Nitrogen-Centered Radicals Derived from Azidonucleosides
Abstract
1. Introduction
1.1. 2′-Azido-2′-Deoxy Pyrimidine Nucleosides and Nucleotides: Inhibition of Ribonucleotide Reductases and Importance of Nitrogen-Centered Radical Chemistry
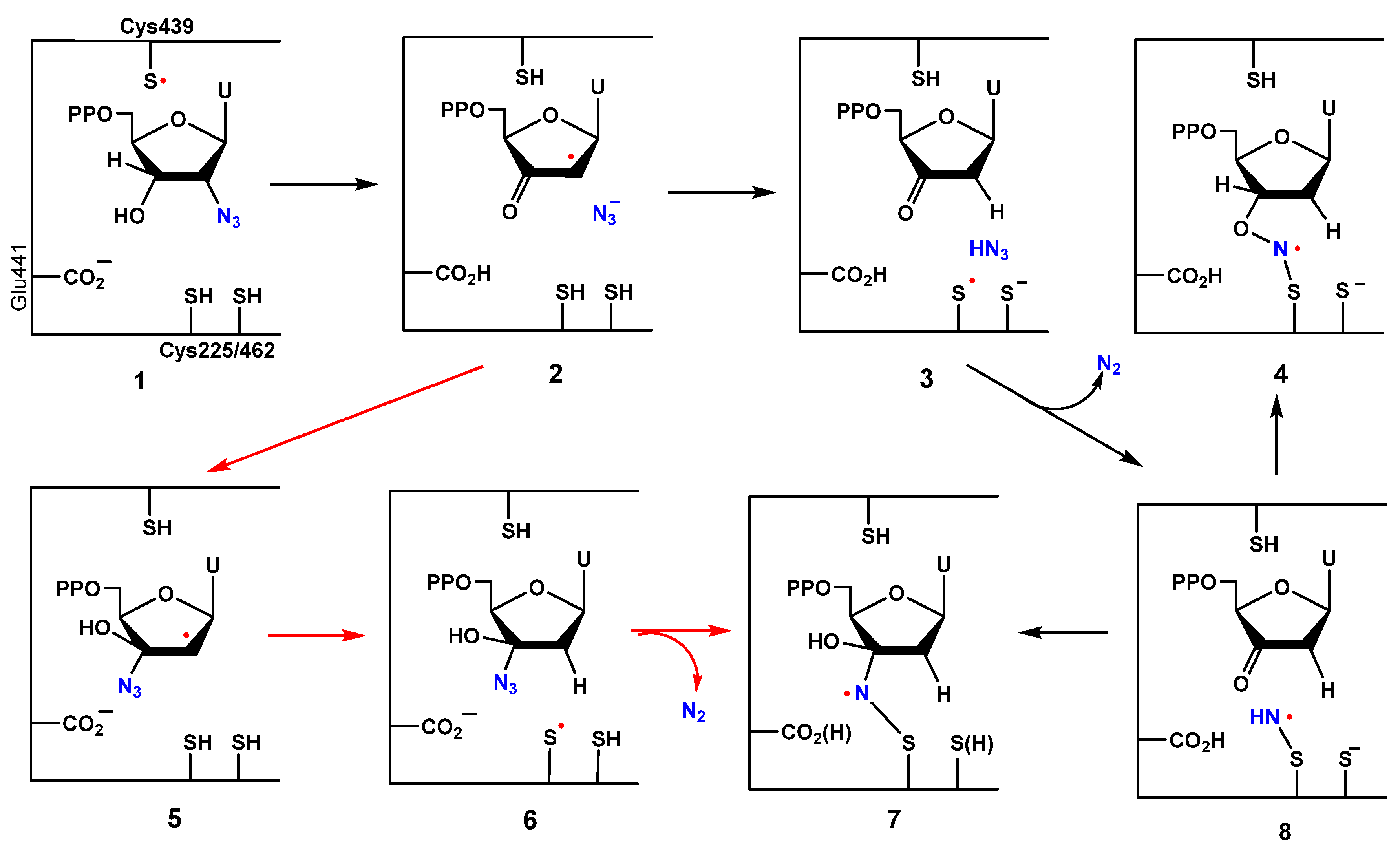
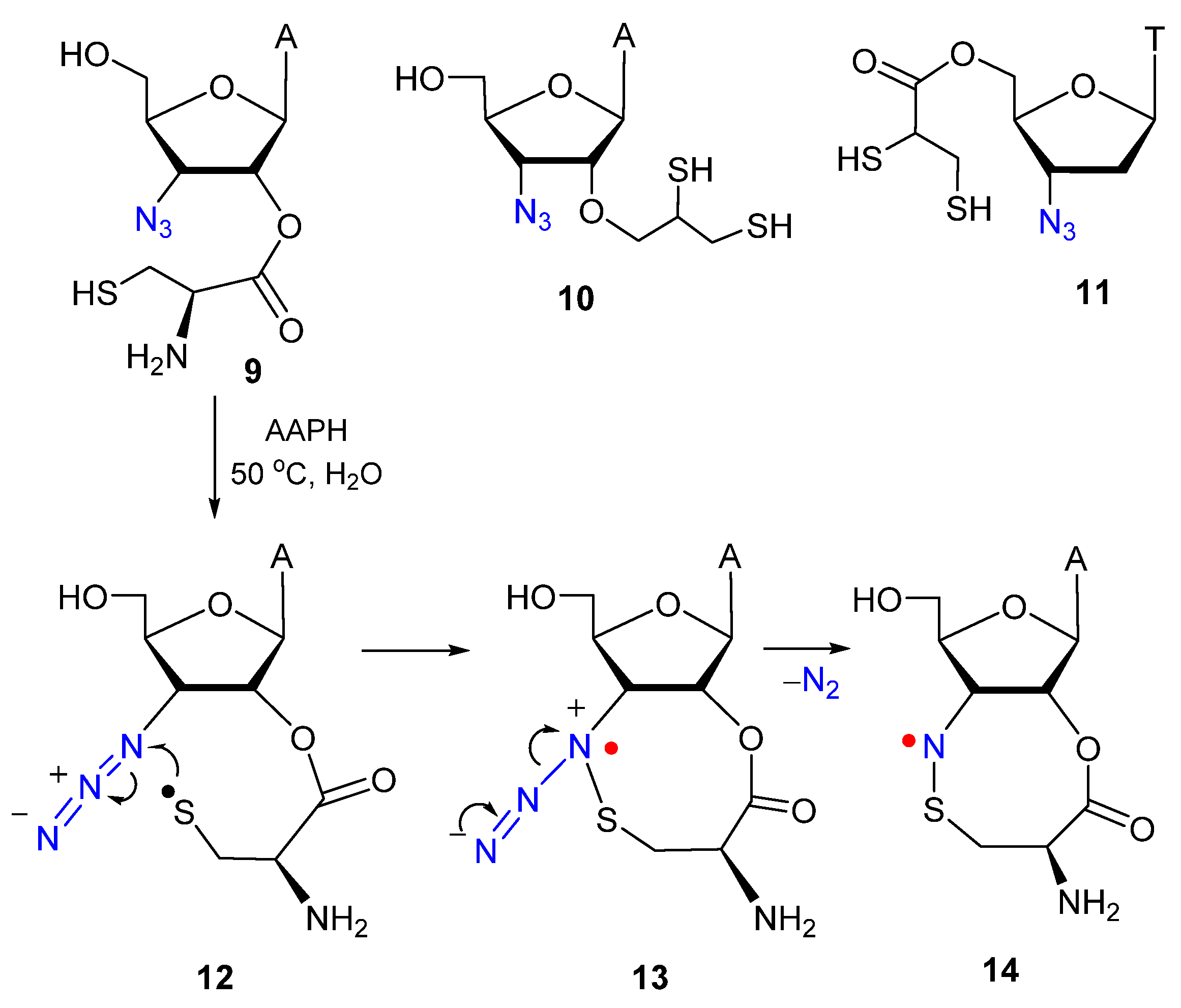
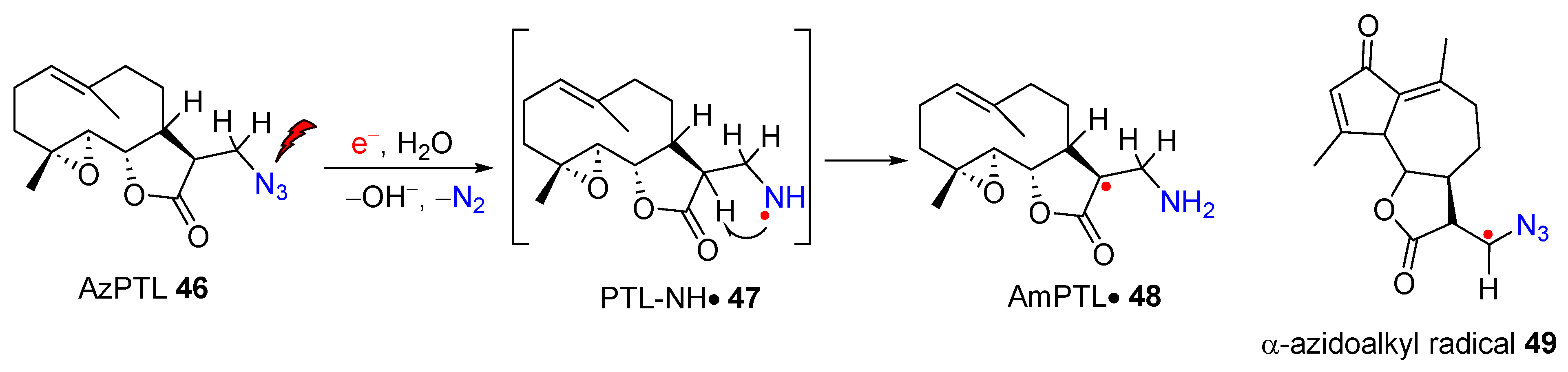
1.2. Biomimetic Studies
2. Nitrogen-Centered Radicals Generated from Azidonucleosides by Radiation-Produced Electrons
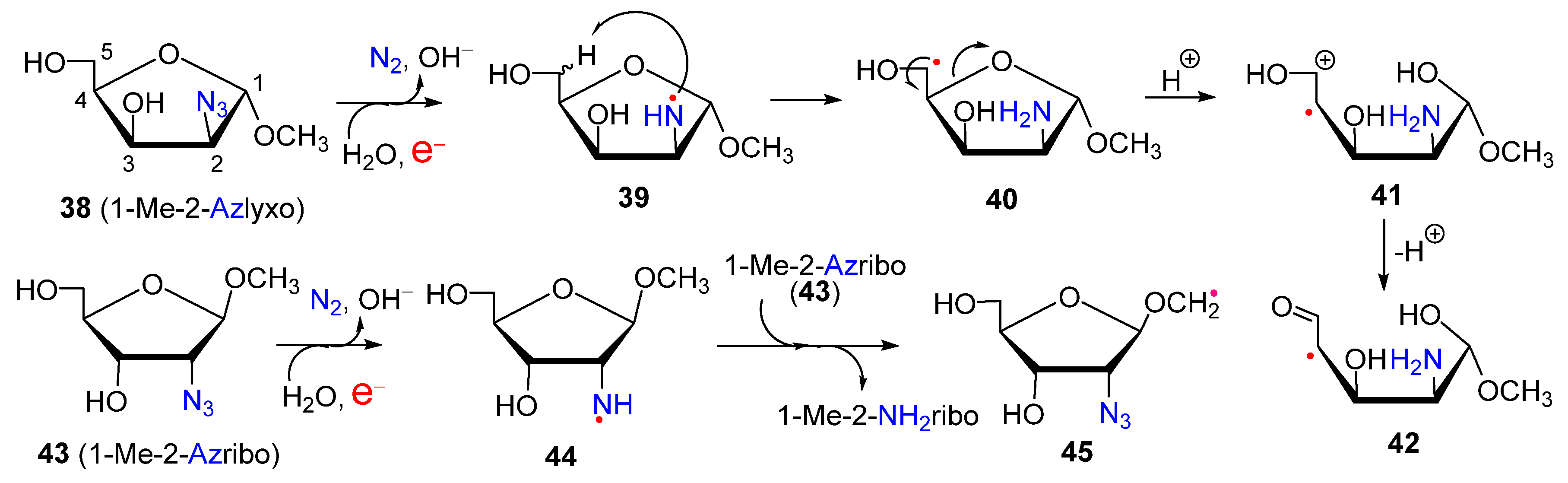
2.1. From Azido-Modified Nucleoside Sugars
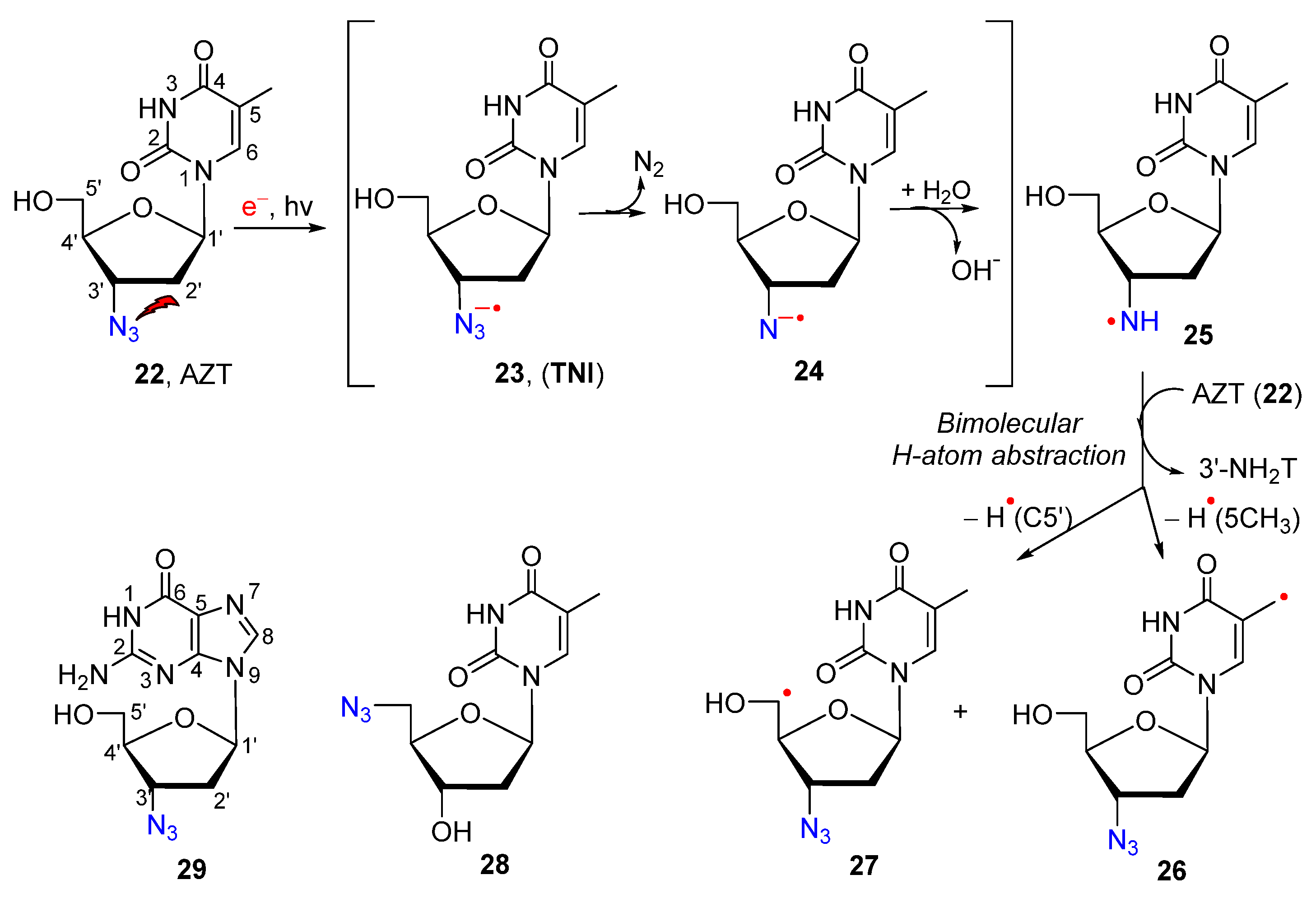
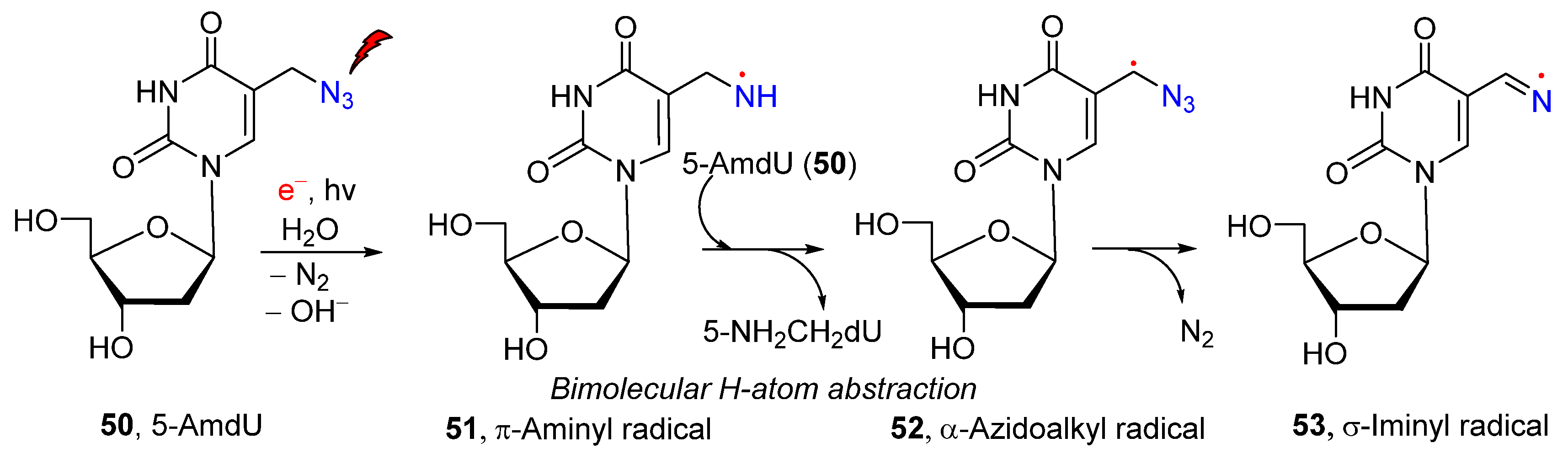
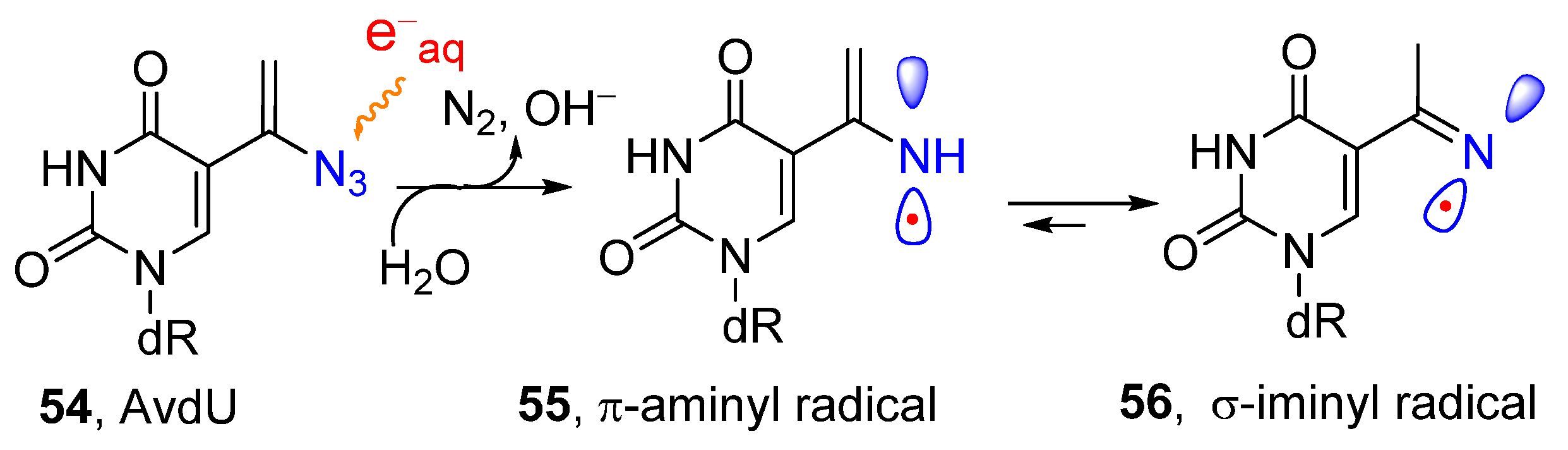
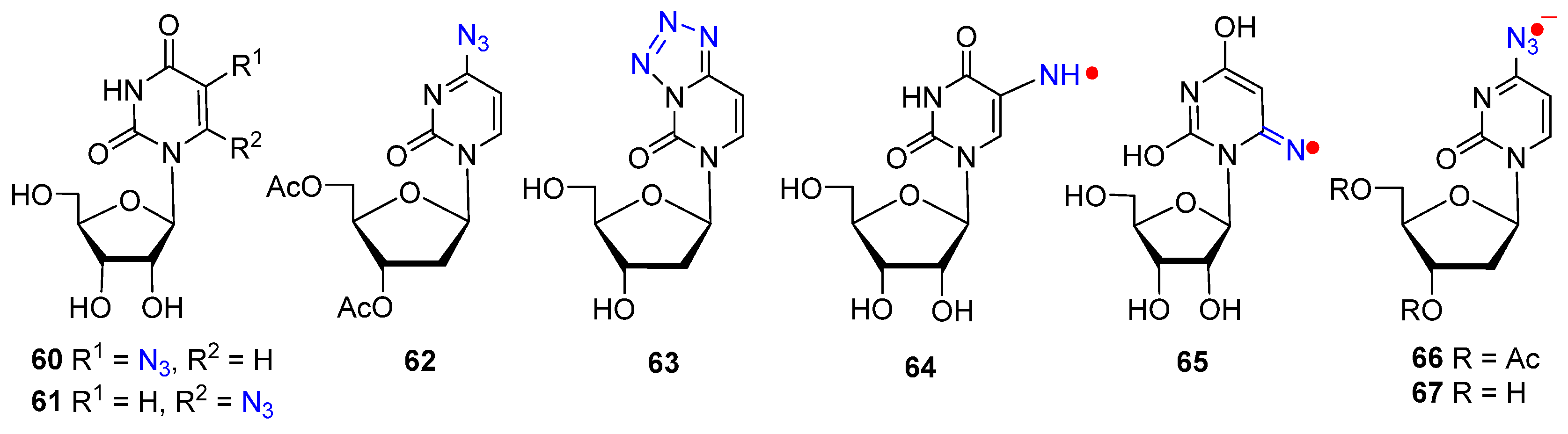
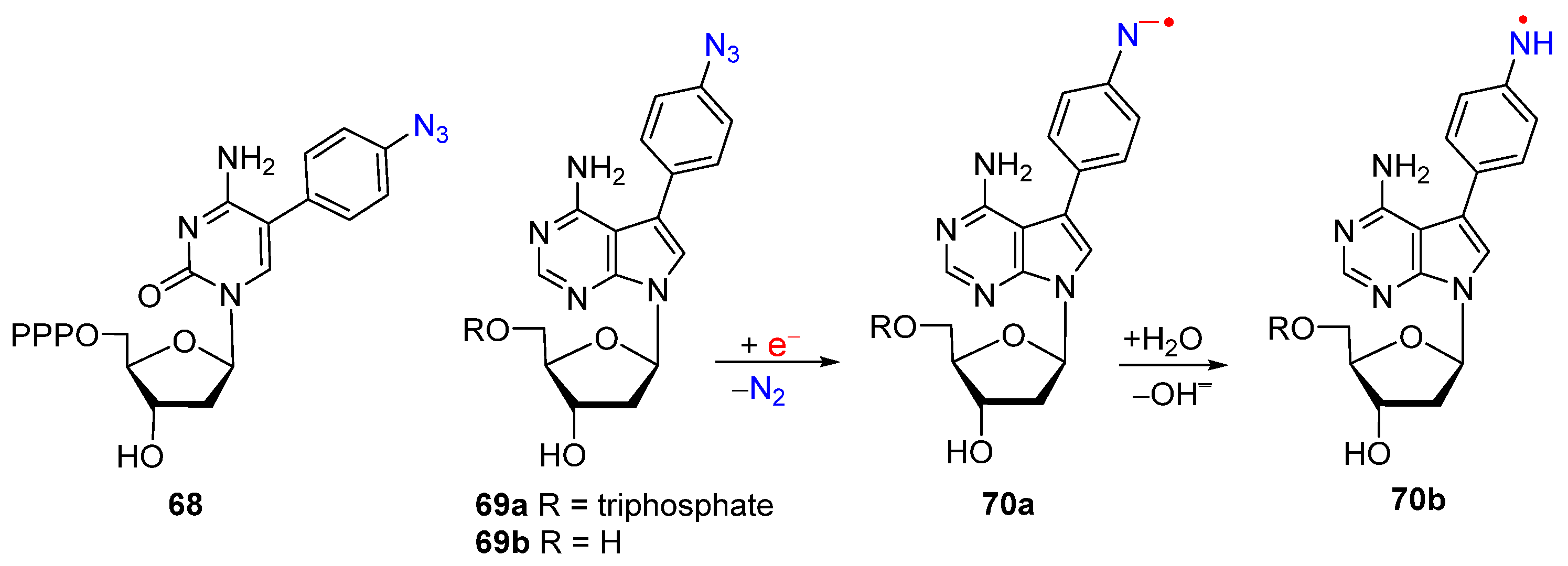
2.2. From Azido-Modified Nucleobases
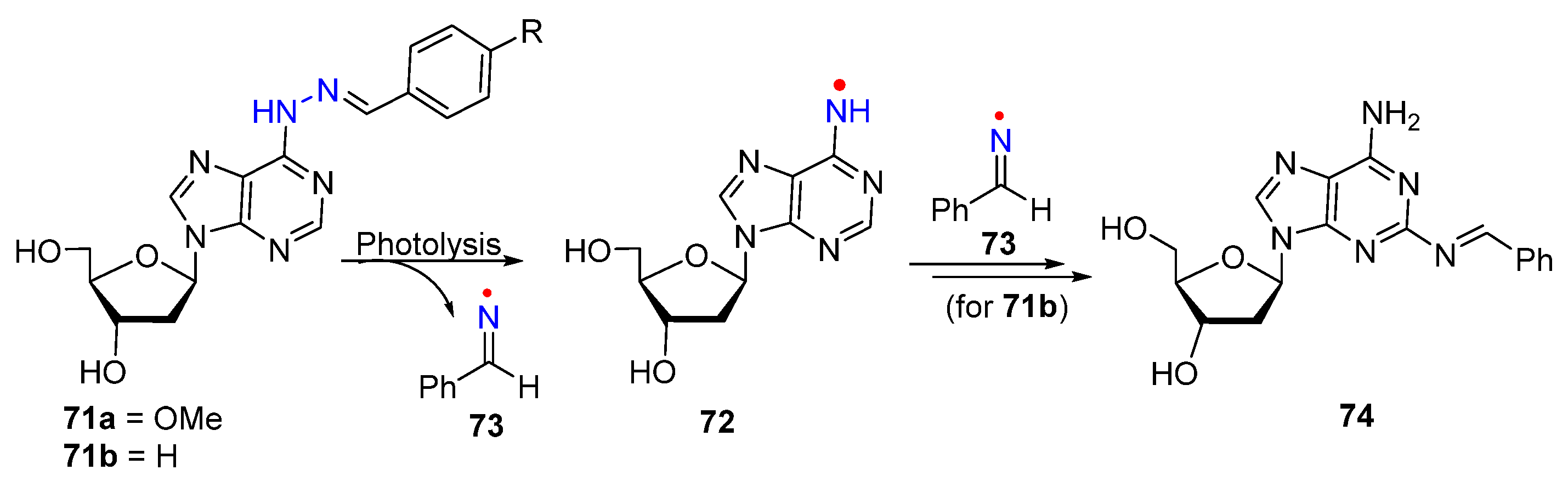
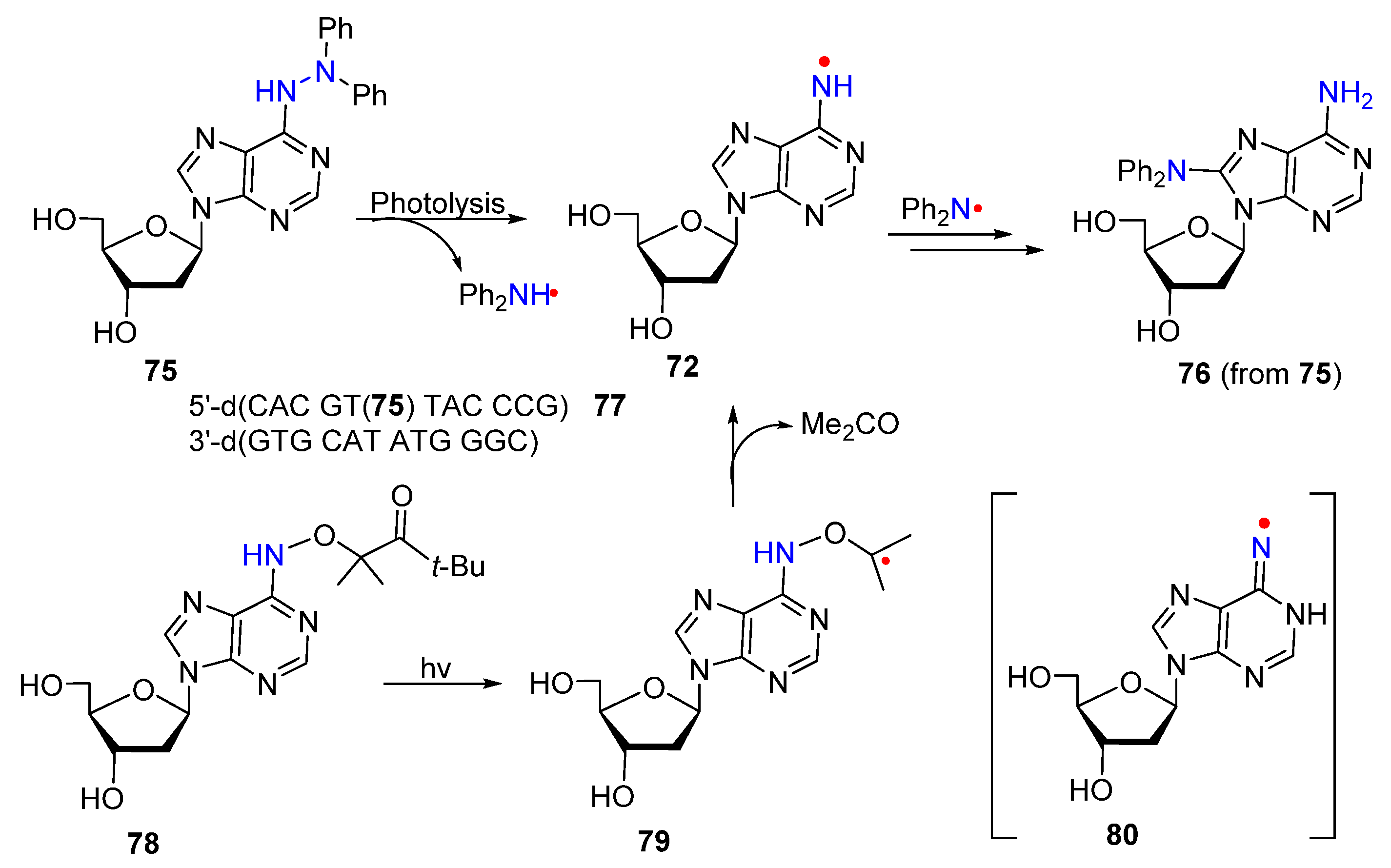
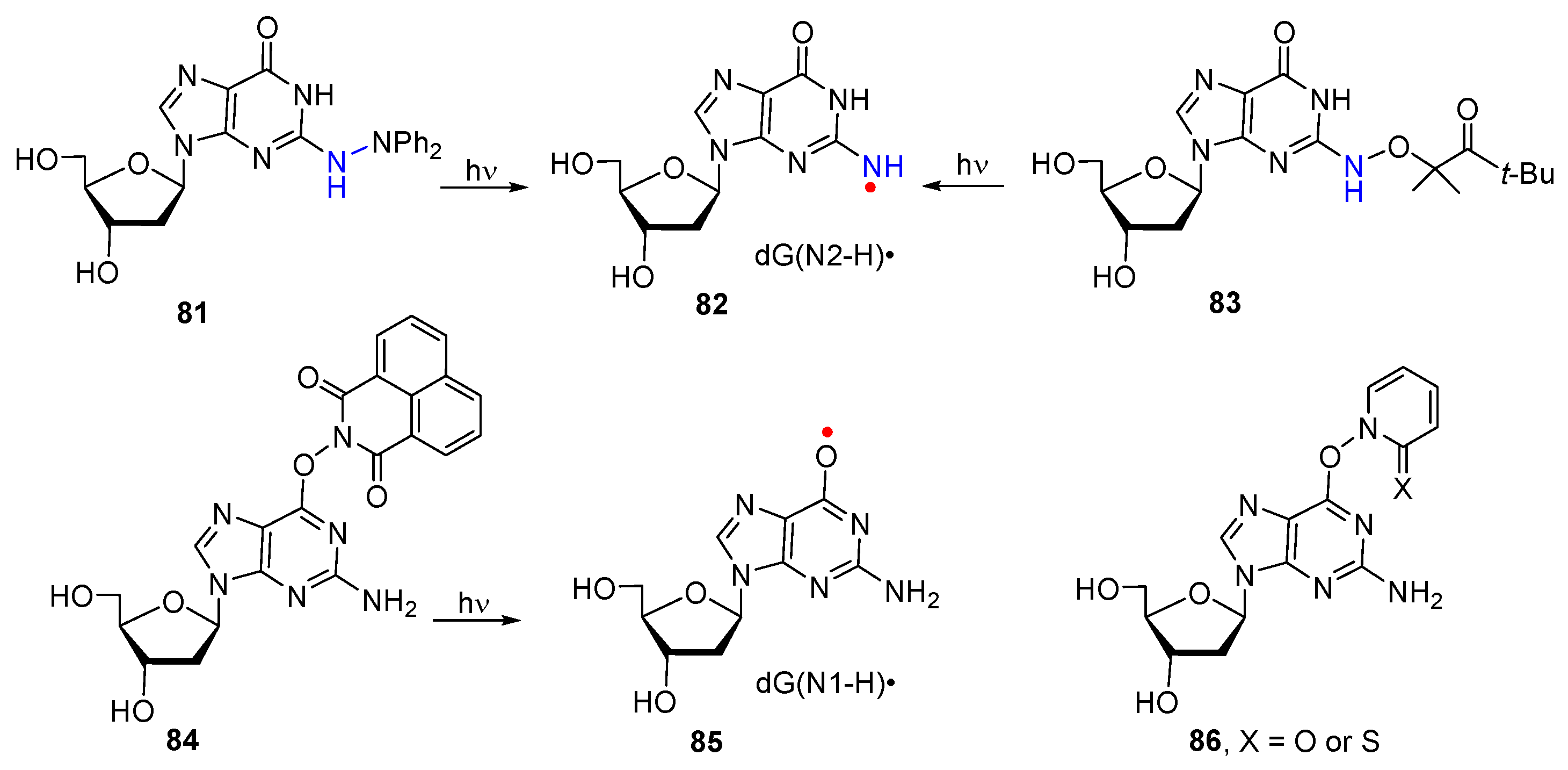
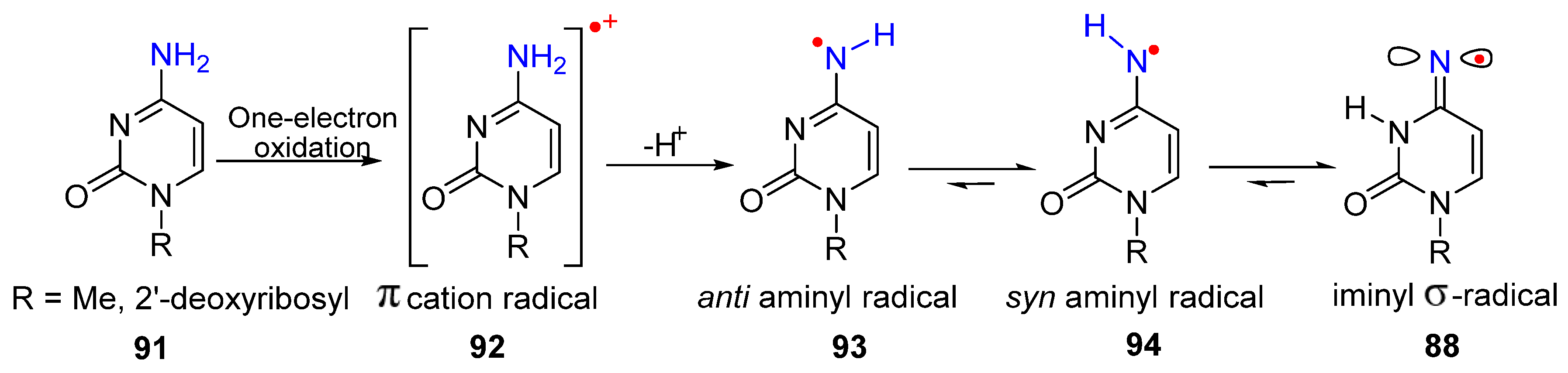
3. Nitrogen-Centered Radicals Generated on Non-Azido Nucleobases
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A Review on Recent Advances in Nitrogen-Containing Molecules and Their Biological Applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Dedon, P.C.; Tannenbaum, S.R. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004, 423, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.; Franco, M.C.; Estevez, A.G. Reactive nitrogen species in cellular signaling. Exp. Biol. Med. 2015, 240, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Pratley, C.; Fenner, S.; Murphy, J.A. Nitrogen-Centered Radicals in Functionalization of sp2 Systems: Generation, Reactivity, and Applications in Synthesis. Chem. Rev. 2022, 122, 8181–8260. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Studer, A. Chemistry With N-Centered Radicals Generated by Single-Electron Transfer-Oxidation Using Photoredox Catalysis. CCS Chem. 2019, 1, 38–49. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Q.; Xiao, W.; Chen, J. Recent advances in visible-light photoredox-catalyzed nitrogen radical cyclization. Green Synth. Catal. 2020, 1, 42–51. [Google Scholar] [CrossRef]
- Xiong, P.; Xu, H.-C. Chemistry with Electrochemically Generated N-Centered Radicals. Acc. Chem. Res. 2019, 52, 3339–3350. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Y.; Zhao, Q.-Q.; Chen, J.; Xiao, W.-J.; Chen, J.-R. When Light Meets Nitrogen-Centered Radicals: From Reagents to Catalysts. Acc. Chem. Res. 2020, 53, 1066–1083. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Chen, D.; Li, Y.; Xiang, J.-N.; Li, J.-H. Fluoroamide-driven intermolecular hydrogen-atom-transfer-enabled intermolecular 1,2-alkylarylation of alkenes. Org. Chem. Front. 2023, 10, 943–950. [Google Scholar] [CrossRef]
- Sirinimal, H.S.; Hebert, S.P.; Samala, G.; Chen, H.; Rosenhauer, G.J.; Schlegel, H.B.; Stockdill, J.L. Synthetic and Computational Study of Tin-Free Reductive Tandem Cyclizations of Neutral Aminyl Radicals. Org. Lett. 2018, 20, 6340–6344. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ward, R.M.; Schomaker, J.M. Regioselective Intramolecular Allene Amidation Enabled by an EDA Complex. Chem. Eur. J. 2020, 26, 13783–13787. [Google Scholar] [CrossRef]
- Azzi, E.; Ghigo, G.; Sarasino, L.; Parisotto, S.; Moro, R.; Renzi, P.; Deagostino, A. Photoinduced Chloroamination Cyclization Cascade with N-Chlorosuccinimide: From N-(Allenyl)sulfonylamides to 2-(1-Chlorovinyl)pyrrolidines. J. Org. Chem. 2022, 88, 6420–6433. [Google Scholar] [CrossRef]
- Yamada, K.; Sato, M.; Tanaka, K.; Wakabayashi, A.; Igarashi, T.; Sakurai, T. Nitrile-forming radical elimination reactions of 1-naphthaldehyde O-(4-substituted benzoyl)oximes activated by triplet benzophenone. J. Photochem. Photobiol. A Chem. 2006, 183, 205–211. [Google Scholar] [CrossRef]
- Shee, M.; Singh, N.D.P. Chemical versatility of azide radical: Journey from a transient species to synthetic accessibility in organic transformations. Chem. Soc. Rev. 2022, 51, 2255–2312. [Google Scholar] [CrossRef]
- Minozzi, M.; Nanni, D.; Spagnolo, P. From Azides to Nitrogen-Centered Radicals: Applications of Azide Radical Chemistry to Organic Synthesis. Chem. Eur. J. 2009, 15, 7830–7840. [Google Scholar] [CrossRef]
- Horwitz, J.P.; Chua, J.; Noel, M.; Nucleosides, V. The Monomesylates of 1-(2’-Deoxy-β-D-lyxofuranosyl)thymine1,2. J. Org. Chem. 1964, 29, 2076–2078. [Google Scholar] [CrossRef]
- Mitsuya, H.; Weinhold, K.J.; Furman, P.A.; St Clair, M.H.; Lehrman, S.N.; Gallo, R.C.; Bolognesi, D.; Barry, D.W.; Broder, S. 3′-Azido-3′-deoxythymidine (BW A509U): An antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc. Natl. Acad. Sci. USA 1985, 82, 7096–7100. [Google Scholar] [CrossRef]
- Pathak, T. Azidonucleosides: Synthesis, Reactions, and Biological Properties. Chem. Rev. 2002, 102, 1623–1668. [Google Scholar] [CrossRef]
- Müggenburg, F.; Müller, S. Azide-modified Nucleosides as Versatile Tools for Bioorthogonal Labeling and Functionalization. Chem. Rec. 2022, 22, e202100322. [Google Scholar] [CrossRef]
- Perrone, D.; Marchesi, E.; Preti, L.; Navacchia, M.L. Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications. Molecules 2021, 26, 3100. [Google Scholar] [CrossRef]
- Robins, M.J.; Hawrelak, S.D.; Hernández, A.E.; Wnuk, S.F. Nucleic Acid Related Compounds. LXXXI. Efficient General Synthesis of Purine (Amino, Azido, and Triflate)-Sugar Nucleosides. Nucleosides Nucleotides 1992, 11, 821–834. [Google Scholar] [CrossRef]
- Tera, M.; Glasauer, S.M.K.; Luedtke, N.W. In Vivo Incorporation of Azide Groups into DNA by Using Membrane-Permeable Nucleotide Triesters. ChemBioChem 2018, 19, 1939–1943. [Google Scholar] [CrossRef]
- Stubbe, J.; van der Donk, W.A. Ribonucleotide reductases: Radical enzymes with suicidal tendencies. Chem. Biol. 1995, 2, 793–801. [Google Scholar] [CrossRef]
- Stubbe, J.; Nocera, D.G. Radicals in Biology: Your Life Is in Their Hands. J. Am. Chem. Soc. 2021, 143, 13463–13472. [Google Scholar] [CrossRef]
- Meisenheimer, K.M.; Koch, T.H. Photocross-Linking of Nucleic Acids to Associated Proteins. Crit. Rev. Biochem. Mol. Biol. 1997, 32, 101–140. [Google Scholar] [CrossRef]
- Sirivolu, V.R.; Vernekar, S.K.V.; Ilina, T.; Myshakina, N.S.; Parniak, M.A.; Wang, Z. Clicking 3′-Azidothymidine into Novel Potent Inhibitors of Human Immunodeficiency Virus. J. Med. Chem. 2013, 56, 8765–8780. [Google Scholar] [CrossRef]
- Wu, J.; Yu, W.; Fu, L.; He, W.; Wang, Y.; Chai, B.; Song, C.; Chang, J. Design, synthesis, and biological evaluation of new 2′-deoxy-2′-fluoro-4′-triazole cytidine nucleosides as potent antiviral agents. Eur. J. Med. Chem. 2013, 63, 739–745. [Google Scholar] [CrossRef]
- Kore, A.R.; Charles, I. Click Chemistry Based Functionalizations of Nucleoside, Nucleotide and Nucleic Acids. Curr. Org. Chem. 2013, 17, 2164–2191. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click chemistry with DNA. Chem. Soc. Rev. 2010, 39, 1388–1405. [Google Scholar] [CrossRef]
- Fomich, M.A.; Kvach, M.V.; Navakouski, M.J.; Weise, C.; Baranovsky, A.V.; Korshun, V.A.; Shmanai, V.V. Azide phosphoramidite in direct synthesis of azide-modified oligonucleotides. Org. Lett. 2014, 16, 4590–4593. [Google Scholar] [CrossRef]
- Krause, A.; Hertl, A.; Muttach, F.; Jäschke, A. Phosphine-Free Stille–Migita Chemistry for the Mild and Orthogonal Modification of DNA and RNA. Chem. Eur. J. 2014, 20, 16613–16619. [Google Scholar] [CrossRef]
- Fauster, K.; Hartl, M.; Santner, T.; Aigner, M.; Kreutz, C.; Bister, K.; Ennifar, E.; Micura, R. 2′-Azido RNA, a versatile tool for chemical biology: Synthesis, X-ray structure, siRNA applications, click labeling. ACS Chem. Biol. 2012, 7, 581–589. [Google Scholar] [CrossRef]
- Beyer, C.; Wagenknecht, H.-A. In situ azide formation and “click” reaction of nile red with DNA as an alternative postsynthetic route. Chem. Commun. 2010, 46, 2230–2231. [Google Scholar] [CrossRef]
- Wada, T.; Mochizuki, A.; Higashiya, S.; Tsuruoka, H.; Kawahara, S.-i.; Ishikawa, M.; Sekine, M. Synthesis and properties of 2-azidodeoxyadenosine and its incorporation into oligodeoxynucleotides. Tetrahedron Lett. 2001, 42, 9215–9219. [Google Scholar] [CrossRef]
- Weisbrod, S.H.; Baccaro, A.; Marx, A. DNA Conjugation by Staudinger Ligation. Nucleic Acids Symp. Ser. 2008, 52, 383–384. [Google Scholar] [CrossRef]
- Jawalekar, A.M.; Meeuwenoord, N.; Cremers, J.S.; Overkleeft, H.S.; van der Marel, G.A.; Rutjes, F.P.; van Delft, F.L. Conjugation of nucleosides and oligonucleotides by [3 + 2] cycloaddition. J. Org. Chem. 2008, 73, 287–290. [Google Scholar] [CrossRef]
- Coppola, C.; Simeone, L.; De Napoli, L.; Montesarchio, D. On the Compatibility of Azides in Phosphoramidite-Based Couplings: Synthesis of a Novel, Convertible Azido-Functionalized CyPLOS Analogue. Eur. J. Org. Chem. 2011, 2011, 1155–1165. [Google Scholar] [CrossRef]
- Ozols, K.; Cīrule, D.; Novosjolova, I.; Stepanovs, D.; Liepinsh, E.; Bizdēna, Ē.; Turks, M. Development of N6-methyl-2-(1,2,3-triazol-1-yl)-2′-deoxyadenosine as a novel fluorophore and its application in nucleotide synthesis. Tetrahedron Lett. 2016, 57, 1174–1178. [Google Scholar] [CrossRef]
- Zayas, J.; Annoual, M.; Das, J.K.; Felty, Q.; Gonzalez, W.G.; Miksovska, J.; Sharifai, N.; Chiba, A.; Wnuk, S.F. Strain Promoted Click Chemistry of 2- or 8-Azidopurine and 5-Azidopyrimidine Nucleosides and 8-Azidoadenosine Triphosphate with Cyclooctynes. Application to Living Cell Fluorescent Imaging. Bioconjugate Chem. 2015, 26, 1519–1532. [Google Scholar] [CrossRef]
- Wen, Z.; Tuttle, P.R.; Howlader, A.H.; Vasilyeva, A.; Gonzalez, L.; Tangar, A.; Lei, R.; Laverde, E.E.; Liu, Y.; Miksovska, J.; et al. Fluorescent 5-Pyrimidine and 8-Purine Nucleosides Modified with an N-Unsubstituted 1,2,3-Triazol-4-yl Moiety. J. Org. Chem. 2019, 84, 3624–3631. [Google Scholar] [CrossRef]
- Tera, M.; Luedtke, N.W. Chapter Nineteen—Cross-linking cellular nucleic acids via a target-directing double click reagent. In Methods in Enzymology; Chenoweth, D.M., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 641, pp. 433–457. [Google Scholar]
- Kotra, L.P.; Wang, P.; Bartlett, M.G.; Shanmuganathan, K.; Xu, Z.; Cavalcanti, S.; Newton, M.G.; Chu, C.K. 4-Azido-2-pyrimidinone Nucleosides and Related Chemistry. J. Org. Chem. 1997, 62, 7267–7271. [Google Scholar] [CrossRef]
- Lakshman, M.K.; Singh, M.K.; Parrish, D.; Balachandran, R.; Day, B.W. Azide-tetrazole equilibrium of C-6 azidopurine nucleosides and their ligation reactions with alkynes. J. Org. Chem. 2010, 75, 2461–2473. [Google Scholar] [CrossRef]
- Ren, Z.; Luo, H.; Yu, Z.; Song, J.; Liang, L.; Wang, L.; Wang, H.; Cui, G.; Liu, Y.; Wang, J.; et al. A Randomized, Open-label, Controlled Clinical Trial of Azvudine Tablets in the Treatment of Mild and Common COVID-19, A Pilot Study. Adv. Sci. 2020, 7, 2001435. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Chang, J. Azvudine (FNC): A promising clinical candidate for COVID-19 treatment. Signal transduct. target. ther. 2020, 5, 236. [Google Scholar] [CrossRef]
- Klumpp, K.; Lévêque, V.; Le Pogam, S.; Ma, H.; Jiang, W.R.; Kang, H.; Granycome, C.; Singer, M.; Laxton, C.; Hang, J.Q.; et al. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 2006, 281, 3793–3799. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.P.; Manouilov, K.K.; Cretton-Scott, E.; Sommadossi, J.P.; Boudinot, F.D.; Schinazi, R.F.; Chu, C.K. Synthesis, Biotransformation, and Pharmacokinetic Studies of 9-(β-d-Arabinofuranosyl)-6-azidopurine: A Prodrug for Ara-A Designed To Utilize the Azide Reduction Pathway. J. Med. Chem. 1996, 39, 5202–5207. [Google Scholar] [CrossRef] [PubMed]
- Cosyn, L.; Palaniappan, K.K.; Kim, S.-K.; Duong, H.T.; Gao, Z.-G.; Jacobson, K.A.; Van Calenbergh, S. 2-Triazole-Substituted Adenosines: A New Class of Selective A3 Adenosine Receptor Agonists, Partial Agonists, and Antagonists. J. Med. Chem. 2006, 49, 7373–7383. [Google Scholar] [CrossRef] [PubMed]
- Fabrega, C.; Garcia, R.G.; Diaz, A.R.; Eritja, R. Studies on the Synthesis of Oligonucleotides Containing Photoreactive Nucleosides: 2-Azido-2’-Deoxyinosine and 8-Azido-2′-Deoxyadenosine. Biol. Chem. 1998, 379, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Lakshman, M.K.; Kumar, A.; Balachandran, R.; Day, B.W.; Andrei, G.; Snoeck, R.; Balzarini, J. Synthesis and Biological Properties of C-2 Triazolylinosine Derivatives. J. Org. Chem. 2012, 77, 5870–5883. [Google Scholar] [CrossRef] [PubMed]
- Lioux, T.; Gosselin, G.; Mathé, C. Azido/Tetrazole Tautomerism in 2-Azidoadenineβ-D-Pentofuranonucleoside Derivatives. Eur. J. Org. Chem. 2003, 2003, 3997–4002. [Google Scholar] [CrossRef]
- Housri, N.; Yarchoan, R.; Kaushal, A. Radiotherapy for patients with the human immunodeficiency virus: Are special precautions necessary? Cancer 2010, 116, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Coucke, P.A.; Cottin, E.; Decosterd, L.A. Simultaneous alteration of de novo and salvage pathway to the deoxynucleoside triphosphate pool by (E)-2’-Deoxy-(fluoromethylene)cytidine (FMdC) and zidovudine (AZT) results in increased radiosensitivity in vitro. Acta Oncol. 2007, 46, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.-X.; Liao, Z.-K.; Dai, J.; Xiong, J.; Xie, C.-H.; Luo, Z.-G.; Liu, S.-Q.; Zhou, Y.-F. Radiosensitization effect of zidovudine on human malignant glioma cells. Biochem. Biophys. Res. Commun. 2007, 354, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.-K.; Zhou, F.-X.; Luo, Z.-G.; Zhang, W.-J.; Jie, X.; Bao, J.; Han, G.; Zhang, M.-S.; Xie, C.-H.; Zhou, Y.-F. Radioactivation of hTERT promoter in larynx squamous carcinoma cells: An ‘indirected-activator’ strategy in radio-gene-therapy. Oncol. Rep. 2008, 19, 281–286. [Google Scholar] [PubMed]
- Sjoberg, B.M.; Graslund, A.; Eckstein, F. A substrate radical intermediate in the reaction between ribonucleotide reductase from Escherichia coli and 2’-azido-2’-deoxynucleoside diphosphates. J. Biol. Chem. 1983, 258, 8060–8067. [Google Scholar] [CrossRef]
- Salowe, S.; Bollinger, J.M., Jr.; Ator, M.; Stubbe, J.; McCracken, J.; Peisach, J.; Samano, M.C.; Robins, M.J. Alternative model for mechanism-based inhibition of Escherichia coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry 1993, 32, 12749–12760. [Google Scholar] [CrossRef] [PubMed]
- van der Donk, W.A.; Stubbe, J.; Gerfen, G.J.; Bellew, B.F.; Griffin, R.G. EPR Investigations of the Inactivation of E. coli Ribonucleotide Reductase with 2′-Azido-2′-deoxyuridine 5′-Diphosphate: Evidence for the Involvement of the Thiyl Radical of C225-R1. J. Am. Chem. Soc. 1995, 117, 8908–8916. [Google Scholar] [CrossRef]
- Fritscher, J.; Artin, E.; Wnuk, S.; Bar, G.; Robblee, J.H.; Kacprzak, S.; Kaupp, M.; Griffin, R.G.; Bennati, M.; Stubbe, J. Structure of the Nitrogen-Centered Radical Formed during Inactivation of E. coli Ribonucleotide Reductase by 2′-Azido-2′-deoxyuridine-5′-diphosphate: Trapping of the 3′-Ketonucleotide. J. Am. Chem. Soc. 2005, 127, 7729–7738. [Google Scholar] [CrossRef] [PubMed]
- Zipse, H.; Artin, E.; Wnuk, S.; Lohman, G.J.S.; Martino, D.; Griffin, R.G.; Kacprzak, S.; Kaupp, M.; Hoffman, B.; Bennati, M.; et al. Structure of the Nucleotide Radical Formed during Reaction of CDP/TTP with the E441Q-a2b2 of E. coli Ribonucleotide Reductase. J. Am. Chem. Soc. 2009, 131, 200–211. [Google Scholar] [CrossRef]
- Wnuk, S.F.; Chowdhury, S.M.; Garcia, P.I., Jr.; Robins, M.J. Stereo-defined Synthesis of O3’-Labeled Uracil Nucleosides. 3′-[17O]-2′-Azido-2′-deoxyuridine 5′-Diphosphate as a Probe for the Mechanism of Inactivation of Ribonucleotide Reductases. J. Org. Chem. 2002, 67, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Joe, G.H.; Do, J.Y. Highly efficient intramolecular addition of aminyl radicals to carbonyl groups: A new ring expansion reaction leading to lactams. J. Am. Chem. Soc. 1993, 115, 3328–3329. [Google Scholar] [CrossRef]
- Kim, S.; Joe, G.H.; Do, J.Y. Novel Radical Cyclizations of Alkyl Azides. A New Route to N-Heterocycles. J. Am. Chem. Soc. 1994, 116, 5521–5522. [Google Scholar] [CrossRef]
- Ashley, G.W.; Lawrence, C.C.; Stubbe, J.; Robins, M.J. Mechanism based inactivation of the adenosylcobalamin-dependent ribonucleotide reductase from L. leichmannii by 2′-ara-2′-azido-2′-deoxy adenosine-5′-triphosphate: Observation of paramagnetic intermediates. Tetrahedron 1997, 53, 12005–12016. [Google Scholar] [CrossRef]
- Pereira, S.; Fernandes, P.A.; Ramos, M.J. Theoretical study of ribonucleotide reductase mechanism-based inhibition by 2’-azido-2’-deoxyribonucleoside 5’-diphosphates. J. Comput. Chem. 2004, 25, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Bräse, S.; Gil, C.; Knepper, K.; Zimmermann, V. Organic Azides: An Exploding Diversity of a Unique Class of Compounds. Angew. Chem. Int. Ed. 2005, 44, 5188–5240. [Google Scholar] [CrossRef] [PubMed]
- Benati, L.; Bencivenni, G.; Leardini, R.; Minozzi, M.; Nanni, D.; Scialpi, R.; Spagnolo, P.; Zanardi, G. Thermal Reactions of Tributyltin Hydride with a-Azido Esters: Unexpected Intervention of Tin Triazene Adducts under Both Nonradical and Radical Conditions. J. Org. Chem. 2005, 70, 3046–3053. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.P.; Winter, J.N. Electron spin resonance studies of radicals derived from organic azides. J. Chem. Soc. Perk. Trans. 2 1979, 1353–1361. [Google Scholar] [CrossRef]
- Benati, L.; Bencivenni, G.; Leardini, R.; Minozzi, M.; Nanni, D.; Scialpi, R.; Spagnolo, P.; Zanardi, G. Radical Reduction of Aromatic Azides to Amines with Triethylsilane. J. Org. Chem. 2006, 71, 5822–5825. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.; Kopsov, S.; Ferreri, C.; Chatgilialoglu, C. Radical Reactions in Aqueous Medium Using (Me3Si)3SiH. Org. Lett. 2007, 9, 5159–5162. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.P.; Sobczak, A.J.; Mebel, A.M.; Chatgilialoglu, C.; Wnuk, S.F. Investigation of reactions postulated to occur during inhibition of ribonucleotide reductases by 2′-azido-2′-deoxynucleotides. Tetrahedron 2012, 68, 5655–5667. [Google Scholar] [CrossRef] [PubMed]
- Robins, M.J.; Guo, Z.; Samano, M.C.; Wnuk, S.F. Biomimetic Simulation of Free Radical-Initiated Cascade Reactions Postulated To Occur at the Active Site of Ribonucleotide Reductases. J. Am. Chem. Soc. 1999, 121, 1425–1433. [Google Scholar] [CrossRef]
- Robins, M.J.; Wnuk, S.F.; Hernandez-Thirring, A.E.; Samano, M.C. Nucleic Acid-Related Compounds. 91. Biomimetic Reactions Are in Harmony with Loss of 2’-Substituents as Free Radicals (Not Anions) during Mechanism-Based Inactivation of Ribonucleotide Reductases. Differential Interactions of Azide, Halogen, and Alkylthio Groups with Tributylstannane and Triphenylsilane. J. Am. Chem. Soc. 1996, 118, 11341–11348. [Google Scholar]
- Samano, M.C.; Robins, M.J. Efficient reduction of azides to amines with tributylstannane. high-yield syntheses of amino and diamino deoxynucleosides. Tetrahedron Lett. 1991, 32, 6293–6296. [Google Scholar] [CrossRef]
- Hayon, E.M. Formation and Reactions of Azide Radicals; Accesssion Number: AD0750330; Army Natick Laboratories, National Technical Information Service, U.S. Department of Commerce: Natick, MA, USA, 1972; pp. 45–56. [Google Scholar]
- Adjei, D.; Reyes, Y.; Kumar, A.; Ward, S.; Denisov, S.A.; Alahmadi, M.; Sevilla, M.D.; Wnuk, S.F.; Mostafavi, M.; Adhikary, A. Pathways of the Dissociative Electron Attachment Observed in 5- and 6-Azidomethyluracil Nucleosides: Nitrogen (N2) Elimination vs Azide Anion (N3−) Elimination. J. Phys. Chem. B 2023, 127, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Živanov, S.; Ibanescu, B.C.; Paech, M.; Poffet, M.; Baettig, P.; Sergenton, A.C.; Grimme, S.; Allan, M. Dissociative Electron Attachment and Electron Energy-Loss Spectra of Phenyl Azide. J. Phys. B At. Mol. Opt. Phys. 2007, 40, 101–107. [Google Scholar] [CrossRef]
- Narayanan, S.J.J.; Tripathi, D.; Verma, P.; Adhikary, A.; Dutta, A.K. Secondary Electron Attachment-Induced Radiation Damage to Genetic Materials. ACS Omega 2023, 8, 10669–10689. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Khanduri, D.; Pottiboyina, V.; Rice, C.T.; Sevilla, M.D. Formation of Aminyl Radicals on Electron Attachment to AZT: Abstraction from the Sugar Phosphate Backbone versus One-Electron Oxidation of Guanine. J. Phys. Chem. B 2010, 114, 9289–9299. [Google Scholar] [CrossRef]
- Mudgal, M.; Dang, T.P.; Sobczak, A.J.; Lumpuy, D.A.; Dutta, P.; Ward, S.; Ward, K.; Alahmadi, M.; Kumar, A.; Sevilla, M.D.; et al. Site of Azido Substitution in the Sugar Moiety of Azidopyrimidine Nucleosides Influences the Reactivity of Aminyl Radicals Formed by Dissociative Electron Attachment. J. Phys. Chem. B 2020, 124, 11357–11370. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Dai, X.; Su, H.; Greenberg, M.M. Independent Generation and Time-Resolved Detection of 2′-Deoxyguanosin-N2-yl Radicals. Angew. Chem. Int. Ed. 2020, 59, 13406–13413. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; D’Angelantonio, M.; Guerra, M.; Kaloudis, P.; Mulazzani, Q.G. A reevaluation of the ambident reactivity of the guanine moiety towards hydroxyl radicals. Angew. Chem. Int. Ed. 2009, 48, 2214–2217. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Kumar, A.; Becker, D.; Sevilla, M.D. The Guanine Cation Radical: Investigation of Deprotonation States by ESR and DFT. J. Phys. Chem. B 2006, 110, 24171–24180. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Kumar, A.; Munafo, S.A.; Khanduri, D.; Sevilla, M.D. Prototropic equilibria in DNA containing one-electron oxidized GC: Intra-duplex vs. duplex to solvent deprotonation. Phys. Chem. Chem. Phys. 2010, 12, 5353–5368. [Google Scholar] [CrossRef] [PubMed]
- Mudgal, M.; Rishi, S.; Lumpuy, D.A.; Curran, K.A.; Verley, K.L.; Sobczak, A.J.; Dang, T.P.; Sulimoff, N.; Kumar, A.; Sevilla, M.D.; et al. Prehydrated One-Electron Attachment to Azido-Modified Pentofuranoses: Aminyl Radical Formation, Rapid H-Atom Transfer, and Subsequent Ring Opening. J. Phys. Chem. B 2017, 121, 4968–4980. [Google Scholar] [CrossRef] [PubMed]
- Reyes, Y.; Barbieri, M.; Pathak, R.; Ward, S.; Sevilla, M.D.; Adhikary, A.; Wnuk, S.F. Studies of amino and azido derivatives of sesquiterpene lactones and their potential augmentation for radiation damage to cancer cells. In Proceedings of the 263rd ACS National Meeting, Medicinal Chemistry Division, Abstract 3647326, San Diego, CA, USA, 20–24 March 2022. [Google Scholar]
- Liang, Y.; Wen, Z.; Cabrera, M.; Howlader, A.H.; Wnuk, S.F. Purines. In Science of Synthesis: Houben-Weyl Methods of Molecular Transformations: Knowledge Updates 2020/1; Christmann, M., Huang, Z., Jiang, X., Li, J.J., Oestreich, M., Petersson, E.J., Schaumann, E., Wang, M., Eds.; Georg Thieme Verlag: Stuttgart, Germany, 2020; pp. 195–384. [Google Scholar]
- Wen, Z.; Peng, J.; Tuttle, P.R.; Ren, Y.; Garcia, C.; Debnath, D.; Rishi, S.; Hanson, C.; Ward, S.; Kumar, A.; et al. Electron-Mediated Aminyl and Iminyl Radicals from C5 Azido-Modified Pyrimidine Nucleosides Augment Radiation Damage to Cancer Cells. Org. Lett. 2018, 20, 7400–7404. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; DiMagno, S.G. Reactivities of vinyl azides and their recent applications in nitrogen heterocycle synthesis. Org. Biomol. Chem. 2015, 13, 3844–3855. [Google Scholar] [CrossRef] [PubMed]
- Neef, A.B.; Luedtke, N.W. An azide-modified nucleoside for metabolic labeling of DNA. ChemBioChem 2014, 15, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; El-Sagheer, A.H.; Brown, T. Azide and trans-cyclooctene dUTPs: Incorporation into DNA probes and fluorescent click-labelling. Analyst 2015, 140, 2671–2678. [Google Scholar] [CrossRef] [PubMed]
- von Sonntag, C. Free-Radical-Induced DNA Damage and Its Repair; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Reyes, Y.; Mebel, A.; Wnuk, S.F. 6-azido and 6-azidomethyl uracil nucleosides. Nucleosides Nucleotides Nucl. Acids 2023, 43, 453–471. [Google Scholar] [CrossRef] [PubMed]
- de Cabrera, M.; Reyes, Y.; Ward, S.; Alahmadi, M.; Barbolovici, A.; Kumar, A.; Sevilla, M.D.; Wnuk, S.F.; Adhikary, A. Characterization of nitrogen-centered radicals formed via dissociative electron attachment from 4-, 5-, and 6-azido substituted pyrimidine nucleosides. In Proceedings of the 263rd ACS National Meeting, Physical Chemistry Division, Abstract 3649777, San Diego, CA, USA, 20–24 March 2022. [Google Scholar]
- De Cabrera, M. Nucleoside analogues for positron emission tomography imaging and to study radiation mediated generation of radicals from azides. Ph.D. Thesis, Florida international University, Miami, FL, USA, 2019. [Google Scholar]
- Balintová, J.; Špaček, J.; Pohl, R.; Brázdová, M.; Havran, L.; Fojta, M.; Hocek, M. Azidophenyl as a click-transformable redox label of DNA suitable for electrochemical detection of DNA–protein interactions. Chem. Sci. 2015, 6, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Danhel, A.; Trosanova, Z.; Balintova, J.; Havran, L.; Hocek, M.; Barek, J.; Fojta, M. Voltammetric analysis of 5-(4-Azidophenyl)-2′-deoxycytidine nucleoside and azidophenyl-labelled single- and double-stranded DNAs. Electrochim. Acta 2016, 215, 72–83. [Google Scholar] [CrossRef]
- Daňhel, A.; Trošanová, Z.; Balintová, J.; Simonová, A.; Pospíšil, L.; Cvačka, J.; Hocek, M.; Fojta, M. Electrochemical reduction of azidophenyl-deoxynucleoside conjugates at mercury surface. Electrochim. Acta 2018, 259, 377–385. [Google Scholar] [CrossRef]
- Kuttappan-Nair, V.; Samson-Thibault, F.; Wagner, J.R. Generation of 2′-Deoxyadenosine N6-Aminyl Radicals from the Photolysis of Phenylhydrazone Derivatives. Chem. Res. Toxicol. 2010, 23, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lin, L.; Qu, K.; Adhikary, A.; Sevilla, M.D.; Greenberg, M.M. Independent Photochemical Generation and Reactivity of Nitrogen-Centered Purine Nucleoside Radicals from Hydrazines. Org. Lett. 2017, 19, 6444–6447. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Griesser, M.; Pratt, D.A.; Greenberg, M.M. Aminyl Radical Generation via Tandem Norrish Type I Photocleavage, β-Fragmentation: Independent Generation and Reactivity of the 2’-Deoxyadenosin- N6-yl Radical. J. Org. Chem. 2017, 82, 3571–3580. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Greenberg, M.M. DNA Damage Emanating From a Neutral Purine Radical Reveals the Sequence Dependent Convergence of the Direct and Indirect Effects of γ-Radiolysis. J. Am. Chem. Soc. 2017, 139, 17751–17754. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Greenberg, M.M. Traceless Tandem Lesion Formation in DNA from a Nitrogen-Centered Purine Radical. J. Am. Chem. Soc. 2018, 140, 6400–6407. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zheng, L.; Greenberg, M.M. Independent Generation of Reactive Intermediates Leads to an Alternative Mechanism for Strand Damage Induced by Hole Transfer in Poly(dA–T) Sequences. J. Am. Chem. Soc. 2018, 140, 11308–11316. [Google Scholar] [CrossRef] [PubMed]
- Naumov, S.; von Sonntag, C. Guanine-derived radicals: Dielectric constant-dependent stability and UV/Vis spectral properties: A DFT study. Radiat. Res. 2008, 169, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Greenberg, M.M. Independent Generation and Reactivity of 2′-Deoxyguanosin-N1-yl Radical. J. Org. Chem. 2020, 85, 8665–8672. [Google Scholar] [CrossRef] [PubMed]
- Vrantza, D.; Kaloudis, P.; Leondiadis, L.; Gimisis, T.; Vougioukalakis, G.C.; Orfanopoulos, M.; Gasparutto, D.; Cadet, J.; Encinas, S.; Paris, C.; et al. Modification of Guanine with Photolabile N-Hydroxypyridine-2(1H)-thione: Monomer Synthesis, Oligonucleotide Elaboration, and Photochemical Studies. Helv. Chim. Acta 2006, 89, 2371–2386. [Google Scholar] [CrossRef]
- Kaloudis, P.; Paris, C.; Vrantza, D.; Encinas, S.; Pérez-Ruiz, R.; Miranda, M.A.; Gimisis, T. Photolabile N-hydroxypyrid-2(1H)-one derivatives of guanine nucleosides: A new method for independent guanine radical generation. Org. Biomol. Chem. 2009, 7, 4965–4972. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Jie, J.; Mortimer, I.P.; Ma, Z.; Su, H.; Greenberg, M.M. Reactivity and DNA Damage by Independently Generated 2′-Deoxycytidin-N4-yl Radical. J. Am. Chem. Soc. 2021, 143, 14738–14747. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Vu, S.; Retes, P.; Ward, S.; Kumar, A.; Sevilla, M.D.; Adhikary, A.; Greenberg, M.M. Photochemical and Single Electron Transfer Generation of 2′-Deoxycytidin-N4-yl Radical from Oxime Esters. J. Org. Chem. 2023, 88, 7381–7390. [Google Scholar] [CrossRef]
- Adhikary, A.; Kumar, A.; Bishop, C.T.; Wiegand, T.J.; Hindi, R.M.; Adhikary, A.; Sevilla, M.D. π-Radical to σ-Radical Tautomerization in One-Electron-Oxidized 1-Methylcytosine and Its Analogs. J. Phys. Chem. B 2015, 119, 11496–11505. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.A. Light-Induced Iminyl Radicals: Generation and Synthetic Applications. In Molecular Photochemistry: Various Aspects; Satayen, S., Ed.; IntechOpen: London, UK, 2012; pp. 265–282. [Google Scholar]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes, Y.; Adhikary, A.; Wnuk, S.F. Nitrogen-Centered Radicals Derived from Azidonucleosides. Molecules 2024, 29, 2310. https://doi.org/10.3390/molecules29102310
Reyes Y, Adhikary A, Wnuk SF. Nitrogen-Centered Radicals Derived from Azidonucleosides. Molecules. 2024; 29(10):2310. https://doi.org/10.3390/molecules29102310
Chicago/Turabian StyleReyes, Yahaira, Amitava Adhikary, and Stanislaw F. Wnuk. 2024. "Nitrogen-Centered Radicals Derived from Azidonucleosides" Molecules 29, no. 10: 2310. https://doi.org/10.3390/molecules29102310
APA StyleReyes, Y., Adhikary, A., & Wnuk, S. F. (2024). Nitrogen-Centered Radicals Derived from Azidonucleosides. Molecules, 29(10), 2310. https://doi.org/10.3390/molecules29102310






