Exploring the Biological Pathways of Siderophores and Their Multidisciplinary Applications: A Comprehensive Review
Abstract
1. Introduction
2. Results
2.1. Classification of Siderophores
2.2. Synthesis of Siderophores
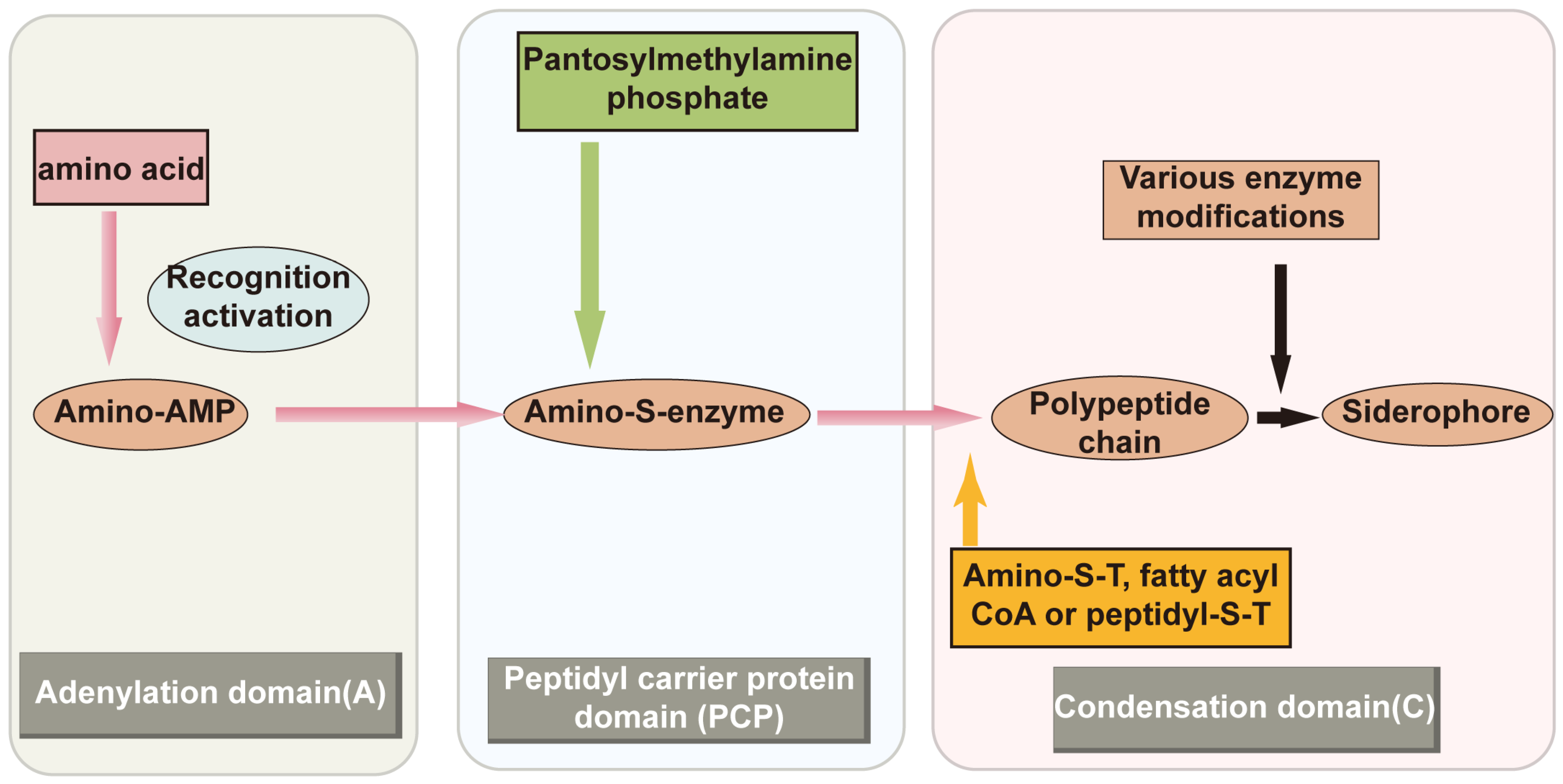
2.2.1. NRPS Pathway Synthesis of Siderophores
2.2.2. Synthesis of Siderophores Independent of the NRPS Pathway (NIS)
2.3. Sources of Siderophore Secretion
2.3.1. Plants
2.3.2. Microorganisms
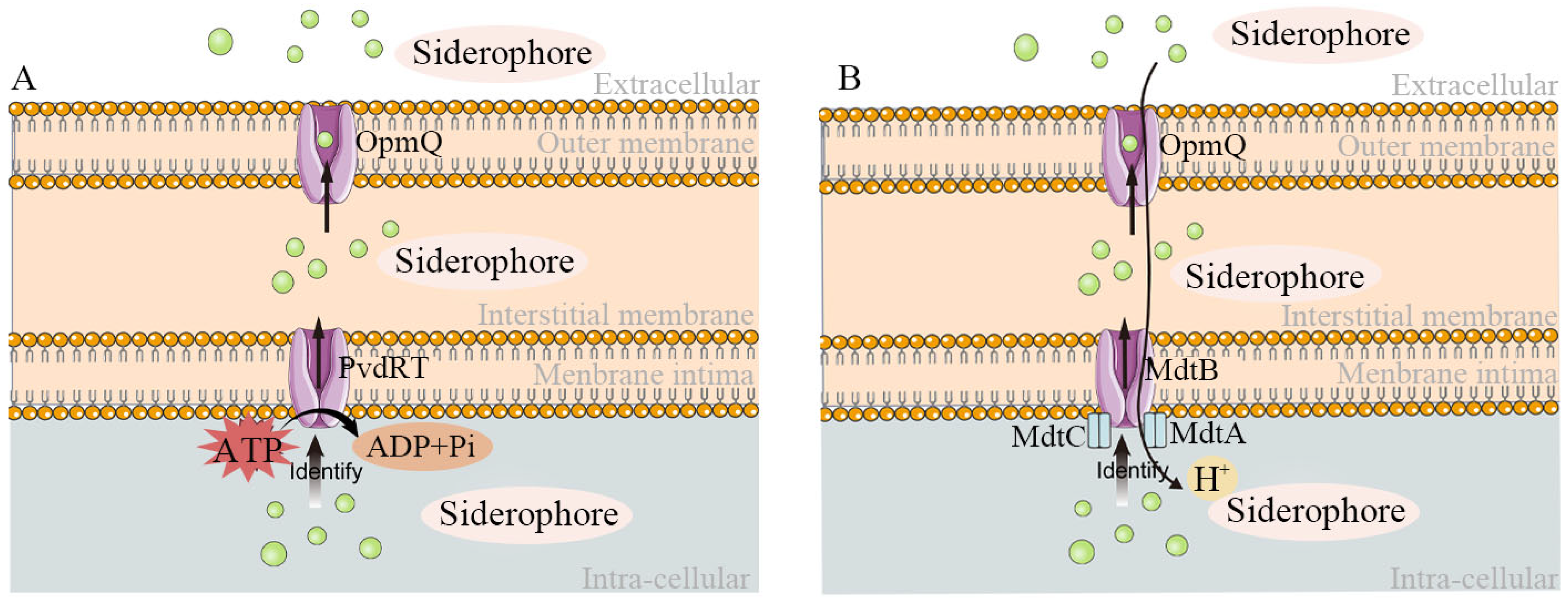
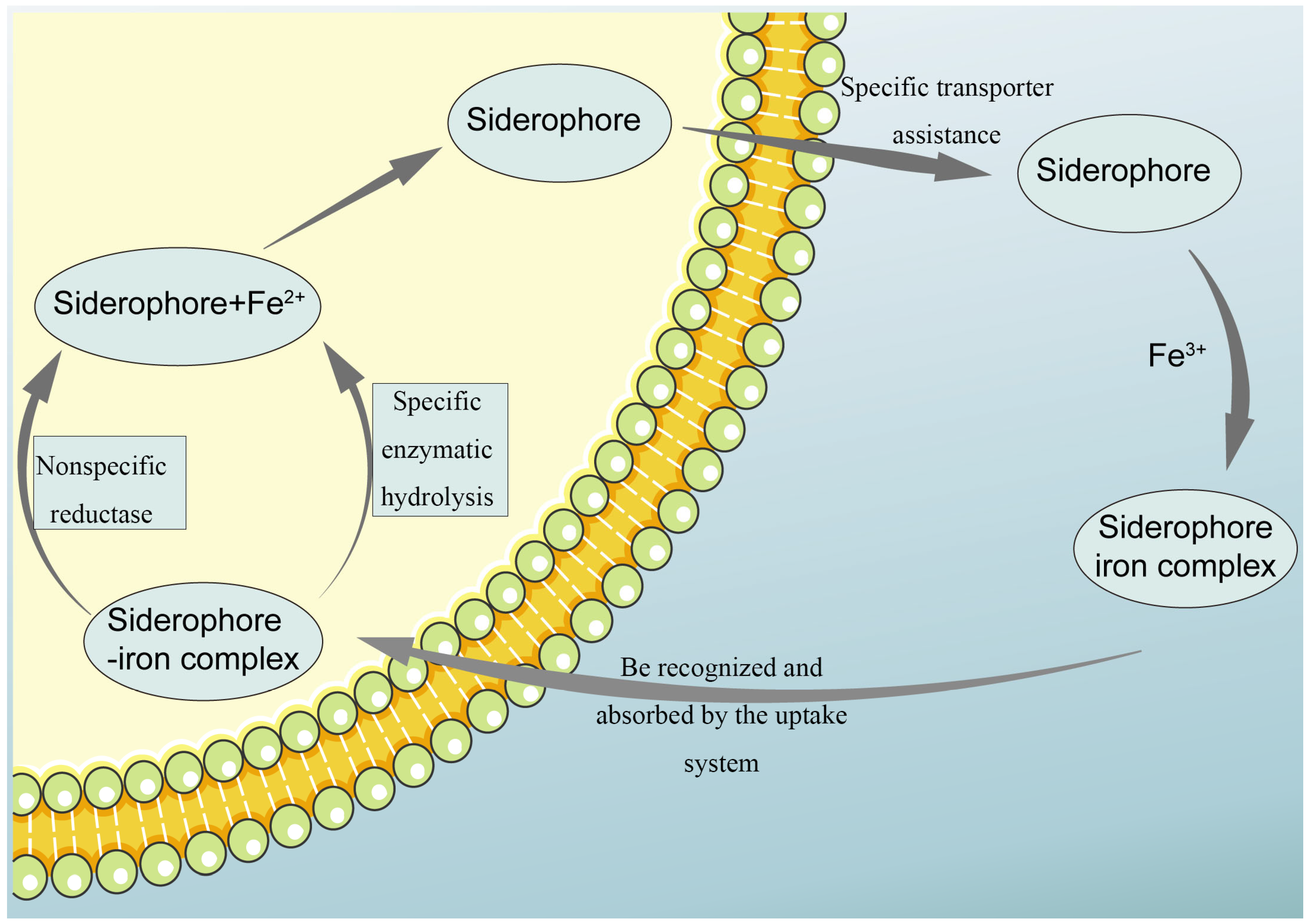
2.4. Secretion and Mechanism of Action of Siderophores
2.5. Functions of Siderophores
2.5.1. Sustaining Normal Biological Activities
2.5.2. Biological Control
2.5.3. Environmental Protection
2.5.4. Disease Treatment
2.5.5. Remediation of Hydrocarbon Pollution
2.5.6. Remediation of Heavy Metal Pollution
2.5.7. Additional Functions
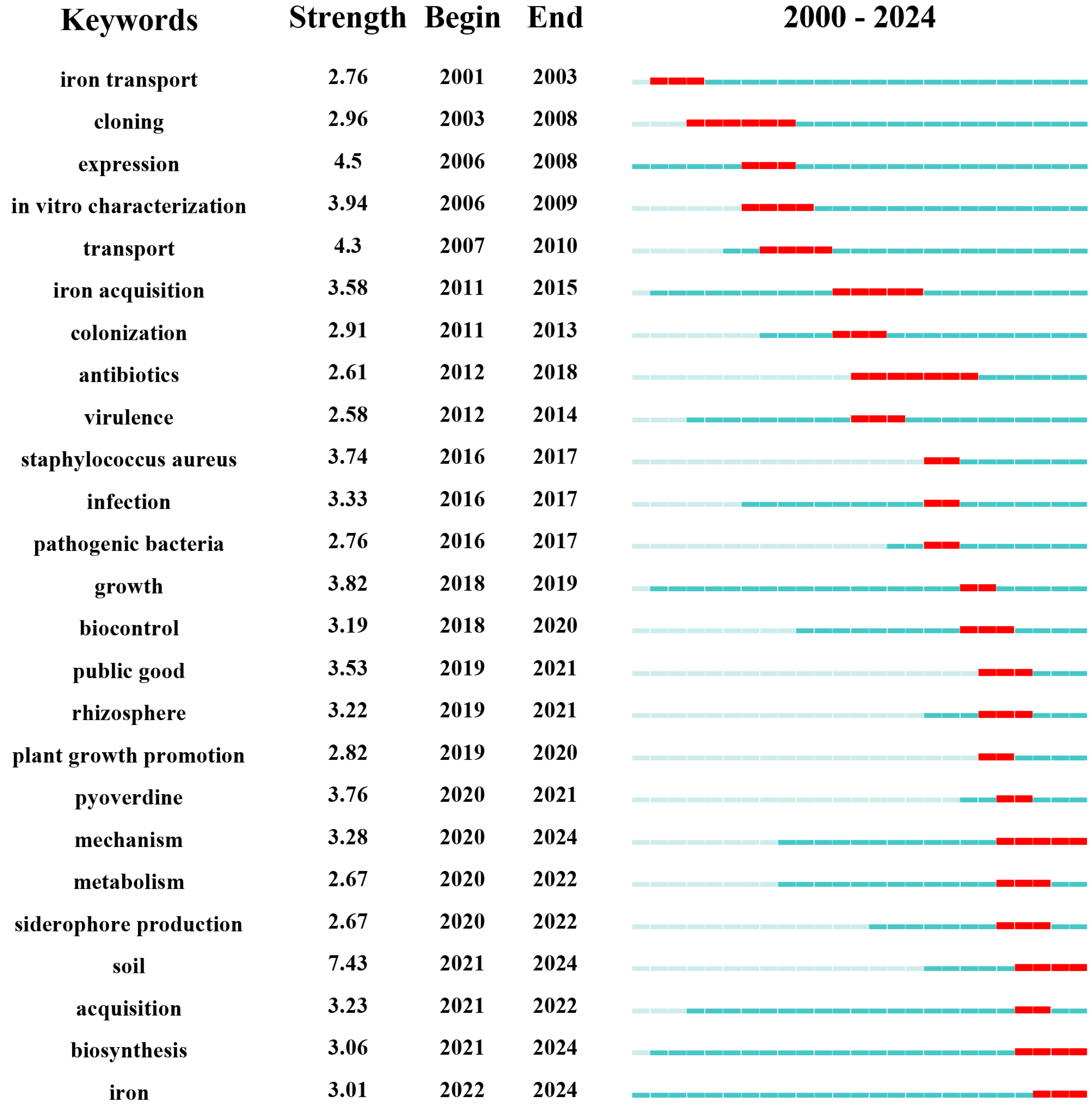
3. Research Trends and Hotspots in the Field of Siderophore Secretion
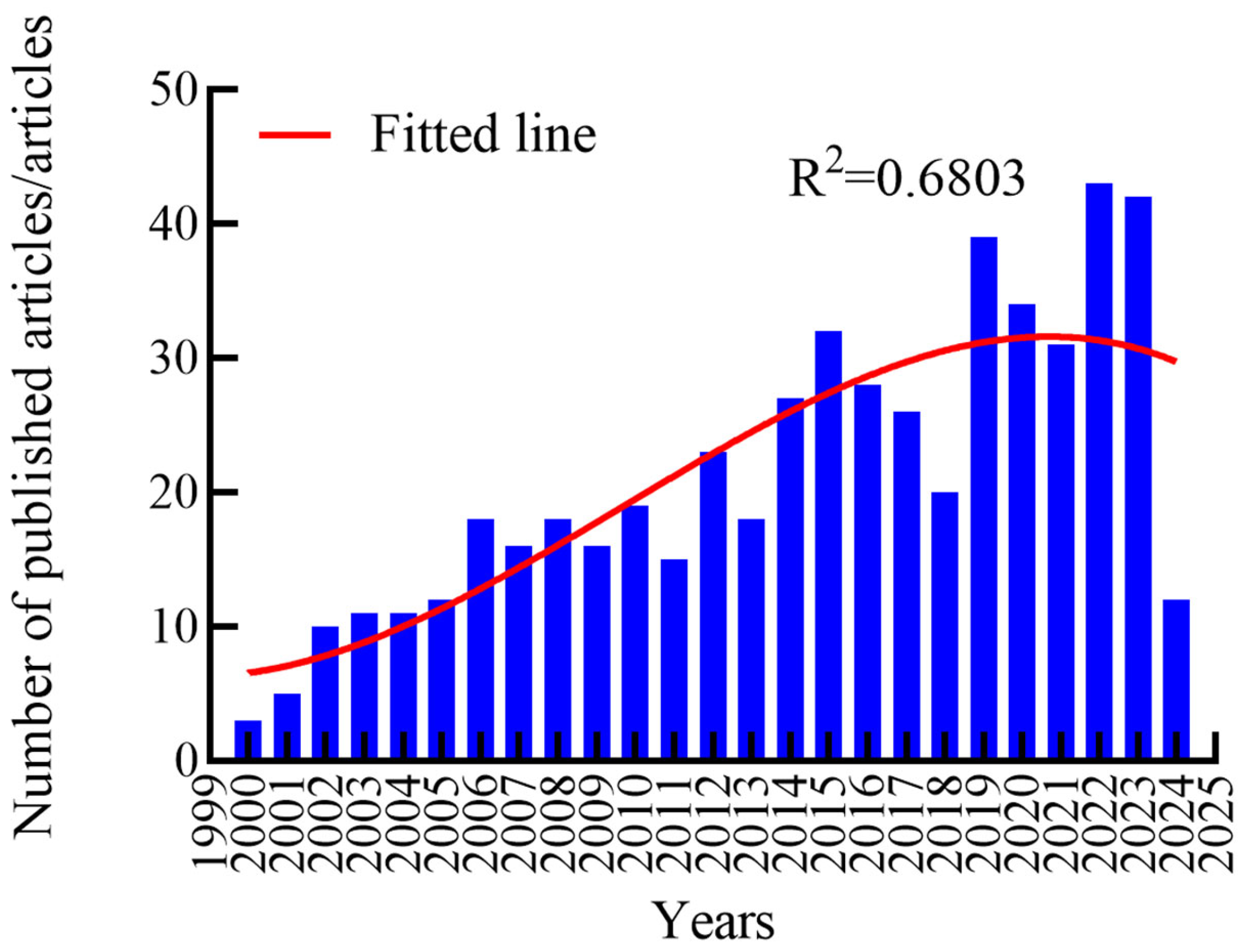
3.1. Annual Publication Volume
3.2. Analysis of Research Hotspots
3.3. Trends in Theme Evolution
4. Future Prospects
- (1)
- Further Exploration of Siderophore Secretion Mechanisms: Deepen the understanding of the molecular mechanisms and regulatory pathways controlling siderophore production. This includes examining molecular interactions and the complex regulatory networks that influence siderophore production and secretion, aiming to uncover new details about these essential processes.
- (2)
- Enhanced Study of Siderophores in Animals and Plants: Expand research into the roles and dynamics of siderophores in non-microbial organisms. By broadening the scope to include animal and plant systems, researchers can gain a holistic view of siderophore activities across various biological domains, potentially uncovering unique uses and functions.
- (3)
- Investigation of Siderophore-Related Genes and Toxicity: Focus on identifying and characterizing the genes involved in siderophore synthesis and secretion, as well as assessing the toxicity of siderophores. This research could provide critical insights necessary for the safe and effective application of siderophores in agriculture, medicine, and environmental management.
- (4)
- Development of New Techniques and Approaches: Pursue innovative research methods, utilizing advances in bioinformatics, genomics, and other cutting-edge technologies. This approach should also incorporate interdisciplinary collaboration, bringing together experts from chemistry, soil science, ecology, and other fields to foster a comprehensive and integrated understanding of siderophores. Such collaborative efforts can accelerate discoveries and applications of siderophores in various environmental and biological contexts.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saha, R.; Saha, N.; Donofrio, R.S.; Bestervelt, L.L. Microbial siderophores: A mini review. J. Basic Microbiol. 2012, 53, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Hasnain, A.; Kanwal, S. Biological Uses and Importance of Iron Regulation. MARKHOR (J. Zool.) 2020, 1, 11–13. [Google Scholar] [CrossRef]
- Raymond, K.; Dertz, E.A.; Kim, S.S. Enterobactin-an-archetype-for-microbial-iron-transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Li, L.; Dong, W.; He, L.; Sheng, X. Impact of the Iron-Response Regulator Genes on the Release of Iron and Aluminum from Biotite by Rhizobium pusense S41. Geomicrobiol. J. 2024, 41, 298–307. [Google Scholar] [CrossRef]
- Liu, L.; Wang, W.; Wu, S.; Gao, H. Recent Advances in the Siderophore Biology of Shewanella. Front. Microbiol. 2022, 13, 823758. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, Z.P.; Lin, Z.; Wei, G.; Wen, X.; Li, S.; Yang, X.; Zhang, Q.; Jing, C.; Dai, Y.; et al. Drug Repurposing by Siderophore Conjugation: Synthesis and Biological Evaluation of Siderophore-Methotrexate Conjugates as Antibiotics. Angew. Chem. Int. Ed. 2022, 61, e202204139. [Google Scholar] [CrossRef] [PubMed]
- Khasheii, B.; Mahmoodi, P.; Mohammadzadeh, A. Siderophores: Importance in bacterial pathogenesis and applications in medicine and industry. Microbiol. Res. 2021, 250, 126790. [Google Scholar] [CrossRef]
- Meikle, V.; Zhang, L.; Niederweis, M.; Dooley, K.E. Intricate link between siderophore secretion and drug efflux in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2023, 67, e0162922. [Google Scholar] [CrossRef]
- Rizzi, A.; Leroux, J.; Charron-Lamoureux, V.; Roy, S.; Beauregard, P.B.; Bellenger, J.-P.; Nojiri, H. Bacillus subtilis Modulates Its Usage of Biofilm-Bound Iron in Response to Environmental Iron Availability. Appl. Environ. Microbiol. 2020, 86, e00944-20. [Google Scholar] [CrossRef]
- Nandre, V.; Kumbhar, N.; Battu, S.; Kale, Y.; Bagade, A.; Haram, S.; Kodam, K. Siderophore mediated mineralization of struvite: A novel greener route of sustainable phosphate management. Water Res. 2021, 203, 117511. [Google Scholar] [CrossRef]
- David, S.R.; Jaouen, A.; Ihiawakrim, D.; Geoffroy, V.A. Biodeterioration of asbestos cement by siderophore-producing Pseudomonas. J. Hazard. Mater. 2021, 403, 123699. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial Siderophores: Classification, Biosynthesis, Perspectives of Use in Agriculture. Plants 2022, 11, 3065. [Google Scholar] [CrossRef] [PubMed]
- Chandran, H.; Meena, M.; Swapnil, P. Plant Growth-Promoting Rhizobacteria as a Green Alternative for Sustainable Agriculture. Sustainability 2021, 13, 10986. [Google Scholar] [CrossRef]
- Chaudhary, P.; Singh, S.; Chaudhary, A.; Sharma, A.; Kumar, G. Overview of biofertilizers in crop production and stress management for sustainable agriculture. Front. Plant Sci. 2022, 13, 930340. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.J.; Moore, M.M. Screening method to identify inhibitors of siderophore biosynthesis in the opportunistic fungal pathogen, Aspergillus fumigatus. Lett. Appl. Microbiol. 2009, 49, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, H.; Lv, H. Metabolomics Assay Identified a Novel Virulence-Associated Siderophore Encoded by the High-Pathogenicity Island in Uropathogenic Escherichia coli. J. Proteome Res. 2019, 18, 2331–2336. [Google Scholar] [CrossRef]
- He, R.; Gu, S.; Xu, J.; Li, X.; Chen, H.; Shao, Z.; Wang, F.; Shao, J.; Yin, W.B.; Qian, L.; et al. SIDERITE: Unveiling hidden siderophore diversity in the chemical space through digital exploration. iMeta 2024, 3, e192. [Google Scholar] [CrossRef]
- Prabhakar, P.K. Bacterial Siderophores and Their Potential Applications: A Review. Curr. Mol. Pharmacol. 2020, 13, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Moynié, L.; Hoegy, F.; Milenkovic, S.; Munier, M.; Paulen, A.; Gasser, V.; Faucon, A.L.; Zill, N.; Naismith, J.H.; Ceccarelli, M.; et al. Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa. ACS Infect. Dis. 2022, 8, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wang, Y.; Xie, F.; Liu, H.; Dai, H. Identification of siderophores blocking infection of Pseudomonas aeruginosa from Kitasatospora sp. LS1784. J. Antibiot. 2023, 77, 4–12. [Google Scholar] [CrossRef]
- Khilyas, I.V.; Markelova, M.I.; Valeeva, L.R.; Ivoilova, T.M.; Shagimardanova, E.; Laikov, A.V.; Elistratova, A.A.; Berkutova, E.S.; Lochnit, G.; Sharipova, M.R. Genomic insights and anti-phytopathogenic potential of siderophore metabolome of endolithic Nocardia mangyaensis NH1. Sci. Rep. 2024, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Abanoz-Seçgin, B.; Otur, Ç.; Okay, S.; Kurt-Kızıldoğan, A. The regulatory role of Fur-encoding SCLAV_3199 in iron homeostasis in Streptomyces clavuligerus. Gene 2023, 878, 147594. [Google Scholar] [CrossRef] [PubMed]
- Will, V.; Gasser, V.; Kuhn, L.; Fritsch, S.; Heinrichs, D.E.; Schalk, I.J. Siderophore specificities of the Pseudomonas aeruginosa TonB-dependent transporters ChtA and ActA. FEBS Lett. 2023, 597, 2963–2974. [Google Scholar] [CrossRef] [PubMed]
- Rehan, M.; Barakat, H.; Almami, I.S.; Qureshi, K.A.; Alsohim, A.S. Production and Potential Genetic Pathways of Three Different Siderophore Types in Streptomyces tricolor Strain HM10. Fermentation 2022, 8, 346. [Google Scholar] [CrossRef]
- Vindeirinho, J.M.; Soares, H.M.V.M.; Soares, E.V. Modulation of Siderophore Production by Pseudomonas fluorescens Through the Manipulation of the Culture Medium Composition. Appl. Biochem. Biotechnol. 2020, 193, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Baakza, A.; Vala, A.K.; Dave, B.P.; Dube, H.C. A comparative study of siderophore production by fungi from marine and terrestrial habitats. J. Exp. Mar. Biol. Ecol. 2004, 311, 1–9. [Google Scholar] [CrossRef]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 2015, 23, 3984–3999. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med. 2016, 22, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Oberegger, H.; Schoeser, M.; Zadra, I.; Abt, B.; Haas, H. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 2008, 41, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Dubey, R. Characterization of protein involve in nitrogen fixation and estimation of CO factor. Int. J. Adv. Biotechnol. Res. 2014, 5, 582–597. [Google Scholar]
- Youzhou, L.; Jiahui, S.; Junqing, Q.; Yang, Z.; Yongfeng, L. Research progress of siderophore produced by Bacillus spp. Jiangsu J. Agr. Sci. 2023, 39, 266–276. [Google Scholar]
- Liang, J. Identification of siderophore-producing bacterium HZ-2 and its ability to produce siderophore. J. Zhejiang Agric. Sci. 2021, 62, 1849–1852. [Google Scholar]
- Blanco Nouche, C.; Paris, C.; Dhalleine, T.; Oger, P.; Turpault, M.-P.; Uroz, S.; Spear, J.R. The non-ribosomal peptide synthetase-independent siderophore (NIS) rhizobactin produced by Caballeronia mineralivorans PML1(12) confers the ability to weather minerals. Appl. Environ. Microbiol. 2023, 89, e0045323. [Google Scholar] [CrossRef] [PubMed]
- Berti, A.D.; Thomas, M.G. Analysis of Achromobactin Biosynthesis by Pseudomonas syringaepv. syringae B728a. J. Bacteriol. 2009, 191, 4594–4604. [Google Scholar] [CrossRef] [PubMed]
- Kadi, N.; Oves-Costales, D.; Barona-Gomez, F.; Challis, G.L. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat. Chem. Biol. 2007, 3, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.-o.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef]
- Nakib, D.; Slatni, T.; Di Foggia, M.; Rombolà, A.D.; Abdelly, C. Changes in organic compounds secreted by roots in two Poaceae species (Hordeum vulgare and Polypogon monspenliensis) subjected to iron deficiency. J. Plant Res. 2020, 134, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Northover, G.H.R.; Mao, Y.; Ahmed, H.; Blasco, S.; Vilar, R.; Garcia-España, E.; Weiss, D.J. Effect of salinity on the zinc(II) binding efficiency of siderophore functional groups and implications for salinity tolerance mechanisms in barley. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Yoneyama, T. Iron delivery to the growing leaves associated with leaf chlorosis in mugineic acid family phytosiderophores-generating graminaceous crops. Soil Sci. Plant Nutr. 2021, 67, 415–426. [Google Scholar] [CrossRef]
- Spiridon, A.; Oburger, E.; Valadbeigi, Y.; Kloimböck, T.; Stanetty, C.; Kratena, N.; Draskovits, M.; Causon, T.; Hann, S. Surveying the mugineic acid family: Ion mobility—Quadrupole time-of-flight mass spectrometry (IM-QTOFMS) characterization and tandem mass spectrometry (LC-ESI-MS/MS) quantification of all eight naturally occurring phytosiderophores. Anal. Chim. Acta 2023, 1278, 341718. [Google Scholar] [CrossRef]
- Kratena, N.; Draskovits, M.; Biedermann, N.; Oburger, E.; Stanetty, C. Total synthesis of [13C2]-labeled phytosiderophores of the mugineic and avenic acid families. J. Label. Compd. Radiopharm. 2023, 66, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, T.S.; Ramkissoon, S.; Garrison, V.H.; Ramsubhag, A.; Thies, J.E. Siderophore production of African dust microorganisms over Trinidad and Tobago. Aerobiologia 2012, 28, 391–401. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Sinha, A.K.; Parli, B.V. Siderophore production by bacteria isolated from mangrove sediments: A microcosm study. J. Exp. Mar. Biol. Ecol. 2020, 524, 151290. [Google Scholar] [CrossRef]
- Müller, L.; Müller, D.C.; Kammerecker, S.; Fluri, M.; Neutsch, L.; Remus Emsermann, M.; Pelludat, C.; Alexandre, G. Priority Effects in the Apple Flower Determine If the Siderophore Desferrioxamine Is a Virulence Factor for Erwinia amylovora CFBP1430. Appl. Environ. Microbiol. 2022, 88, e0243321. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, W.; Li, Y.; Yu, F.; Penttinen, P. Isolation, characterization, and evaluation of a high-siderophore-yielding bacterium from heavy metal–contaminated soil. Environ. Sci. Pollut. Res. 2021, 29, 3888–3899. [Google Scholar] [CrossRef] [PubMed]
- Okoth, D.A.; Hug, J.J.; Garcia, R.; Müller, R. Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020. Microorganisms 2022, 10, 1959. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tian, Z.; Li, J. A Streptomyces morookaensis strain promotes plant growth and suppresses Fusarium wilt of banana. Trop. Plant Pathol. 2020, 46, 175–185. [Google Scholar] [CrossRef]
- Ramos, J.-L.; Sol Cuenca, M.; Molina-Santiago, C.; Segura, A.; Duque, E.; Gómez-García, M.R.; Udaondo, Z.; Roca, A.; Filloux, A. Mechanisms of solvent resistance mediated by interplay of cellular factors in Pseudomonas putida. FEMS Microbiol. Rev. 2015, 39, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Stein, N.V.; Eder, M.; Burr, F.; Stoss, S.; Holzner, L.; Kunz, H.-H.; Jung, H.; Rampioni, G. The RND efflux system ParXY affects siderophore secretion in Pseudomonas putida KT2440. Microbiol. Spectr. 2023, 11, e0230023. [Google Scholar] [CrossRef] [PubMed]
- Henríquez, T.; Stein, N.V.; Jung, H. PvdRT-OpmQ and MdtABC-OpmB efflux systems are involved in pyoverdine secretion in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 2018, 11, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Dong, H.; Feng, Z.; Wang, X.; Yao, Q.; Zhu, H. A Disturbed Siderophore Transport Inhibits Myxobacterial Predation. Cells 2022, 11, 3718. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [PubMed]
- Ellermann, M.; Arthur, J.C. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radic. Biol. Med. 2017, 105, 68–78. [Google Scholar] [CrossRef]
- Ahmadi, H.; Motesharezadeh, B.; Dadrasnia, A. Iron chlorosis in fruit stone trees with emphasis on chlorosis correction mechanisms in orchards: A review. J. Plant Nutr. 2022, 46, 782–800. [Google Scholar] [CrossRef]
- Gao, B.; Chai, X.; Huang, Y.; Wang, X.; Han, Z.; Xu, X.; Wu, T.; Zhang, X.; Wang, Y. Siderophore production in pseudomonas SP. strain SP3 enhances iron acquisition in apple rootstock. J. Appl. Microbiol. 2022, 133, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, N.; Alikhani, H.A.; Pourbabaei, A.A.; Besharati, H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J. Plant Nutr. 2017, 40, 736–746. [Google Scholar] [CrossRef]
- Ghavami, N.; Alikhani, H.A.; Pourbabaee, A.A.; Besharati, H. Study the Effects of Siderophore-Producing Bacteria on Zinc and Phosphorous Nutrition of Canola and Maize Plants. Commun. Soil Sci. Plant Anal. 2016, 47, 1517–1527. [Google Scholar] [CrossRef]
- Liang, J.; Hao, Z.; Wang, L.; Tao, R.; Jingwu, Z. Research Progress on the Function of Siderophore. Chin. Agric. Sci. Bull. 2011, 27, 284–287. [Google Scholar]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Schiessl, K.T.; Janssen, E.M.L.; Kraemer, S.M.; McNeill, K.; Ackermann, M. Magnitude and Mechanism of Siderophore-Mediated Competition at Low Iron Solubility in the Pseudomonas aeruginosa Pyochelin System. Front. Microbiol. 2017, 8, 1964. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.-M. Pyoverdines: Pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000, 174, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Górska, A.; Sloderbach, A.; Marszałł, M.P. Siderophore–drug complexes: Potential medicinal applications of the ‘Trojan horse’ strategy. Trends Pharmacol. Sci. 2014, 35, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Negash, K.H.; Norris, J.K.S.; Hodgkinson, J.T. Siderophore–Antibiotic Conjugate Design: New Drugs for Bad Bugs. Molecules 2019, 24, 3314. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Butala, S.; Khan, T.; Suvarna, V.; Sherje, A.; Dravyakar, B. Mycobacterial siderophore: A review on chemistry and biology of siderophore and its potential as a target for tuberculosis. Eur. J. Med. Chem. 2018, 157, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Gauglitz, J.M.; Zhou, H.; Butler, A. A suite of citrate-derived siderophores from a marine Vibrio species isolated following the Deepwater Horizon oil spill. J. Inorg. Biochem. 2012, 107, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, T.; Nakase, M.; Liu, J.; Dotsuta, Y.; Satou, Y.; Kitagaki, T.; Kozai, N. Degradation of nuclear fuel debris analog by siderophore-releasing microorganisms. J. Nucl. Sci. Technol. 2023, 61, 1–13. [Google Scholar] [CrossRef]
- Wasi, S.; Tabrez, S.; Ahmad, M. Toxicological effects of major environmental pollutants: An overview. Environ. Monit. Assess. 2012, 185, 2585–2593. [Google Scholar] [CrossRef]
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef]
- Wichard, T.; Bellenger, J.-P.; Morel, F.M.M.; Kraepiel, A.M.L. Role of the Siderophore Azotobactin in the Bacterial Acquisition of Nitrogenase Metal Cofactors. Environ. Sci. Technol. 2009, 43, 7218–7224. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Gupta, A.; Singh, P.; Mishra, A.K.; Ranjan, R.K.; Srivastava, A. Siderophore-assisted cadmium hyperaccumulation in Bacillus subtilis. Int. Microbiol. 2019, 23, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Heng, S.; Munis, M.F.H.; Fahad, S.; Yang, X. Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: A review. Environ. Exp. Bot. 2015, 117, 28–40. [Google Scholar] [CrossRef]
- Kumari, S.; Khan, A.; Singh, P.; Dwivedi, S.K.; Ojha, K.K.; Srivastava, A. Mitigation of As toxicity in wheat by exogenous application of hydroxamate siderophore of Aspergillus origin. Acta Physiol. Plant. 2019, 41, 107. [Google Scholar] [CrossRef]
- Singh, A.; Kaushik, M.S.; Srivastava, M.; Tiwari, D.N.; Mishra, A.K. Siderophore mediated attenuation of cadmium toxicity by paddy field cyanobacterium Anabaena oryzae. Algal Res. 2016, 16, 63–68. [Google Scholar] [CrossRef]
- Bajpai, P. Biological Bleaching of Chemical Pulps. Crit. Rev. Biotechnol. 2008, 24, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Goodell, B. Mechanisms of wood degradation by brown-rot fungi: Chelator-mediated cellulose degradation and binding of iron by cellulose. J. Biotechnol. 2001, 87, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Arantes, V.; Milagres, A.M.F. The effect of a catecholate chelator as a redox agent in Fenton-based reactions on degradation of lignin-model substrates and on COD removal from effluent of an ECF kraft pulp mill. J. Hazard. Mater. 2007, 141, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Carlson Jr, P.E.; Dixon, S.D.; Janes, B.K.; Carr, K.A.; Nusca, T.D.; Anderson, E.C.; Keene, S.E.; Sherman, D.H.; Hanna, P.C. Genetic analysis of petrobactin transport in Bacillus anthracis. Mol. Microbiol. 2010, 75, 900–909. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator—Siderophore: A review. Microbiol. Res. 2018, 212–213, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.K.; Gonneau, C.; Salamatipour, A.; Pietrofesa, R.A.; Casper, B.; Christofidou-Solomidou, M.; Willenbring, J.K. Siderophore-mediated iron removal from chrysotile: Implications for asbestos toxicity reduction and bioremediation. J. Hazard. Mater. 2018, 341, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Puja, H.; Mislin, G.L.A.; Rigouin, C. Engineering Siderophore Biosynthesis and Regulation Pathways to Increase Diversity and Availability. Biomolecules 2023, 13, 959. [Google Scholar] [CrossRef] [PubMed]
- Bruns, H.; Crüsemann, M.; Letzel, A.-C.; Alanjary, M.; McInerney, J.O.; Jensen, P.R.; Schulz, S.; Moore, B.S.; Ziemert, N. Function-related replacement of bacterial siderophore pathways. ISME J. 2017, 12, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, W.-H.; Bruner, S.D. Microbial siderophore-based iron assimilation and therapeutic applications. BioMetals 2016, 29, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Peduzzi, J.; Thomas, X.; Djediat, C.; Rebuffat, S. Parasitism of Iron-siderophore Receptors of Escherichia Coli by the Siderophore-peptide Microcin E492m and its Unmodified Counterpart. BioMetals 2006, 19, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Leventhal, G.E.; Ackermann, M.; Schiessl, K.T. Why microbes secrete molecules to modify their environment: The case of iron-chelating siderophores. J. R. Soc. Interface 2019, 16, 20180674. [Google Scholar] [CrossRef]
- Condon, B.J.; Oide, S.; Gibson, D.M.; Krasnoff, S.B.; Turgeon, B.G. Reductive Iron Assimilation and Intracellular Siderophores Assist Extracellular Siderophore-Driven Iron Homeostasis and Virulence. Mol. Plant-Microbe Interact. 2014, 27, 793–808. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Olejnickova, K.; Hola, V.; Ruzicka, F. Catheter-related infections caused by Pseudomonas aeruginosa: Virulence factors involved and their relationships. Pathog. Dis. 2014, 72, 87–94. [Google Scholar] [PubMed]
- Roop, R.M.; Silva-Bailão, M.G.; Bailão, E.F.L.C.; Lechner, B.E.; Gauthier, G.M.; Lindner, H.; Bailão, A.M.; Haas, H.; de Almeida Soares, C.M. Hydroxamate Production as a High Affinity Iron Acquisition Mechanism in Paracoccidioides spp. PLoS ONE 2014, 9, e105805. [Google Scholar]
- Mucha, J.; Gabała, E.; Zadworny, M. The effects of structurally different siderophores on the organelles of Pinus sylvestris root cells. Planta 2019, 249, 1747–1760. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, H.; Heng, J.; Wang, D.; Bian, K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 2019, 218, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Mudhulkar, R.; Rajapitamahuni, S.; Srivastava, S.; Bharadwaj, S.V.V.; Boricha, V.P.; Mishra, S.; Chatterjee, P.B. Identification of a New Siderophore Acinetoamonabactin Produced by a Salt-Tolerant Bacterium Acinetobacter Soli. ChemistrySelect 2018, 3, 8207–8211. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Batagov, A.; Ward, J.; Baganz, F.; Krabben, P. Identification of erythrobactin, a hydroxamate-type siderophore produced by Saccharopolyspora erythraea. Lett. Appl. Microbiol. 2006, 42, 375–380. [Google Scholar] [CrossRef]
- Valdebenito, M.; Crumbliss, A.L.; Winkelmann, G.; Hantke, K. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 2006, 296, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Furrer, J.L.; Sanders, D.N.; Hook-Barnard, I.G.; McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 2002, 44, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Wang, Q.; Liu, J.; Wang, D.; Li, J.; Li, T. Effects of deletion of siderophore biosynthesis gene in Pseudomonas fragi on quorum sensing and spoilage ability. Int. J. Food Microbiol. 2023, 396, 110196. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Zhang, B.; Yang, B.-S.; Shi, W.-T.; Li, Y.-F.; Wang, Y.; Zhang, P.; Jiao, J.; Tian, C.-F.; Hirsch, A.M.; et al. Rhizobiales Specific RirA Represses a Naturally “Synthetic” Foreign Siderophore Gene Cluster To Maintain Sinorhizobium Legume Mutualism. mBio 2022, 13, e0290021. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Ro, H.-S. Generation of Iron-Independent Siderophore-Producing Agaricus bisporus through the Constitutive Expression of hapX. Genes 2021, 12, 724. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Niu, M.; Liu, S.; Mao, J.; Yu, Y.; Du, Y. The effect of siderophore virulence genes entB and ybtS on the virulence of Carbapenem-resistant Klebsiella pneumoniae. Microb. Pathog. 2022, 171, 105746. [Google Scholar] [CrossRef]
- Jeong, G.-J.; Khan, F.; Khan, S.; Tabassum, N.; Mehta, S.; Kim, Y.-M. Pseudomonas aeruginosa virulence attenuation by inhibiting siderophore functions. Appl. Microbiol. Biotechnol. 2023, 107, 1019–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kent, J.E.; Whitaker, M.; Young, D.C.; Herrmann, D.; Aleshin, A.E.; Ko, Y.-H.; Cingolani, G.; Saad, J.S.; Moody, D.B.; et al. A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]



| Type of Siderophore | Characteristic Functional Group | Characteristic | Siderophore-Producing Microorganism |
|---|---|---|---|
| Hydroxamic salt-type | 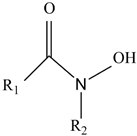 | The most common in nature, the structure is more complex; it is more hydrophilic and prone to photooxidation | Pseudomonas fluorescens [26] Aspergillus nidulans [29] |
| Catechol salt-type | 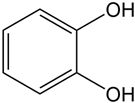 | Strong lipophilicity and high affinity with Fe; strong resistance to environmental pH changes | Escherichia coli [28] Klebsiella pneumoniae [28] |
| Carboxylate-type | 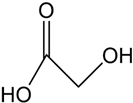 | Potential ligands for the ocean iron cycle; some of them are photoactive | Rhizobium meliloti [30] Staphylococcus aureus [30] |
| Mixed-type | Mixed functional groups | It presents different characteristics according to different functional groups | Rhodococcus erythropolis [28] Escherichia coli [28] |
| Plant Species Name | Type of Siderophore | Year | References DOI |
|---|---|---|---|
| Hordeum vulgare L. | hydroxamate-type | 2021 | https://doi.org/10.1038/s41598-021-95736-7 [38] |
| Hordeum vulgare L. | catecholate-type | 2021 | https://doi.org/10.1080/00380768.2021.1947735 [39] |
| Poaceae | catecholate-type | 2023 | https://doi.org/10.1016/j.aca.2023.341718 [40] |
| Poaceae | catecholate-type | 2023 | https://doi.org/10.1002/jlcr.4064 [41] |
| Polypogon monspenliensis | unidentified | 2020 | https://doi.org/10.1007/s10265-020-01237-5 [37] |
| Microbial Species Name | Type of Siderophore | Year | References DOI |
|---|---|---|---|
| Escherichia vulneris | hydroxamate-type | 2020 | https://doi.org/10.1016/j.jembe.2019.151290 [44] |
| Enterobacter cancerogenus | hydroxamate-type | 2020 | https://doi.org/10.1016/j.jembe.2019.151290 [44] |
| Pantoea agglomerans | hydroxamate-type | 2020 | https://doi.org/10.1016/j.jembe.2019.151290 [44] |
| Enterobacter bugandensis | hydroxamate-type | 2020 | https://doi.org/10.1016/j.jembe.2019.151290 [44] |
| Erwinia amylovora CFBP1430 | hydroxamate-type | 2022 | https://doi.org/10.1128/aem.02433-21 [45] |
| Burkholderia sp. SX9 | catecholate-type | 2021 | https://doi.org/10.1007/s11356-021-15996-8 [46] |
| Myxobacterial Strain MSr12020 | catecholate-type | 2022 | https://doi.org/10.3390/microorganisms10101959 [47] |
| Streptomyces tricolor Strain HM10 | carboxylate-type | 2022 | https://doi.org/10.3390/fermentation8080346 [24] |
| Seudomonas aeruginosa TonB | carboxylate-type | 2023 | https://doi.org/10.1002/1873-3468.14740 [23] |
| Streptomyces morookaensiss | carboxylate-type | 2020 | https://doi.org/10.1007/s40858-020-00396-z [48] |
| Years | Quantity |
|---|---|
| 2000–2002 | 8 |
| 2003–2005 | 9 |
| 2006–2008 | 13 |
| 2009–2011 | 15 |
| 2012–2014 | 19 |
| 2015–2017 | 592 |
| 2018–2020 | 818 |
| 2021–2023 | 1024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, B.; Wei, X.; Wan, C.; Zhao, W.; Song, R.; Xin, S.; Song, K. Exploring the Biological Pathways of Siderophores and Their Multidisciplinary Applications: A Comprehensive Review. Molecules 2024, 29, 2318. https://doi.org/10.3390/molecules29102318
Xie B, Wei X, Wan C, Zhao W, Song R, Xin S, Song K. Exploring the Biological Pathways of Siderophores and Their Multidisciplinary Applications: A Comprehensive Review. Molecules. 2024; 29(10):2318. https://doi.org/10.3390/molecules29102318
Chicago/Turabian StyleXie, Benkang, Xinpei Wei, Chu Wan, Wei Zhao, Renfeng Song, Shuquan Xin, and Kai Song. 2024. "Exploring the Biological Pathways of Siderophores and Their Multidisciplinary Applications: A Comprehensive Review" Molecules 29, no. 10: 2318. https://doi.org/10.3390/molecules29102318
APA StyleXie, B., Wei, X., Wan, C., Zhao, W., Song, R., Xin, S., & Song, K. (2024). Exploring the Biological Pathways of Siderophores and Their Multidisciplinary Applications: A Comprehensive Review. Molecules, 29(10), 2318. https://doi.org/10.3390/molecules29102318






