The Identification of SQS/SQE/OSC Gene Families in Regulating the Biosynthesis of Triterpenes in Potentilla anserina
Abstract
1. Introduction
2. Results
2.1. The Identification and Characterization of SQS/SQE/OSC Gene Families in Pan
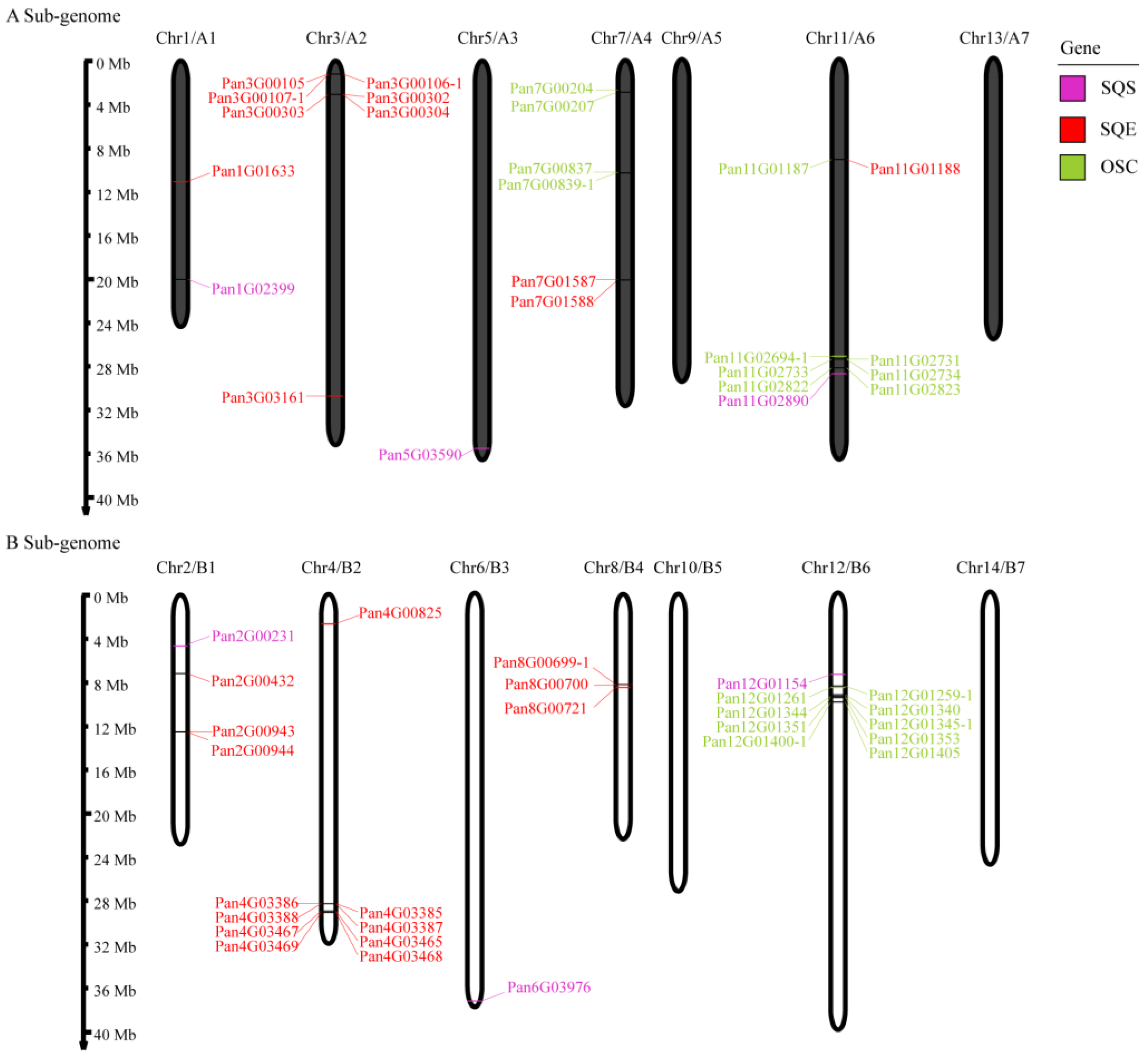
2.2. The Chromosomal Locations and Collinearity Analysis of SQSs/SQEs/OSCs
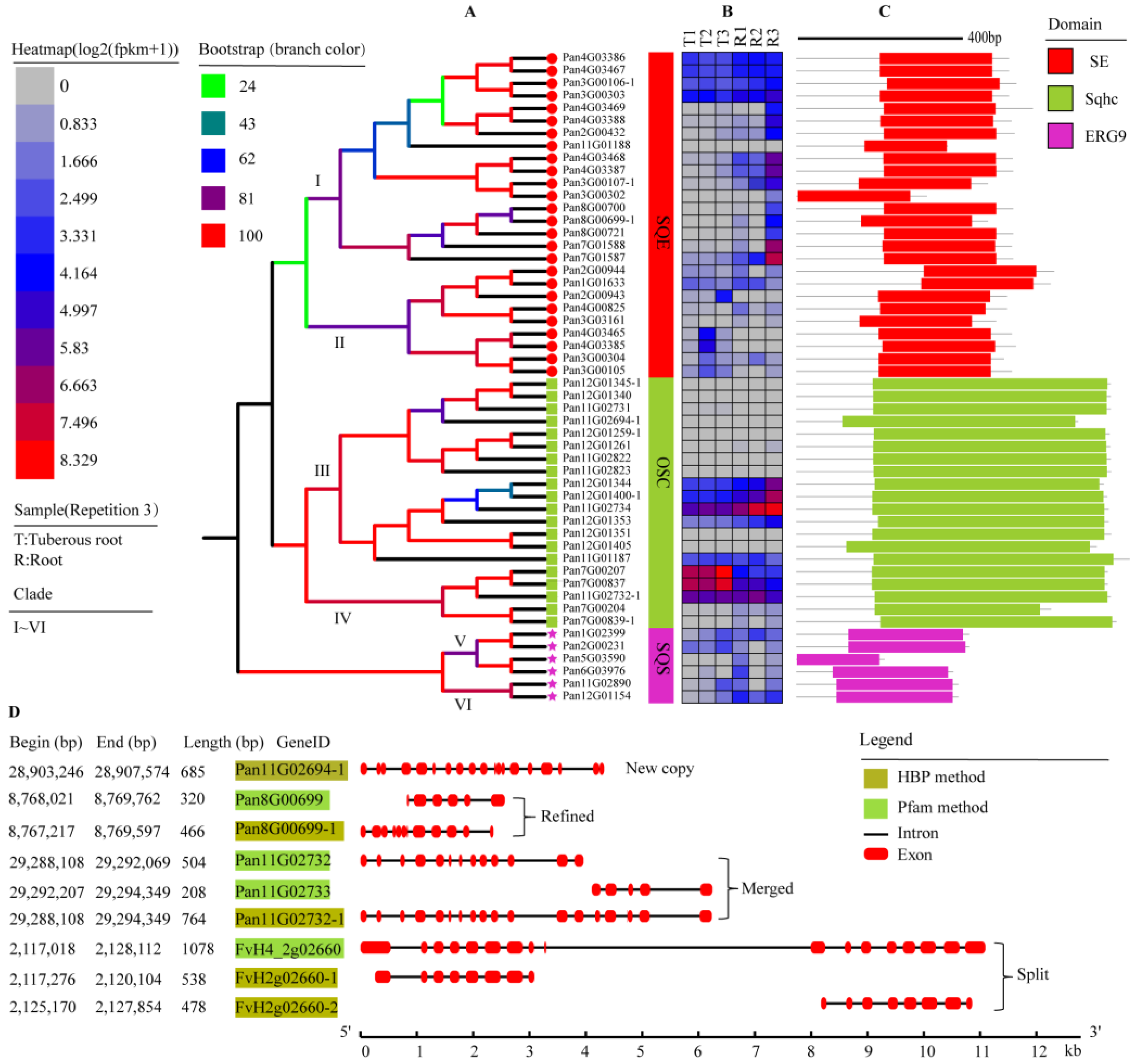
2.3. The Phylogenetic Analysis of SQSs/SQEs
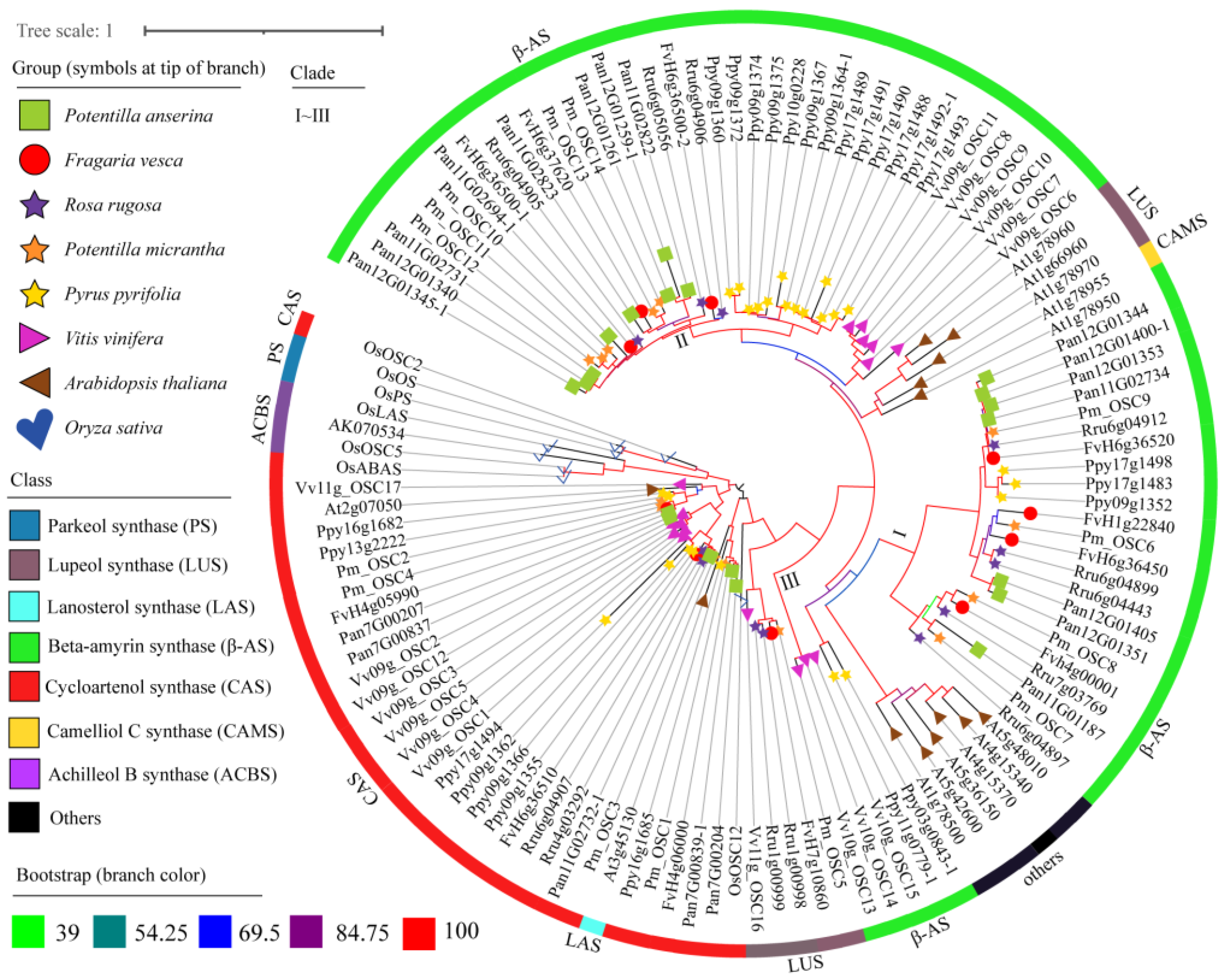
2.4. Phylogenetic Analysis of OSCs
3. Discussion
3.1. Gene Families Identified by the Pfam Database and the HBP Method
3.2. SQS/SQE/OSC Gene Family Expression and Evolution
3.3. The Functional Prediction of OCSs and Triterpene Synthesis in Pan
4. Materials and Methods
4.1. SQS/SQE/OSC Gene Family Identification
4.2. Physicochemical Properties of OSCs/SQEs/SQSs from Pan
4.3. Sequence Alignment and Phylogenetic Analysis
4.4. RNA-seq Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, M.F.; Shen, Y.Q. Research advances on effects of ursolic acid and oleanolic acid against hepatic steatosis and fibrosis. Drug Eval. Res. 2017, 40, 270–278. [Google Scholar]
- Ji, Z.X.; Li, J.Q.; Wang, J.B. Research status on pharmacological effects of ginsenoside Rh3. SH. J. TCM. 2021, 55, 97–100. [Google Scholar]
- Wang, Y.; Zhang, H.; Ri, H.C.; An, Z.Y.; Wang, X.; Zhou, N.J.; Zheng, D.R.; Wu, H.; Wang, P.C.; Yang, J.F.; et al. Deletion and tandem duplications of biosynthetic genes drive the diversity of triterpenoids in Aralia elata. Nat. Commun. 2022, 13, 2224. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, Y.H.; Yin, X.; Wang, X.N.; Qi, X.Q.; Xue, Z.Y. Diverse triterpene skeletons are derived from the expansion and divergent evolution of 2,3-oxidosqualene cyclases in plants. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 113–132. [Google Scholar] [CrossRef]
- Xue, Z.Y.; Duan, L.X.; Liu, D.; Guo, J.; Ge, S.; Dicks, J.; ÓMáille, P.; Osbourn, A.; Qi, X.Q. Divergent evolution of oxidosqualene cyclases in plants. New. Phytol. 2012, 193, 1022–1038. [Google Scholar] [CrossRef]
- Chen, C.Y.; Pang, Y.R.; Chen, Q.B.; Li, C.; Lü, B. Oxidosqualene cyclases in triterpenoids biosynthesis: A review. Chin. J. Biotech. 2022, 38, 443–459. [Google Scholar]
- Hoshino, T. β-Amyrin biosynthesis: Catalytic mechanism and substrate recognition. Org. Biomol. Chem. 2017, 15, 2869–2891. [Google Scholar] [CrossRef]
- Tao, H.; Lauterbach, L.; Bian, G.K.; Chen, R.; Hou, A.; Mori, T.; Cheng, S.; Hu, B.; Lu, L.; Mu, X.; et al. Discovery of non-squalene triterpenes. Nature 2022, 606, 414–419. [Google Scholar] [CrossRef]
- Morikawa, T.; Ninomiya, K.; Imura, K.; Yamaguchi, T.; Akagi, Y.; Yoshikawa, M.; Hayakawa, T.; Muraoka, O. Hepatoprotective triterpenes from traditional Tibetan medicine Potentilla anserina. Phytochemistry 2014, 102, 169–181. [Google Scholar] [CrossRef]
- Morikawa, T.; Imura, K.; Akagi, Y.; Muraoka, O.; Ninomiya, K. Ellagic acid glycosides with hepatoprotective activity from traditional Tibetan medicine Potentilla anserina. J. Nat. Med. 2018, 72, 317–325. [Google Scholar] [CrossRef]
- Chen, J.R.; Yang, Z.Q.; Hu, T.J.; Yan, Z.T.; Niu, T.X.; Wang, L.; Cui, D.A.; Wang, M. Immunomodulatory activity in vitro and in vivo of polysaccharide from Potentilla anserina. Fitoterapia 2010, 81, 1117–1124. [Google Scholar] [CrossRef]
- Guo, T.; Wei, J.Q.; Ma, J.P. Antitussive and expectorant activities of Potentilla anserina. Pharm. Biol. 2016, 54, 807–811. [Google Scholar] [CrossRef]
- Yang, D.; Han, N.; Wang, Y.W.; Zhai, J.X.; Liu, Z.H.; Li, S.K.; Yin, J. Pentacyclic triterpenoid saponins, poterinasides A–J, from Silverweed Cinquefoil Roots promising hepatoprotective and anti-inflammatory Agents. J. Org. Chem. 2021, 86, 11220–11236. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Z.Q.; Yu, H.H.; Huang, F.B.; Chen, Z.L.; Chu, L.L.; Wang, W. Research progress on chemical constituents and pharmacological activities of Potentilla. China J. Chin. Mater. Med. 2022, 47, 1509–1538. [Google Scholar]
- Wu, J.; Zhang, Z.Q.; Zhou, X.D.; Yao, Q.Y.; Chen, Z.L.; Chu, L.L.; Yu, H.H.; Yang, Y.P.; Li, B.; Wang, W. New terpenoids from Potentilla freyniana Bornm. and their cytotoxic activities. Molecules 2022, 27, 3665. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.L.; Li, S.M.; Zong, Y.; Cao, D.; Li, Y.; Liu, R.J.; Cheng, S.; Liu, B.L.; Zhang, H.G. Chromosome-level genome assembly provides new insights into genome evolution and tuberous root formation of Potentilla anserina. Genes 2021, 12, 1993. [Google Scholar] [CrossRef] [PubMed]
- Summers, C.; Karst, F.; Charles, A.D. Cloning, expression and characterisation of the cDNA encoding human hepatic squalene synthase, and its relationship to phytoene synthase. Gene 1993, 136, 185–192. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.C.; Qiao, Q.; Du, X.; Zhang, X.; Hou, Y.; Wei, X.; Sun, C.; Zhang, R.G.; Yun, Q.Z.; Crabbe, M.C.; et al. Cultivated hawthorn (Crataegus pinnatifida var. major) genome sheds light on the evolution of Maleae (apple tribe). J. Integr. Plant Biol. 2022, 64, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.D.; Beridze, T.; Cantu, D. The population genetics of structural variants in grapevine domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Chen, F.; Su, L.Y.; Hu, S.Y.; Xue, J.Y.; Liu, H.; Liu, G.H.; Jiang, Y.F.; Du, J.K.; Qiao, Y.S.; Fan, Y.N.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic. Res. 2021, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.P.; Kong, W.W.; Cao, X.Y.; Chen, C.; Liu, Q.; Li, X.M.; Wang, Z.Z. Transcriptome analysis of Dioscorea zingiberensis identifies genes involved in diosgenin biosynthesis. Genes Genom. 2017, 39, 509–520. [Google Scholar] [CrossRef]
- Song, Y.D.; Chen, S.; Wang, X.J.; Zhang, R.; Tu, L.C.; Hu, T.Y.; Liu, X.H.; Zhang, Y.F.; Huang, L.Q.; Gao, W. A novel strategy to enhance terpenoids production using cambial meristematic cells of Tripterygium wilfordii Hook. f. Plant Methods 2019, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.Y.; Su, P.; Zhao, Y.J.; Zhang, X.N.; Dai, Z.B.; Guo, J.; Tong, Y.R.; Liu, Y.J.; Hu, T.Y.; Yin, Y.; et al. Cloning and functional analysis of two sterol-C24-methyltransferase 1 (SMT1) genes from Paris polyphylla. J. Asian Nat. Prod. Res. 2018, 20, 595–604. [Google Scholar] [CrossRef]
- Lu, Y.D.; Zhou, W.X.; Wei, L.; Li, J.; Jia, J.; Li, F.; Smith, S.M.; Xu, J. Regulation of the cholesterol biosynthetic pathway and its integration with fatty acid biosynthesis in the oleaginous microalga Nannochloropsis oceanica. Biotechnol. Biofuels 2014, 7, 81. [Google Scholar] [CrossRef]
- Christ, B.; Xu, C.C.; Xu, M.L.; Li, F.S.; Wada, N.; Mitchell, A.J.; Han, X.L.; Wen, M.L.; Fujita, M.; Weng, J.K. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 2019, 10, 3206. [Google Scholar] [CrossRef]
- Xue, Z.Y.; Tan, Z.W.; Huang, A.C.; Zhou, Y.; Sun, J.C.; Wang, X.N.; Thimmappa, R.B.; Stephenson, M.J.; Osbourn, A.; Qi, X.Q. Identification of key amino acid residues determining product specificity of 2,3-oxidosqualene cyclase in Oryza species. New Phytol. 2018, 218, 1076–1088. [Google Scholar] [CrossRef]
- Sun, J.C.; Xu, X.; Xue, Z.Y.; Snydera, J.H.; Qi, X.Q. Functional analysis of a rice oxidosqualene cyclase through total gene synthesis. Mol. Plant 2013, 6, 1726–1729. [Google Scholar] [CrossRef]
- Lu, J.J.; Luo, M.F.; Wang, L.; Li, K.P.; Yu, Y.Y.; Yang, W.F.; Gong, P.C.; Gao, H.H.; Li, Q.R.; Zhao, J.; et al. The Physalis floridana genome provides insights into the biochemical and morphological evolution of Physalis fruits. Hortic. Res. 2021, 8, 244. [Google Scholar] [CrossRef]
- Buti, M.; Moretto, M.; Barghini, E.; Mascagni, F.; Natali, L.; Brilli, M.; Lomsadze, A.; Sonego, P.; Giongo, L.; Alonge, M.; et al. The genome sequence and transcriptome of Potentilla micrantha and their comparison to Fragaria vesca (the woodland strawberry). GigaScience 2018, 7, giy010. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 2018, 7, gix124. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Itai, A.; Isobe, S. Chromosome-scale genome assembly of Japanese pear (Pyrus pyrifolia) variety ‘Nijisseiki’. DNA Res. 2021, 28, dsab001. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Edgar, R.C.; Batzoglou, S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006, 16, 368–373. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Bio. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Xue, Z.Y.; Xu, X.; Zhou, Y.; Wang, X.M.; Zhang, Y.C.; Liu, D.; Zhao, B.B.; Duan, L.X.; Qi, X.Q. Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat. Commun. 2018, 9, 604. [Google Scholar] [CrossRef]
- Inagaki, Y.S.; Etherington, G.; Geisler, K.; Field, B.; Dokarry, M.; Ikeda, K.; Mutsukado, K.; Dicks, J.; Osbourn, A. Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol. 2011, 191, 432–448. [Google Scholar] [CrossRef]
- Sun, R.; Liu, S.; Tang, Z.Z.; Zheng, T.R.; Wang, T.; Chen, H.; Li, C.L.; Wu, Q. β-Amyrin synthase from Conyza blinii expressed in Saccharomyces cerevisiae. FEBS Open Bio. 2017, 7, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Su, W.B.; Jing, Y.; Lin, S.K.; Yue, Z.; Yang, X.H.; Xu, J.B.; Wu, J.C.; Zhang, Z.K.; Rui, X.; Zhu, J.J.; et al. underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc. Natl. Acad. Sci. USA 2021, 118, e2101767118. [Google Scholar] [CrossRef]
- Busquets, A.; Keim, V.; Closa, M.; Arco, A.; Boronat, A.; Arró, M.; Ferrer, A. Arabidopsis thaliana contains a single gene encoding squalene synthase. Plant Mol. Biol. 2008, 67, 25–36. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, L.P.; Chen, X.; Clarke, J.; Salmeron, J.; Nguyen, H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012, 63, 163–175. [Google Scholar] [CrossRef] [PubMed]
- The French–Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Shen, L.Y.; Luo, H.; Wang, X.L.; Wang, X.M.; Qiu, X.J.; Liu, H.; Zhou, S.S.; Jia, K.H.; Nie, S.; Bao, Y.T.; et al. Chromosome-scale genome assembly for Chinese sour jujube and insights into its genome evolution and domestication signature. Front. Plant Sci. 2021, 12, 773090. [Google Scholar] [CrossRef]
- Lang, D.; Ullrich, K.K.; Murat, F.; Fuchs, J.; Jenkins, J.; Haas, F.; Piednoel, M.; Gundlach, H.; Bel, M.; Meyberg, R.; et al. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018, 93, 515–533. [Google Scholar] [CrossRef]
- Wang, X.; Chen, D.J.; Wang, Y.Q.; Xie, J. De novo transcriptome assembly and the putative biosynthetic pathway of steroidal sapogenins of Dioscorea composita. PLoS ONE 2015, 10, e0124560. [Google Scholar] [CrossRef]
- Minx, P.; Cordum, H.; Wilson, R. Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 2005, 15, 1284–1291. [Google Scholar]
- Laranjeira, S.; Amorim-Silva, V.; Esteban, A.; Arró, M.; Ferrer, A.; Tavares, R.M.; Botella, M.A.; Rosado, A.; Azevedo, H. Arabidopsis squalene epoxidase 3 (SQE3) complements SQE1 and is important for embryo development and bulk squalene epoxidase activity. Mol. Plant 2015, 8, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kaul, S.; Rounsley, S.; Shea, T.; Benito, M.; Town, C.; Fujii, C.; Mason, T.; Bowman, C.; Barnstead, M.; et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 1999, 402, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.L.; Cai, C.C.; Liu, Z.Y.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.; Saxena, R.; Schlueter, J.; Donoghue, M.; Azam, S.; Fan, G.; Whaley, A.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.X.; Mitros, T.; Nelson, W.; Hyten, D.; Song, Q.J.; Thelen, J.; Cheng, J.L.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Nakayasu, M.; Shioya, N.; Shikata, M.; Thagun, C.; Abdelkareem, A.; Okabe, Y.; Ariizumi, T.; Arimura, G.; Mizutani, M.; Ezura, H.; et al. JRE4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. Plant J. 2018, 94, 975–990. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Li, X.; Huang, X.; Liu, F.; Zhang, Z.; Cao, L. The Identification of SQS/SQE/OSC Gene Families in Regulating the Biosynthesis of Triterpenes in Potentilla anserina. Molecules 2023, 28, 2782. https://doi.org/10.3390/molecules28062782
Jiao Y, Li X, Huang X, Liu F, Zhang Z, Cao L. The Identification of SQS/SQE/OSC Gene Families in Regulating the Biosynthesis of Triterpenes in Potentilla anserina. Molecules. 2023; 28(6):2782. https://doi.org/10.3390/molecules28062782
Chicago/Turabian StyleJiao, Yangmiao, Xu Li, Xueshuang Huang, Fan Liu, Zaiqi Zhang, and Liang Cao. 2023. "The Identification of SQS/SQE/OSC Gene Families in Regulating the Biosynthesis of Triterpenes in Potentilla anserina" Molecules 28, no. 6: 2782. https://doi.org/10.3390/molecules28062782
APA StyleJiao, Y., Li, X., Huang, X., Liu, F., Zhang, Z., & Cao, L. (2023). The Identification of SQS/SQE/OSC Gene Families in Regulating the Biosynthesis of Triterpenes in Potentilla anserina. Molecules, 28(6), 2782. https://doi.org/10.3390/molecules28062782





