, s, H-17), 7.00 (1H, d, J = 7.5 Hz, H-12), 7.18 (1H, d, J = 7.5 Hz, H-11), 7.95 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 179.6 (C-18), 171.2 (CO), 169.4 (CO), 169.3 (CO), 146.8 (C-9), 145.6 (C-13), 134.9 (C-8), 126.9 (C-17), 124.1 (C-11), 123.8 (C-12), 61.3, 54.4, 52.6, 52.4, 45.3, 42.6, 41.2, 37.6, 35.8, 33.4, 32.5, 30.1, 27.1, 26.6, 25.2, 23.9, 18.6, 17.9, 14.1. Analysis calculated for C29H42N2O6: C, 67.68; H, 8.23; N, 5.44. Found: C, 67.71; H, 8.20; N, 5.40. MS(APCI) m/z 514.3 [M]+, calculated for C29H42N2O6: 514.66.
3.3.3. Ethyl N-(12-isopropyl-6,9a-dimethyl-1,3-dioxo-3,3a,4,5,5a,6,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-3b,11-ethenophenanthro[1,2-c]furan-6-carbonyl)-N-(2-methoxy-2-oxoethyl)glycylglycinate (5b)
Yield 67%, mp: 60 °C, [α]20D -15 (c 0.05, CHCl3), Rf 0.40. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.55 (3H, s, CH3), 0.81–0.99 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.10 (3H, s, CH3), 1.24–1.35 (4H, m, CH, CH2), 1.42 (3H, s, CH3), 1.44–1.95 (3H, m, CH, CH2), 2.25–2.45 (5H, m, CH, CH2), 2.55 (1H, d, J = 6.8 Hz, H-1a), 2.95–3.05 (2H, m, CH2), 3.10 (1H, s, H-12), 3.45–3.79 (5H, m, CH2, CH3), 3.95–4.50 (6H, m, CH2), 5.50 (1H, br. s, H-14), 7.95 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 179.4 (C-22), 172.9 (C-24), 171.4 (C-23), 169.8 (CO), 169.4 (CO), 169.3 (CO), 148.0 (C-13), 125.2 (C-14), 61.3, 54.4, 53.6, 53.0, 49.7, 46.6, 45.7, 39.1, 38.0, 37.6, 35.8, 35.7, 35.6, 34.9, 32.7, 29.6, 27.1, 22.0, 21.9, 20.6, 19.9, 18.6, 17.2, 15.8, 14.1. Analysis calculated for C33H46N2O9: C, 64.48; H, 7.54; N, 4.56. Found: C, 64.46; H, 7.53; N, 4.54. MS(APCI) m/z 615.4 [M + H]+, calculated for C33H47N2O9: 615.74.
3.3.4. Ethyl N-(13-isopropyl-7,10a-dimethyl-1,4-dioxo-2,3,4,4a,5,6,6a,7,8,9,10,10a,10b,11,12,12a-hexadecahydro-1H-4b,12-ethenochrysene-7-carbonyl)-N-(2-methoxy-2-oxoethyl)glycylglycinate (5c)
Yield 70%, mp: 67 °C, [α]20D +26 (c 0.05, CHCl3), Rf 0.41. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.60 (3H, s, CH3), 0.61–0.90 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.18 (3H, s, CH3), 1.22 (3H, s, CH3), 1.28–1.92 (6H, m, CH, CH2), 2.14–2.25 (9H, m, CH, CH2), 2.62–2.88 (4H, m, CH, CH2), 3.15 (1H, s, H-12), 3.65–3.73 (5H, m, CH2, CH3), 3.85–4.40 (6H, m, CH2), 5.49 (1H, br. s, H-14), 6.75 (1H, br.s, NH), 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 209.6 (C-4), 208.7 (C-1), 179.1 (C-20), 170.6 (CO), 169.8 (CO), 167.4 (CO), 149.4 (C-13), 125.5 (C-14), 62.7, 60.5, 56.2, 55.2, 54.8, 52.5, 49.1, 48.9, 48.7, 41.3, 39.2, 38.6, 37.9, 37.0, 35.7, 34.9, 32.9, 32.6, 27.5, 25.4, 24.7, 22.3, 20.8, 19.9, 16.8, 16.0, 15.8. Analysis calculated for C35H50N2O8: C, 67.07; H, 8.04; N, 4.47. Found: C, 67.03; H, 8.01; N, 4.44. MS(APCI) m/z 627.4 [M + H]+, calculated for C35H51N2O8: 627.79.
3.3.5. Methyl N-(2-((2-ethoxy-2-oxoethyl)amino)-2-oxoethyl)-N-(7-isopropyl-1,4a-dimethyl-1,2,3,4,4a,9,10,10a-octahydrophenanthrene-1-carbonyl)methioninate (6a)
Yield 69%, mp: 50 °C, [α]20D -57 (c 0.05, CHCl3), Rf 0.35. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.78–1.10 (4H, m, CH, CH2), 1.18 (3H, s, H-20), 1.20 (3H, s, H-16), 1.22 (3H, s, H-15), 1.24 (3H, s, H-19), 1.38 (3H, s, CH3), 1.40–2.00 (4H, m, CH, CH2), 2.01 (3H, s, CH3), 2.20–3.00 (4H, m, H-6, H-7), 3.65–3.73 (6H, m, CH, CH2, CH3), 3.90–4.20 (6H, m, CH2), 4.50–4.70 (2H, m, CH2), 6.85 (1H, s, H-17), 7.00 (1H, d, J = 7.5 Hz, H-12), 7.18 (1H, d, J = 7.5 Hz, H-11), 8.20 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 178.1 (C-18), 171.9 (CO), 169.8 (CO), 167.4 (CO), 146.9 (C-9), 145.6 (C-13), 134.9 (C-8), 126.9 (C-17), 124.1 (C-11), 123.8 (C-12), 62.8, 61.3, 52.8, 52.1, 47.8, 46.9, 45.2, 41.2, 37.9, 36.8, 35.8, 34.4, 33.4, 32.5, 30.1, 27.1, 25.2, 23.9, 18.6, 17.9, 15.4, 14.1. Analysis calculated for C32H48N2O6S: C, 65.28; H, 8.22; N, 4.76; S, 5.44. Found: C, 65.30; H, 8.20; N, 5.00; S, 5.40. MS(APCI) m/z 588.3 [M]+, calculated for C32H48N2O6S: 588.80.
3.3.6. Methyl N-(2-((2-ethoxy-2-oxoethyl)amino)-2-oxoethyl)-N-(13-isopropyl-7,10a-dimethyl-1,4-dioxo-2,3,4,4a,5,6,6a,7,8,9,10,10a,10b,11,12,12a-hexadecahydro-1H-4b,12-ethenochrysene-7-carbonyl)methioninate (6c)
Yield 74%, mp: 74 °C, [α]20D -24 (c 0.05, CHCl3), Rf 0.30. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.50 (3H, s, CH3), 0.61–0.90 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.18 (3H, s, CH3), 1.28–1.92 (9H, m, CH, CH2), 2.00 (3H, s, CH3), 2.14–2.25 (8H, m, CH, CH2), 2.62–2.88 (4H, m, CH, CH2), 3.15 (1H, s, H-12), 3.65–3.73 (6H, m, CH, CH2, CH3), 3.85–4.40 (6H, m, CH2), 4.50–4.70 (2H, m, CH2), 5.50 (1H, br. s, H-14), 8.75 (1H, br.s, NH), 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 209.8 (C-4), 208.7 (C-1), 178.3 (C-20), 172.5 (CO), 169.7 (CO), 169.5 (CO), 149.4 (C-13), 125.7 (C-14), 62.9, 60.5, 56.3, 55.2, 54.9, 52.9, 52.2, 49.4, 47.2, 46.6, 41.2, 41.1, 40.9, 39.0, 38.4, 37.1, 37.0, 35.8, 34.7, 32.9, 31.1, 27.6, 21.9, 20.8, 19.9, 17.3, 17.0, 16.4, 16.0, 15.4. Analysis calculated for C38H56N2O8S: C, 65.12; H, 8.05; N, 4.00; S, 4.57. Found: C, 65.20; H, 8.00; N, 4.10; S, 4.60. MS(APCI) m/z 701.4 [M + H]+, calculated for C38H57N2O8S: 701.93.
3.3.7. Methyl N-(12-isopropyl-6,9a-dimethyl-1,3-dioxo-3,3a,4,5,5a,6,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-3b,11-ethenophenanthro[1,2-c]furan-6-carbonyl)-N-(2-((2-morpholinoethyl)amino)-2-oxoethyl)histidinate (7b)
Yield 66%, mp: 55 °C, [α]20D +15 (c 0.05, CHCl3), Rf 0.30. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.55 (3H, s, CH3), 0.81–0.99 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.10 (3H, s, CH3), 1.24–1.95 (12H, m, CH, CH2), 2.25–2.45 (5H, m, CH, CH2), 2.95–3.05 (8H, m, CH2,), 3.15 (1H, s, H-12), 3.57–3.75 (7H, m, CH2, CH3), 4.05–4.10 (2H, br.s., CH2), 5.42 (1H, br. s, H-14), 5.60–5.72 (1H, m, CH), 6.25 (1H, s, CH), 6.50 (1H, s, CH), 7.90 (1H, br.s, NH), 9.11 (1H, br.s, NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 178.4 (C-20), 172.9 (CO), 172.0 (C-24), 171.9 (C-23), 171.1 (CO), 151.1, 148.2 (C-13), 141.1, 125.5 (C-14), 125.4, 60.6, 60.5, 60.3, 58.6, 58.7, 56.0, 55.9, 54.9, 53.6, 53.0, 49.7, 46.6, 45.7, 39.1, 38.0, 37.6, 35.8, 35.7, 34.9, 32.7, 29.6, 27.1, 22.0, 21.9, 20.6, 19.9, 18.6, 17.2, 15.8. 52.0, 50.6, 49.3, 49.0, 41.5, 39.2, 38.9, 38.5, 37.7, 37.0, 36.7, 35.1, 34.7, 32.9, 27.7, 25.4, 21.8, 20.8, 19.9, 16.9, 16.7, 16.4, 15.9. Analysis calculated for C39H55N5O8: C, 64.89; H, 7.68; N, 9.70. Found: C, 64.90; H, 7.77; N, 9.74. MS(APCI) m/z 722.4 [M + H]+, calculated for C39H56N5O8: 722.90.
3.3.8. Methyl N-(13-isopropyl-7,10a-dimethyl-1,4-dioxo-2,3,4,4a,5,6,6a,7,8,9,10,10a,10b,11,12,12a-hexadecahydro-1H-4b,12-ethenochrysene-7-carbonyl)-N-(2-((2-morpholinoethyl)amino)-2-oxoethyl)histidinate (7c)
Yield 69%. Here, mp: 75 °C, [α]20D +14 (c 0.05, CHCl3), Rf 0.33. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.60 (3H, s, CH3), 0.61–0.90 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.18 (3H, s, CH3), 1.28–1.92 (6H, m, CH, CH2), 2.14–2.25 (15H, m, CH, CH2), 2.62–2.88 (8H, m, CH, CH2), 3.15 (1H, s, H-12), 3.60–3.80 (7H, m, CH2, CH3), 4.08–4.11 (2H, br.s., CH2), 5.40 (1H, br. s, H-14), 5.60–5.75 (1H, m, CH), 6.29 (1H, s, CH), 6.45 (1H, s, CH), 7.95 (1H, br.s, NH), 9.15 (1H, br.s, NH),13C NMR (δ, ppm, CDCl3, 125.5 MHz): 210.1 (C-4), 208.9 (C-1), 179.1 (C-20), 172.9 (CO), 170.2 (CO), 151.2, 149.6 (C-13), 141.1, 125.5 (C-14), 125.4, 60.6, 60.5, 60.3, 58.6, 58.7, 56.0, 55.9, 54.9, 52.0, 50.6, 49.3, 49.0, 41.5, 39.2, 38.9, 38.5, 37.7, 37.0, 36.7, 35.1, 34.7, 32.9, 27.7, 25.4, 21.8, 20.8, 19.9, 16.9, 16.7, 16.4, 15.9. Analysis calculated for C41H59N5O7: C, 67.10; H, 8.10; N, 9.54. Found: C, 67.20; H, 8.10; N, 9.60. MS(APCI) m/z 734.4 [M + H]+, calculated for C41H60N5O7: 734.95.
3.3.9. Methyl N-(12-isopropyl-6,9a-dimethyl-1,3-dioxo-3,3a,4,5,5a,6,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-3b,11-ethenophenanthro[1,2-c]furan-6-carbonyl)-N-(2-((2-morpholinoethyl)amino)-2-oxoethyl)phenylalaninate (8b)
Yield 82%, mp: 63 °C, [α]20D -42 (c 0.05, CHCl3), Rf 0.45. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.55 (3H, s, CH3), 0.81–0.99 (2H, m, CH2), 0.98 (3H, d, J = 6.9 Hz, CH3), 1.00 (3H, d, J = 6.9 Hz, CH3), 1.10 (3H, s, CH3), 1.24–1.95 (15H, m, CH, CH2), 2.25–2.95 (8H, m, CH, CH2), 3.15–3.20 (4H, m, H-12, CH3), 3.25–4.08 (8H, m, CH2), 4.50–4.70 (1H, m, CH), 5.50 (1H, br. s, H-14), 7.10–7.22 (5H, m, H-Ar), 8.50 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 178.4 (C-24), 177.2 (C-23), 172.8 (CO), 170.9 (CO), 167.7 (CO), 148.1 (C-13), 137.2, 129.5, 129.4, 129.1, 128.9, 127.1, 125.2 (C-14), 66.7, 66.5, 56.9, 56.1, 53.7, 53.2, 53.1, 49.6, 49.2, 45.7, 39.1, 38.0, 37.4, 36.8, 36.2, 35.7, 34.8, 34.7, 32.7, 29.6, 27.2, 21.9, 20.6, 19.9, 18.6, 17.1, 16.6, 16.0, 15.5. Analysis calculated for C42H57N3O8: C, 68.92; H, 7.85; N, 5.74. Found: C, 69.00; H, 7.90; N, 5.80. MS(APCI) m/z 732.4 [M + H]+, calculated for C42H58N3O8: 732.93.
3.3.10. N-Benzyl-N-(2-(tert-butylamino)-2-oxoethyl)-13-isopropyl-7,10a-dimethyl-1,4-dioxo-4,5,6,6a,7,8,9,10,10a,10b,11,12-dodecahydro-1H-4b,12-ethenochrysene-7-carboxamide (9d)
Yield 83%, mp: 51 °C, [α]20D -14 (c 0.05, CHCl3), Rf 0.41. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.70 (3H, s, H-18), 0.71–0.75 (3H, m, CH, CH2), 0.99 (3H, d, J = 6.9 Hz, H-16/H-17), 1.02 (3H, d, J = 6.9 Hz, H-16/H-17), 1.22 (3H, s, H-19), 1.25 (9H, s, 3CH3), 1.28–1.35 (5H, m, CH2), 1.40–1.68 (6H, m, CH, CH2), 2.35–2.99 (2H, m, CH), 3.70 (2H, s, CH2), 4.95 (2H, dd, J = 6.8 Hz, H-2, H-3); 5.60 (2H, s, CH2), 5.70 (1H, s, H-14), 6.10 (1H, br.s., NH), 7.10–7.22 (5H, m, H-Ar); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 183.9 (C-20), 179.9 (C-1), 178.9 (C-4), 168.4 (CO), 158.9 (C-1a), 153.8 (C-4a), 151.1 (C-2), 148.9 (C-13), 136.5 (C-3), 128.9, 128.8, 127.8, 127.6, 127.5, 127.4, 126.9 (C-14), 56.4, 55.4, 53.2, 53.1, 51.2, 50.2, 49.2, 47.2, 39.0, 36.6, 36.2, 32.0, 31.9, 28.8 (3CH3), 24.5, 22.3, 20.7, 20.4, 18.8, 17.3, 16.7. Analysis calculated for C39H50N2O4: C, 76.69; H, 8.25; N, 4.59. Found: C, 76.71; H, 8.28; N, 4.60. MS(APCI) m/z 611.4 [M + H]+, calculated for C39H51N2O4: 611.84.
3.3.11. Methyl N-(2-(tert-butylamino)-2-oxoethyl)-N-(13-isopropyl-7,10a-dimethyl-1,4-dioxo-4,5,6,6a,7,8,9,10,10a,10b,11,12-dodecahydro-1H-4b,12-ethenochrysene-7-carbonyl)phenylalaninate (10d)
Yield 80%, mp: 59 °C, [α]20D -93 (c 0.05, CHCl3), Rf 0.40. 1H NMR (δ, ppm, CDCl3, 500 MHz): 0.70 (3H, s, H-18), 0.71–0.75 (3H, m, CH, CH2), 0.99 (3H, d, J = 6.9 Hz, H-16/H-17), 1.02 (3H, d, J = 6.9 Hz, H-16/H-17), 1.22 (3H, s, H-19), 1.25 (9H, s, 3CH3), 1.28–1.35 (5H, m, CH2), 1.40–1.68 (6H, m, CH, CH2), 2.35–2.99 (2H, m, CH), 3.23–3.40 (2H, m, CH2), 3.75 (3H, s, CH3), 3.95–4.0 (2H, m, CH2), 5.10 (1H, s, CH), 5.60 (1H, s, H-14), 6.49 (2H, dd, J = 6.8 Hz, 2H, H-2, H-3); 7.10–7.22 (5H, m, H-Ar), 7.95 (1H, br.s., NH); 13C NMR (δ, ppm, CDCl3, 125.5 MHz): 185.2 (C-1), 184.0 (C-4), 178.3 (C-20), 171.6 (CO), 168.1 (CO), 152.8 (C-1a), 150.9 (C-4a), 150.7 (C-13), 137.5 (C-2), 137.4 (C-3), 133.6, 129.4, 128.9, 127.7, 127.4, 127.3, 127.2, 127.1 (C-14), 67.3, 55.4, 55.3, 54.9, 52.8, 51.6, 49.5, 49.3, 49.2, 46.8, 46.5, 39.4, 39.0, 36.6, 36.2, 32.0, 31.9, 29.8, 28.8 (3CH3), 27.2, 24.5, 22.3, 20.7, 20.4, 18.8, 17.3, 16.7. Analysis calculated for C42H54N2O6: C, 73.87; H, 7.97; N, 4.10. Found: C, 73.90; H, 7.95; N, 4.10. MS(APCI) m/z 682.4 [M]+, calculated for C42H54N2O6: 682.90.
3.4. Biological Assay
The in vitro anticancer screening of the tested compounds is given in Supplementary Materials.
4. Conclusions
Thus, the potential of natural and semisynthetic diterpene acids as carboxyl moieties for modification under Ugi and Passerini MCR conditions was demonstrated using dehydroabietic acid and levopimaric acid diene adducts. The first set of P-3CR products was obtained by varying isocyanides, while the second set of U-4CR products used various isocyanides and amines, including amino acid methyl esters. The novel series of diterpene α-acyloxycarboxamide и α-acylamidocarboxamides have been synthesized in good yields and displayed different antiproliferative effects on an NCI-60 panel of tumor cell lines. Among them, eleven compounds inhibited the growth of cancer cells in 4 to 39 cell lines in a one-dose assay. The greatest activity in the five-dose assay was shown by adducts 3d, 9d and 10d, which not only stopped the growth of cancer cells but also caused their death. Compound 3d, in contrast to 9d and 10d, showed moderate positive correlations with standard anticancer drugs that can influence the function of kinase receptors and EGFRs, and thereby effectively reduce the activity of oncogenes that affect the uncontrolled division of tumor cells. The ADMET analysis acknowledges the favorable prognosis using compounds as potential anticancer agents, exceeding doxorubicin in terms of toxicity, absorption and distribution. Based on the obtained data, it can be concluded that among the synthesized diterpene α-acyloxycarboxamide and α-acylamidocarboxamides, the derivatives of 1a,4a-dehydroquinopimaric acid are the most promising compounds for further study of their antitumor activity, and, in general, the decoration of abietane acids with peptide-like substituents leads to submicromolar cytotoxicity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29153532/s1, Pages S1 and S2: Biological assay, Figures S1–S20: One-dose mean graph of NCI-60 cell line screening data for compounds, Figures S21–S38: Anticancer screening data of compounds at five-dose assay, Figure S39: One-dose mean graph of NCI-60 cell line screening data for compound 1d, Figures S40–S75: NMR spectra of products. Reference [51] is cited in the supplementary materials.
Author Contributions
E.T. and A.S. drafted the preparation method and prepared the compounds for screening; D.B. conducted ADMET experiments; and O.K. brought the idea and managed the research. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Russian Science Foundation Grant No. 23-23-00169, https://rscf.ru/project/23-23-00169/ (accessed on 1 January 2023).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank the National Cancer Institute for the screening of cytotoxicity of compounds 2a, 2b, 2d, 3a, 3b, 3c, 3d, 4a, 4b, 4c, 5a, 5b, 5c, 6a, 6c, 7b, 7c, 8b, 9d and 10d. The spectra were recorded at the Center for the Collective Use ‘Chemistry’ of the Ufa Institute of Chemistry of the UFRC RAS and RCCU “Agidel” of the UFRC RAS.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Edwards, B.K. Annual Report to the Nation on the Status of Cancer, 1975–2005, Featuring Trends in Lung Cancer, Tobacco Use, and Tobacco Control. J. Natl. Cancer Inst. 2008, 100, 1672–1694. [Google Scholar] [CrossRef]
- Harris, A.L.; Hochhauser, D. Mechanisms of multidrug resistance in cancer treatment. Acta Oncol. 1992, 31, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, D.; Szewczyk, A.; Radzka, J.; Dubińska-Magiera, M.; Kazimierczak, W.; Daczewska, M.; Migocka-Patrzałek, M. The natural origins of cytostatic compounds used in rhabdomyosarcoma therapy. Adv. Clin. Exp. Med. 2023, 32, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Melfi, F.; Carradori, S.; Mencarelli, N.; Campestre, C.; Gallorini, M.; Di Giacomo, S.; Di Sotto, A. Natural products as a source of new anticancer chemotypes. Expert Opin. Ther. Pat. 2023, 33, 721–744. [Google Scholar] [CrossRef] [PubMed]
- Amiri, M.; Molavi, O.; Sabetkam, S.; Jafari, S.; Montazersaheb, S. Stimulators of immunogenic cell death for cancer therapy: Focusing on natural compounds. Cancer Cell. Int. 2023, 23, 200. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.C.; Kumar, N.V.A.; Thakur, G. Developments in the anticancer activity of structurally modified curcumin: An up-to-date review. Eur. J. Med. Chem. 2019, 177, 76–104. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Verma, N. Nature curing cancer—Review on structural modification studies with natural active compounds having anti-tumor efficiency. Biotechnol. Rep. 2015, 6, 64–78. [Google Scholar] [CrossRef]
- Lewandowska, U.; Fichna, J.; Gorlach, S. Enhancement of anticancer potential of polyphenols by covalent modifications. Biochem. Pharmacol. 2016, 109, 1–13. [Google Scholar] [CrossRef]
- Liu, Z.-Q. Ugi and Passerini Reactions as Successful Models for Investigating Multicomponent Reactions. Cur. Org. Chem. 2014, 18, 719–739. [Google Scholar] [CrossRef]
- De Moliner, F.; Banfi, L.; Riva, R.; Basso, A. Beyond Ugi and Passerini Reactions: Multicomponent Approaches Based on Isocyanides and Alkynes as an Efficient Tool for Diversity Oriented Synthesis. Comb. Chem. High Throughput Screen. 2011, 14, 782–810. [Google Scholar] [CrossRef] [PubMed]
- Ayoup, M.S.; Mansour, A.F.; Abdel-Hamid, H.; Abu-Serie, M.M.; Mohyeldin, S.M.; Teleb, M. Nature-inspired new isoindole-based Passerini adducts as efficient tumor-selective apoptotic inducers via caspase-3/7 activation. Eur. J. Med. Chem. 2023, 245, 114865. [Google Scholar] [CrossRef] [PubMed]
- Avilés, E.; Prudhomme, J.; Le Roch, K.G.; Franzblau, S.G.; Chandrasena, K.; Mayer, A.M.S.; Rodríguez, A.D. Synthesis and preliminary biological evaluation of a small library of hybrid compounds based on Ugi isocyanide multicomponent reactions with a marine natural product scaffold. Bioorganic Med. Chem. Lett. 2015, 25, 5339–5343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomohara, K.; Ohashi, N.; Uchida, T.; Nose, T. Synthesis of natural product hybrids by the Ugi reaction in complex media containing plant extracts. Sci. Rep. 2022, 12, 15568. [Google Scholar] [CrossRef]
- Czollner, L.; Beseda, I.; Jordis, U.; Stanetty, C.; Amer, H.; Del Ruiz-Ruiz, M.C.; Kosma, P.; Classen-Houben, D. Ugi reactions of tertiary carboxylic acids: Combinatorial synthesis of glycyrrhetinic acid derivatives. In Proceedings of the 13th International Electronic Conference on Synthetic Organic Chemistry (ECSOC-13), Sciforum Electronic Conference Series. elektronische Konferenz, 1–30 November 2009; Volume 13, p. b001. [Google Scholar]
- Rodríguez-López, F.; García-Gutiérrez, H.A.; Gámez-Montaño, R. Synthesis of Bis-Amides Employing a Plant-Derived Triterpenoid as Component in the Ugi Reaction. Chem. Proc. 2022, 12, 37. [Google Scholar] [CrossRef]
- Wiemann, J.; Heller, L.; Csuk, R. An access to a library of novel triterpene derivatives with a promising pharmacological potential by Ugi and Passerini multicomponent reactions. Eur. J. Med. Chem. 2018, 150, 176–194. [Google Scholar] [CrossRef]
- Sultani, H.N.; Morgan, I.; Hussain, H.; Roos, A.H.; Haeri, H.H.; Kaluderović, G.N.; Hinderberger, D.; Westermann, B. Access to New Cytotoxic Triterpene and Steroidal Acid-TEMPO Conjugates by Ugi Multicomponent-Reactions. Int. J. Mol. Sci. 2021, 22, 7125. [Google Scholar] [CrossRef]
- Veena, K.S.; Taniya, M.S.; Ravindran, J.; Thangarasu, A.K.; Priya, S.; Lankalapalli, R.S. Semi-synthetic diversification of coronarin D, a labdane diterpene, under Ugi reaction conditions. Nat. Prod. Res. 2020, 36, 334–340. [Google Scholar] [CrossRef]
- Wiemann, J.; Fischer, L.; Kessler, J.; Ströhl, D.; Csuk, R. Ugi multicomponent-reaction: Syntheses of cytotoxic dehydroabietylamine derivatives. Bioorg. Chem. 2018, 81, 567–576. [Google Scholar] [CrossRef]
- Heise, N.V.; Schmidt, A.; Schüler, J.-A.; Csuk, R. Dehydroabietylamine derived bistetrazoles from ultrasound-assisted pseudo-seven-component Ugi reactions act as efficient and selective inhibitors of cholinesterases. Eur. J. Med. Chem. Rep. 2024, 10, 100124. [Google Scholar] [CrossRef]
- Tretyakova, E.V.; Ma, X.; Kazakova, O.B.; Shtro, A.A.; Petukhova, G.D.; Smirnova, A.A.; Xu, H.; Xiao, S. Abietic, maleopimaric and quinopimaric dipeptide Ugi-4CR derivatives and their potency against influenza A and SARS-CoV-2. Nat. Prod. Res. 2023, 37, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, A.A.; Zakirova, L.M.; Smirnova, I.E.; Tretyakova, E.V. Synthesis of Novel Diterpenic Peptides via the Ugi Reaction and Their Anticancer Activities. Molbank 2023, 2023, M1707. [Google Scholar] [CrossRef]
- Smirnova, A.A.; Tretyakova, E.V.; Kazakova, O.B. Inhibiting the growth of cancer cells maleopimarate amidoimide bis-1H-tetrazoles synthesized via azido-Ugi reaction. Mendeleev Commun. 2024, 34, 509–510. [Google Scholar]
- Tolstikov, G.A.; Tolstikova, T.G.; Shults, E.E.; Tolstikov, S.E.; Khvostov, M.V. Resin acids of Russian conifers. In Chemistry, Pharmacology; Trofimov, B.A., Ed.; Geo: Novosibirsk, Russia, 2011; 395 p. (In Russian) [Google Scholar]
- Grever, M.R.; Schepartz, S.A.; Chabner, B.A. The National Cancer Institute: Cancer drug discovery and development program. Semin. Oncol. 1992, 19, 622–638. [Google Scholar] [PubMed]
- Boyd, M.R.; Paull, K.D. Some practical considerations and applications of the national cancer institute in vitro anticancer drug discovery screen. Drug Rev. Res. 1995, 34, 91–109. [Google Scholar] [CrossRef]
- Monks, A.; Scudiero, D.; Skehan, P.; Shoemaker, R.; Paull, K.; Vistica, D.; Hose, C.; Langley, J.; Cronisie, P.; Viagro-Wolff, A.; et al. Feasibility of a High-Flux Anticancer Drug Screen Using a Diverse Panel of Cultured Human Tumor Cell Lines. J. Natl. Cancer Inst. 1991, 83, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Monks, A.; Scudiero, D.; Johnson, G.S.; Paull, K.D.; Sausville, E.A. The NCI anti-cancer drug screen: A smart screen to identify effectors of novel targets. Anti-Cancer Drug Des. 1997, 12, 533–541. [Google Scholar]
- Weinstein, J.N.; Myers, T.G.; O’Connor, P.M.; Friend, S.H., Jr.; Fornace, A.J.; Kohn, K.W.; Fojo, T.; Bates, S.E.; Rubinstein, L.V.; Anderson, N.L.; et al. An Information-Intensive Approach to the Molecular Pharmacology of Cancer. Science 1997, 275, 343–349. [Google Scholar] [CrossRef] [PubMed]
- DTP Databases and Search Tools. Available online: https://dtp.cancer.gov/databases_tools/data_search.htm (accessed on 1 November 2018).
- Rostom, S.A.F. Synthesis and in vitro antitumor evaluation of some indeno [1,2-c]pyrazol(in)es substituted with sulfonamide, sulfonylurea(-thiourea) pharmacophores, and some derived thiazole ring systems. Bioorg. Med. Chem. 2006, 14, 6475–6485. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Available online: https://ioa.cancer.gov/oncologydrugscompare/webpages/ (accessed on 4 March 2021).
- Mukaka, M.M. A guide to appropriate use of Correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar] [PubMed Central]
- Duffy, M.J.; Tang, M.; Rajaram, S.; O’Grady, S.; Crown, J. Targeting Mutant p53 for Cancer Treatment: Moving Closer to Clinical Use? Cancers 2022, 14, 4499. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, K.M.; Zhang, B.Z.; Jackson, T.D.; Ogunkola, M.O.; Nijagal, B.; Milne, J.V.; Sallman, D.A.; Ang, C.-S.; Nikolic, I.; Kearney, C.J.; et al. Eprenetapopt triggers ferroptosis, inhibits NFS1 cysteine desulfurase, and synergizes with serine and glycine dietary restriction. Sci. Adv. 2022, 8, eabm9427. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, P.L. 3-bromopyruvate (3BP) a fast acting, promising, powerful, specific, and effective “small molecule” anti-cancer agent taken from labside to bedside: Introduction to a special issue. J. Bioenerg. Biomembr. 2012, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Barnard, J.P.; Reynafarje, B.; Pedersen, P.L. Glucose catabolism in African trypanosomes. Evidence that the terminal step is catalyzed by a pyruvate transporter capable of facilitating uptake of toxic analogs. J. Biol. Chem. 1993, 268, 3654–3661. [Google Scholar] [CrossRef] [PubMed]
- Mina, R.; Falcone, A.P.; Bringhen, S.; Liberati, A.M.; Pescosta, N.; Petrucci, M.T.; Ciccone, G.; Capra, A.; Patriarca, F.; Rota-Scalabrini, D.; et al. Ixazomib-based induction regimens plus ixazomib maintenance in transplant-ineligible, newly diagnosed multiple myeloma: The phase II, multi-arm, randomized UNITO-EMN10 trial. Blood Cancer J. 2021, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.-C.; Lin, C.-C.; Lee, J.-H.; Yang, J.C.-H. Update on recent preclinical and clinical studies of T790M mutant-specific irreversible epidermal growth factor receptor tyrosine kinase inhibitors. J. Biomed. Sci. 2016, 23, 86. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, L.N.; Larkins, E.; Akinboro, O.; Roy, P.; Amatya, A.K.; Fiero, M.H.; Mishra-Kalyani, P.S.; Helms, W.S.; Myers, C.E.; Skinner, A.M.; et al. FDA Approval Summary: Capmatinib and Tepotinib for the Treatment of Metastatic NSCLC Harboring MET Exon 14 Skipping Mutations or Alterations. Clin. Cancer Res. 2022, 28, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Hutson, T.E.; Tomczak, P.; Michaelson, M.D.; Bukowski, R.M.; Rixe, O.; Oudard, S.; Negrier, S.; Szczylik, C.; Kim, S.T.; et al. Sunitinib versus Interferon Alfa in Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2007, 356, 115–124. [Google Scholar] [CrossRef]
- Tretyakova, E.V.; Smirnova, I.E.; Kazakova, O.B.; Tolstikov, G.A.; Yavorskaya, N.P.; Golubeva, I.S.; Pugacheva, R.B.; Apryshko, G.N.; Poroikov, V.V. Synthesis and anticancer activity of quinopimaric and maleopimaric acids’ derivatives. Bioorg. Med. Chem. 2014, 22, 6481–6489. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Halbrook, N.J.; Lawrence, R.V. The Isolation of Dehydroabietic Acid from Disproportionated Rosin. J. Org. Chem. 1966, 31, 4246–4247. [Google Scholar] [CrossRef]
- Zalkov, L.U.; Ford, R.A.; Cutney, J.P. The Oxidation of Maleopimaric Acid with Alkaline Permanganate. J. Org. Chem. 1962, 27, 3535–3539. [Google Scholar] [CrossRef]
- Herz, W.; Nair, M.G. Resin acids. XIX. Structure and stereochemistry of adducts of levopimaric acid with cyclopentenone and 1-cyclopentene-3,5-dione. Fovarski reaction of an enedione Epoxide. J. Org. Chem. 1969, 34, 4016–4023. [Google Scholar] [CrossRef]
- Shul’ts, E.E.; Oleinikov, D.S.; Nechepurenko, I.V.; Shakirov, M.M.; Tolstikov, G.A. Synthetic transformations of higher terpenoids: XVIII. Synthesis of optically active 9,10-anthraquinone derivatives. Russ. J. Org. Chem. 2009, 45, 102–114. [Google Scholar] [CrossRef]
- Monga, M.; Sausville, E.A. Developmental Therapeutics Program at the NCI: Molecular Target and Drug Discovery Process. Leukemia 2002, 16, 520–526. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).