Analysis and Discrimination of Canadian Honey Using Quantitative NMR and Multivariate Statistical Methods
Abstract
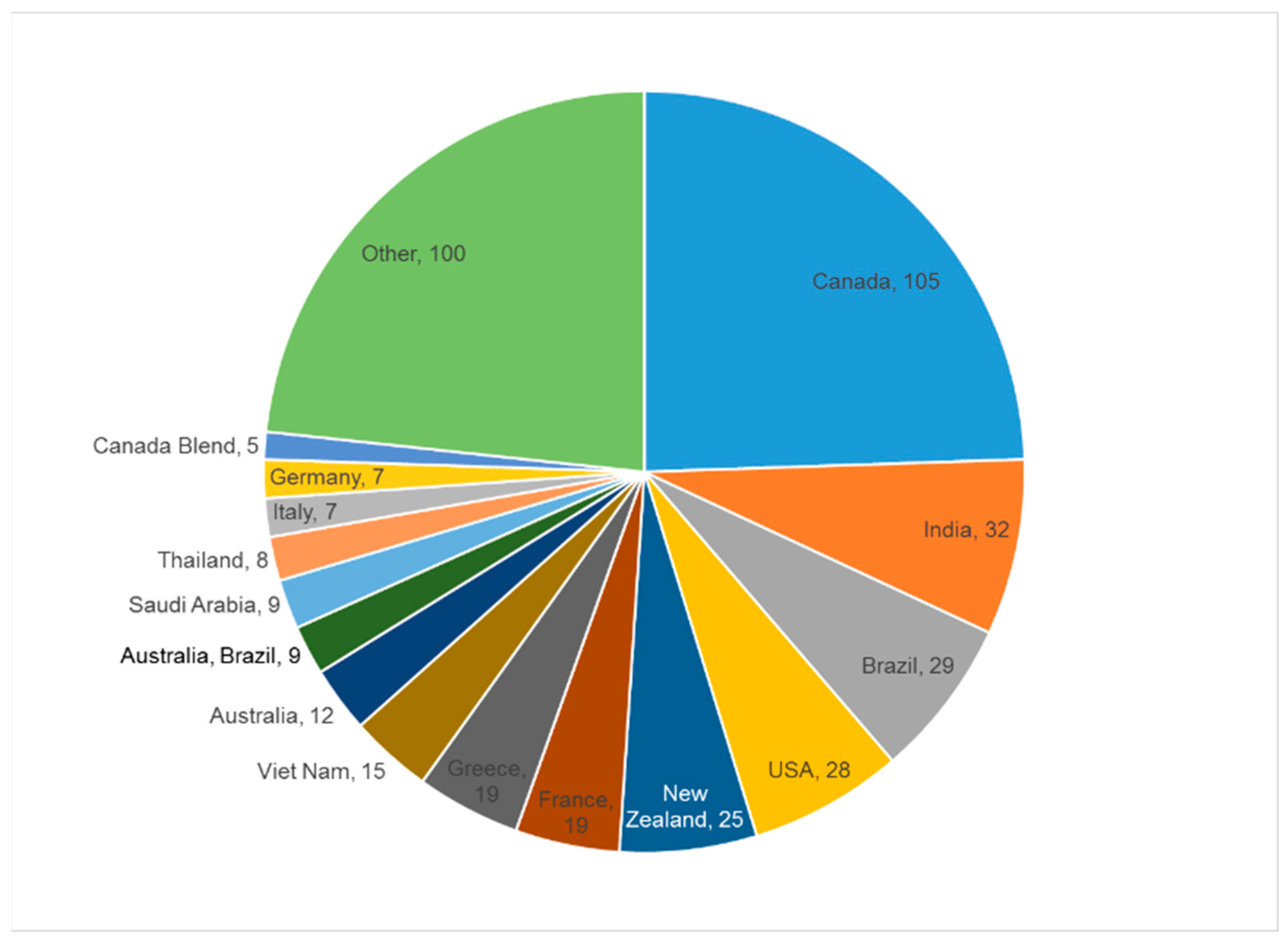
1. Introduction
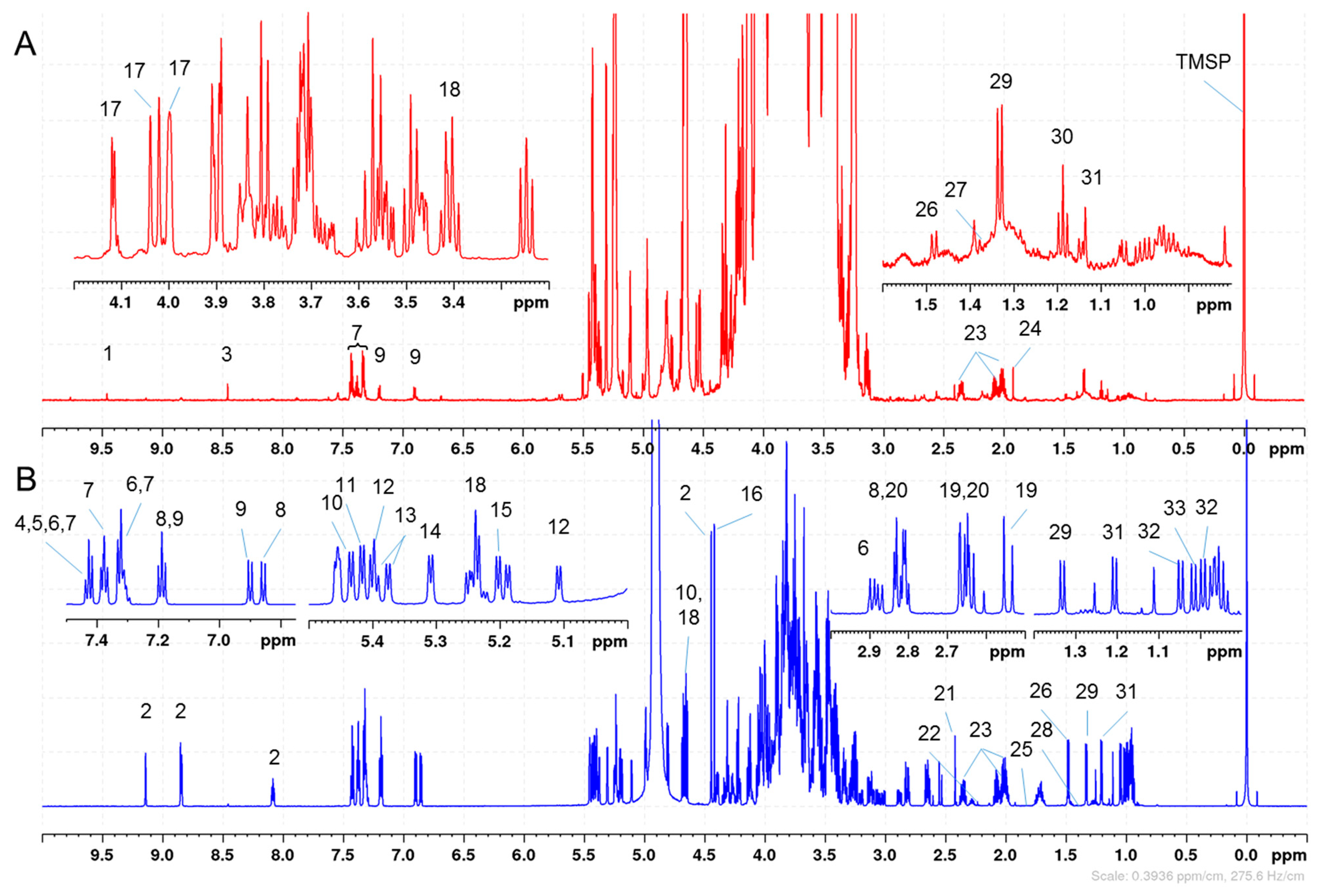
2. Results and Discussion
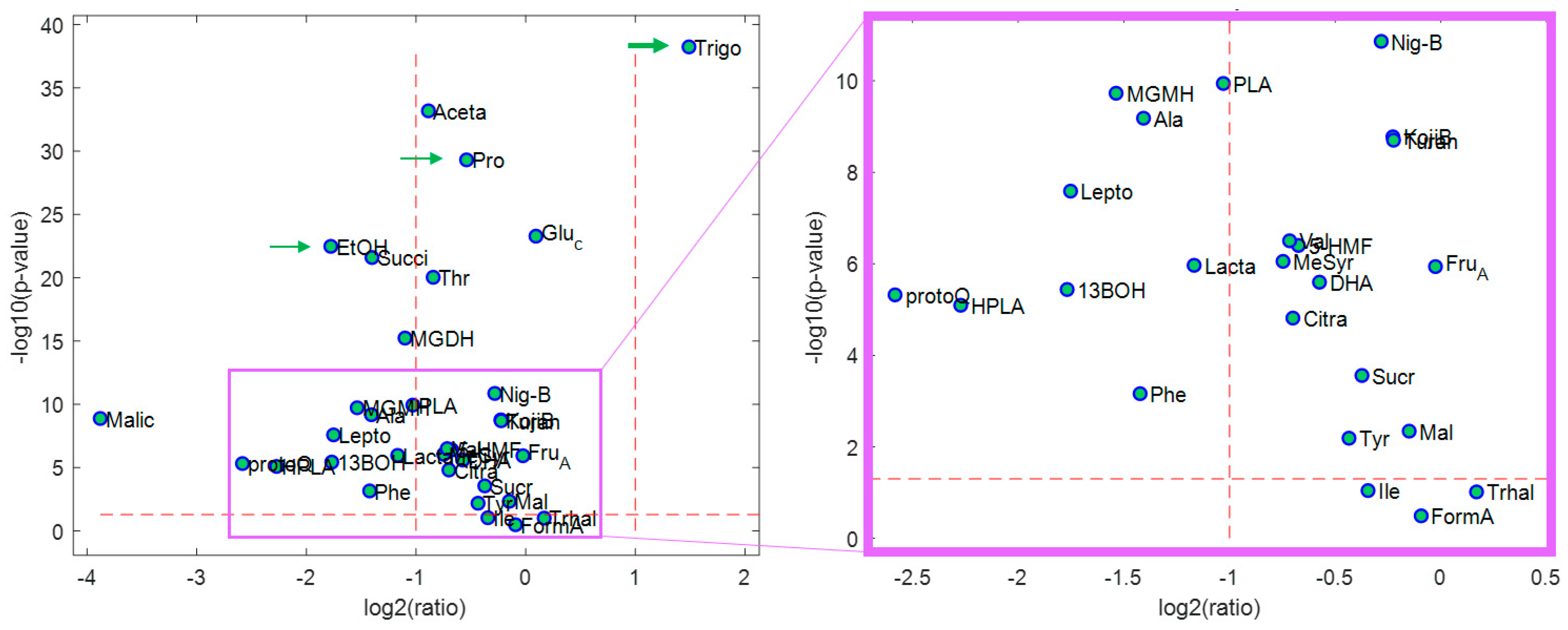
2.1. Quantitation of 33 Honey Components
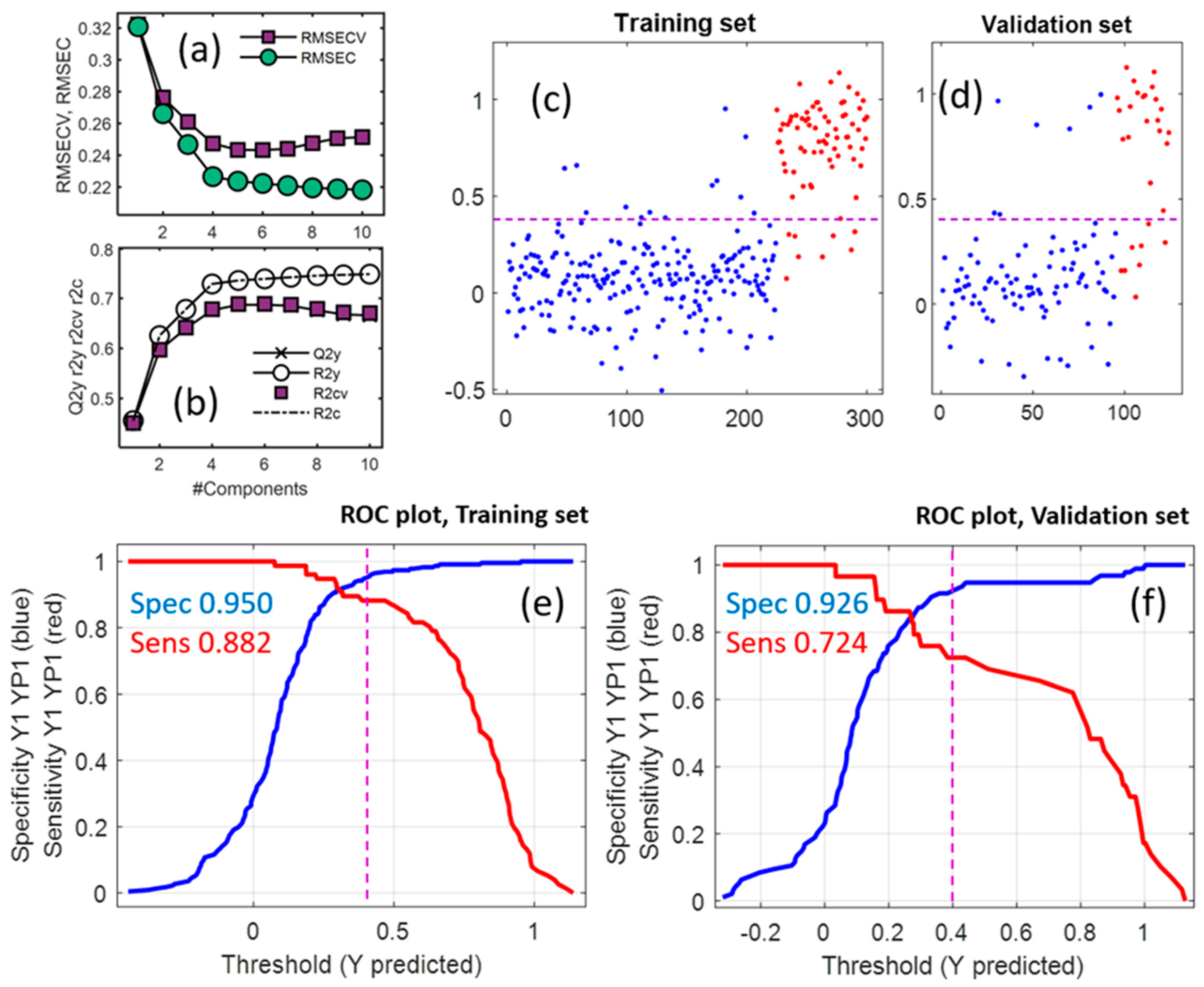
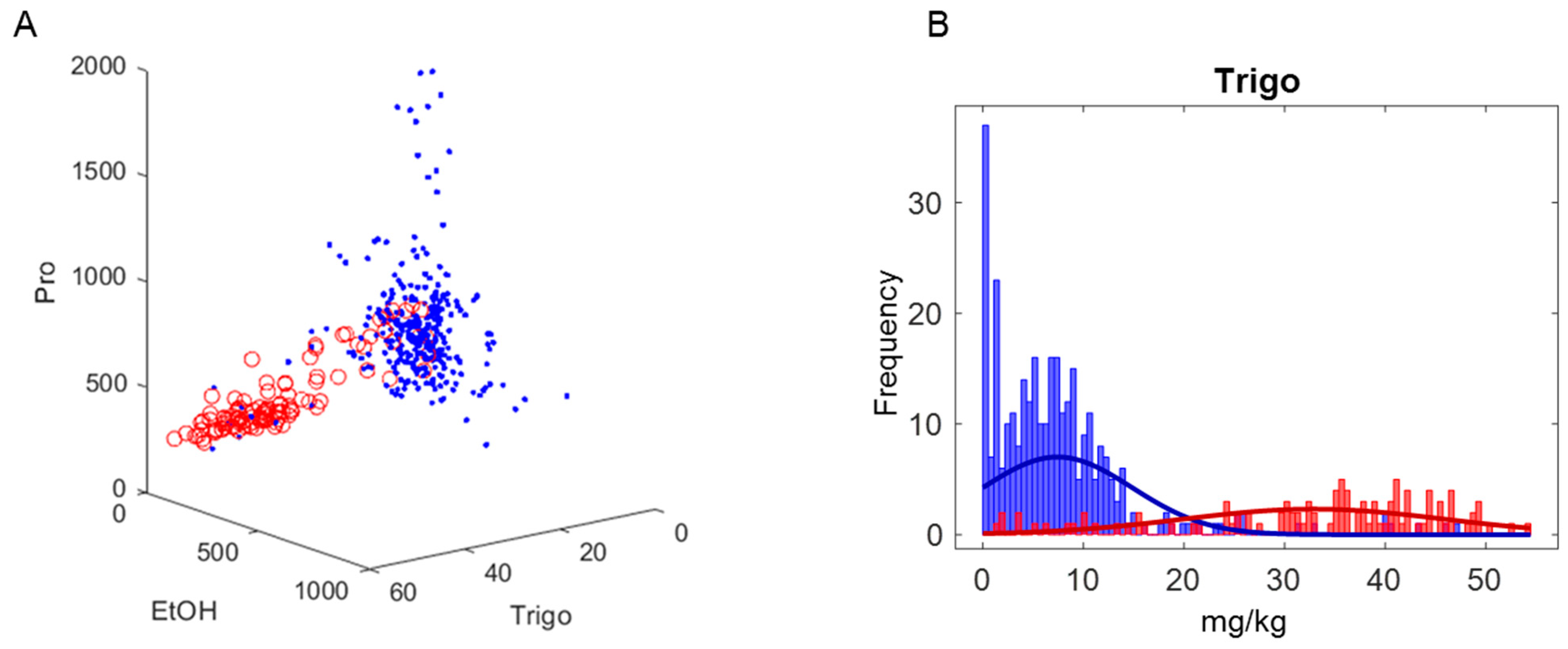
2.2. Multivariate Statistical Results
2.2.1. Principal Component Analysis
2.2.2. PLS-DA on all Variables
2.2.3. PLS-DA on Selected Variables
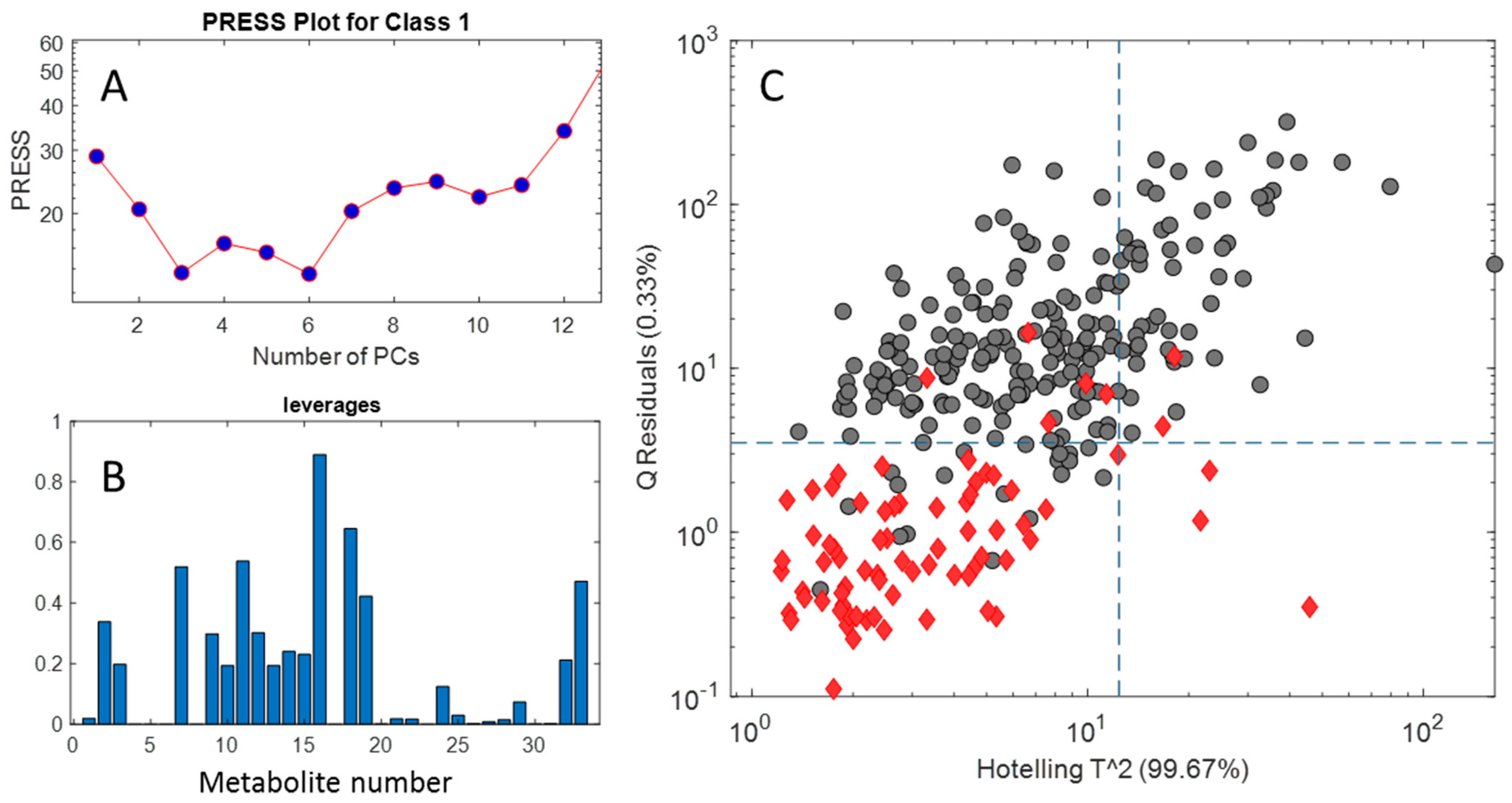
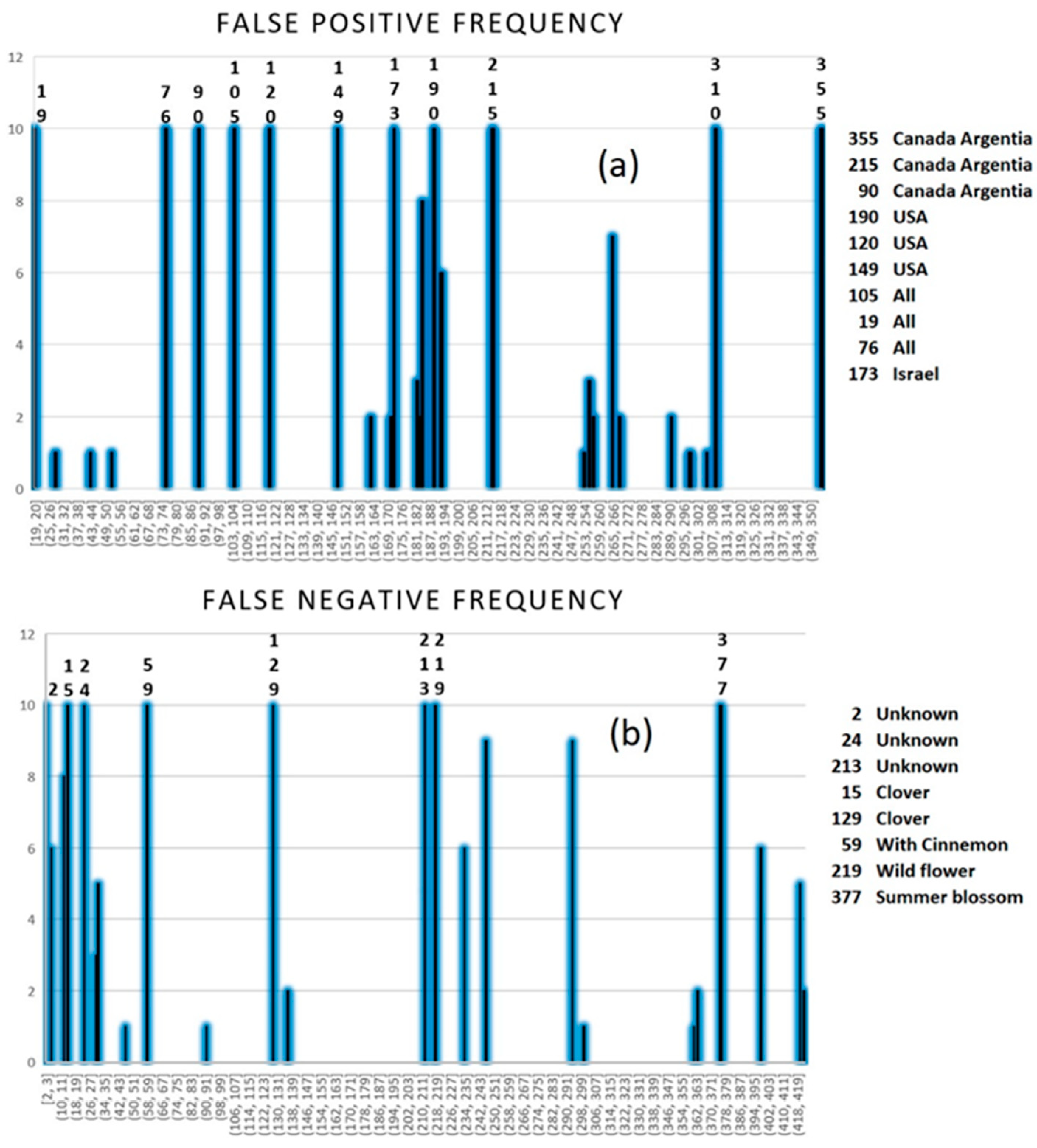
2.2.4. SIMCA Results
3. Materials and Methods
3.1. Sample Preparation
3.2. NMR Spectra
3.3. Peak areas and Concentrations
3.4. Chemometrics Analyses
3.4.1. PLS
3.4.2. SIMCA
3.4.3. Replacement Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cokcetin, N.N.; Pappalardo, M.; Campbell, L.T.; Brooks, P.; Carter, D.A.; Blair, S.E.; Harry, E.J. The Antibacterial Activity of Australian Leptospermum Honey Correlates with Methylglyoxal Levels. PLoS ONE 2016, 11, e0167780. [Google Scholar] [CrossRef]
- Alvarez-Suarez, J.M.; Gasparrini, M.; Forbes-Hernández, T.Y.; Mazzoni, L.; Giampieri, F. The Composition and Biological Activity of Honey: A Focus on Manuka Honey. Foods 2014, 3, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Cianciosi, D.; Forbes-Hernández, T.Y.; Afrin, S.; Gasparrini, M.; Reboredo-Rodriguez, P.; Manna, P.P.; Zhang, J.; Bravo Lamas, L.; Martínez Flórez, S.; Agudo Toyos, P.; et al. Phenolic Compounds in Honey and Their Associated Health Benefits: A Review. Molecules 2018, 23, 2322. [Google Scholar] [CrossRef] [PubMed]
- Spiteri, M.; Rogers, K.M.; Jamin, E.; Thomas, F.; Guyader, S.; Lees, M.; Rutledge, D.N. Combination of 1H NMR and chemometrics to discriminate manuka honey from other floral honey types from Oceania. Food Chem. 2017, 217, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.M.; Miguel, M.G.; Vilas-Boas, M.; Figueiredo, A.C. Honey Volatiles as a Fingerprint for Botanical Origin—A Review on their Occurrence on Monofloral Honeys. Molecules 2020, 25, 374. [Google Scholar] [CrossRef]
- Fakhlaei, R.; Selamat, J.; Khatib, A.; Razis, A.F.; Sukor, R.; Ahmad, S.; Babadi, A.A. The Toxic Impact of Honey Adulteration: A Review. Foods 2020, 9, 1538. [Google Scholar] [CrossRef]
- Burton, I.W.; Martinez Farina, C.F.; Ragupathy, S.; Arunachalam, T.; Newmaster, S.; Berrue, F. Quantitative NMR Methodology for the Authentication of Roasted Coffee and Prediction of Blends. J. Agric Food Chem. 2020, 68, 14643–14651. [Google Scholar] [CrossRef]
- Martinez-Farina, C.; Driscoll, S.; Wicks, C.; Burton, I.; Wentzell, P.; Berrué, F. Chemical Barcoding: A Nuclear-Magnetic-Resonance-Based Approach To Ensure the Quality and Safety of Natural Ingredients. J. Agric. Food Chem. 2019, 67. [Google Scholar] [CrossRef]
- Dong, H.; Xiao, K.; Xian, Y.; Wu, Y. Authenticity determination of honeys with non-extractable proteins by means of elemental analyzer (EA) and liquid chromatography (LC) coupled to isotope ratio mass spectroscopy (IRMS). Food Chem. 2018, 240, 717–724. [Google Scholar] [CrossRef]
- Zhou, X.; Taylor, M.P.; Salouros, H.; Prasad, S. Authenticity and geographic origin of global honeys determined using carbon isotope ratios and trace elements. Sci. Rep. 2018, 8, 14639. [Google Scholar] [CrossRef]
- Qiu, F.; Imai, A.; McAlpine, J.; Lankin, D.; Burton, I.; Karakach, T.; Farnsworth, N.; Chen, S.-N.; Pauli, G. Dereplication, Residual Complexity, and Rational Naming: The Case of the Actaea Triterpenes. J. Nat. Prod. 2012, 75, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Weljie, A.M.; Newton, J.; Jirik, F.R.; Vogel, H.J. Evaluating Low-Intensity Unknown Signals in Quantitative Proton NMR Mixture Analysis. Anal. Chem. 2008, 80, 8956–8965. [Google Scholar] [CrossRef] [PubMed]
- Burton, I.W.; Quilliam, M.A.; Walter, J.A. Quantitative 1H NMR with external standards: Use in preparation of calibration solutions for algal toxins and other natural products. Anal. Chem. 2005, 77, 3123–3131. [Google Scholar] [CrossRef] [PubMed]
- Girelli, C.R.; Schiavone, R.; Vilella, S.; Fanizzi, F.P. Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting. Sustainability 2020, 12, 5009. [Google Scholar] [CrossRef]
- Bertelli, D.; Lolli, M.; Papotti, G.; Bortolotti, L.; Serra, G.; Plessi, M. Detection of Honey Adulteration by Sugar Syrups Using One-Dimensional and Two-Dimensional High-Resolution Nuclear Magnetic Resonance. J. Agric. Food Chem. 2010, 58, 8495–8501. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.; Cogliati, C. NMR Characterization of Saccharides in Italian Honeys of Different Floral Sources. J. Agric. Food Chem. 2012, 60, 4526–4534. [Google Scholar] [CrossRef] [PubMed]
- Lolli, M.; Bertelli, D.; Plessi, M.; Sabatini, A.; Restani, C. Classification of Italian honeys by 2D HR-NMR. J. Agric. Food Chem. 2008, 56, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Ohmenhaeuser, M.; Monakhova, Y.B.; Kuballa, T.; Lachenmeier, D.W. Qualitative and Quantitative Control of Honeys Using NMR Spectroscopy and Chemometrics. ISRN Anal. Chem. 2013, 2013, 825318. [Google Scholar] [CrossRef]
- Schievano, E.; Stocchero, M.; Morelato, E.; Facchin, C.; Mammi, S. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 2011, 8, 679–690. [Google Scholar] [CrossRef]
- CFIA. Government of Canada Protects Canadians against Food Fraud in Honey and Other Products 2020. Available online: https://www.newswire.ca/news-releases/government-of-canada-protects-canadians-against-food-fraud-in-honey-and-other-products-870471000.html (accessed on 15 October 2022).
- CFIA. Report: Enhanced Honey Authenticity Surveillance (2018 to 2019). 2019. Available online: https://inspection.canada.ca/science-and-research/our-research-and-publications/report/eng/1557531883418/1557531883647 (accessed on 15 October 2022).
- Schievano, E.; Tonoli, M.; Rastrelli, F. NMR Quantification of Carbohydrates in Complex Mixtures. A Challenge on Honey. Anal. Chem. 2017, 89, 13405–13414. [Google Scholar] [CrossRef]
- Biswas, Z.; Merkley, N.; Syvitski, R.T. Biomolecular sample considerations essential for optimal performance from cryogenic probes. Metabolomics 2014, 10, 607–615. [Google Scholar] [CrossRef]
- Spiteri, M.; Jamin, E.; Thomas, F.; Rebours, A.; Lees, M.; Rogers, K.M.; Rutledge, D.N. Fast and global authenticity screening of honey using (1)H-NMR profiling. Food Chem. 2015, 189, 60–66. [Google Scholar] [CrossRef]
- Berregi, I.; del Campo, G.; Caracena, R.; Miranda, J.I. Quantitative determination of formic acid in apple juices by 1H NMR spectrometry. Talanta 2007, 72, 1049–1053. [Google Scholar] [CrossRef]
- Babij, N.R.; McCusker, E.O.; Whiteker, G.T.; Canturk, B.; Choy, N.; Creemer, L.C.; Amicis, C.V.D.; Hewlett, N.M.; Johnson, P.L.; Knobelsdorf, J.A.; et al. NMR Chemical Shifts of Trace Impurities: Industrially Preferred Solvents Used in Process and Green Chemistry. Org. Process Res. Dev. 2016, 20, 661–667. [Google Scholar] [CrossRef]
- Donarski, J.A.; Roberts, D.P.T.; Charlton, A.J. Quantitative NMR spectroscopy for the rapid measurement of methylglyoxal in manuka honey. Anal. Methods 2010, 2, 1479–1483. [Google Scholar] [CrossRef]
- Barclay, T.; Ginic-Markovic, M.; Johnston, M.R.; Cooper, P.; Petrovsky, N. Observation of the keto tautomer of D-fructose in D2O using 1H NMR spectroscopy. Carbohydr. Res. 2012, 347, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Boffo, E.; Tavares, L.; Tobias, A.; Ferreira, M.; Ferreira, A. Identification of components of Brazilian honey by 1H NMR and classification of its botanical origin by chemometric methods. LWT Food Sci. Technol. 2012, 49, 55–63. [Google Scholar] [CrossRef]
- Wise, B.M.; Gallagher, N.B.; Bro, R.; Shaver, J.M.; Windig, W.; Koch, R.S. PLS_Toolbox Version 4.0 for use with MATLAB; Eigenvector Research Inc.: Wenatchee, WA, USA, 2006. [Google Scholar]
- Ballabio, D.; Consonni, V. Classification tools in chemistry Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Shahbazy, M.; Moradi, P.; Ertaylan, G.; Zahraei, A.; Kompany-Zareh, M. FTICR mass spectrometry-based multivariate analysis to explore distinctive metabolites and metabolic pathways: A comprehensive bioanalytical strategy toward time-course metabolic profiling of Thymus vulgaris plants responding to drought stress. Plant Sci. 2020, 290, 110257. [Google Scholar] [CrossRef]
- Sjödin, K.; Schroeder, L.M.; Eidmann, H.H.; Norin, T.; Wold, S. Attack rates of scolytids and composition of volatile wood constituents in healthy and mechanically weakened pine trees. Scand. J. For. Res. 1989, 4, 379–391. [Google Scholar] [CrossRef]
- Wold, S. Pattern recognition by means of disjoint principal components models. Pattern Recognit. 1976, 8, 127–139. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, D.; Grisoni, F.; Todeschini, R. Multivariate comparison of classification performance measures. Chemom. Intell. Lab. Syst. 2018, 174, 33–44. [Google Scholar] [CrossRef]
- Pomerantsev, A.L. Acceptance areas for multivariate classification derived by projection methods. J. Chemom. 2008, 22, 601–609. [Google Scholar] [CrossRef]
- Bayat, M.; Kompany-Zareh, M. Non-rotational Tucker3 core simplification. J. Chemom. 2016, 30, 336–345. [Google Scholar] [CrossRef]
- Duchowicz, P.R.; Castro, E.A.; Fernández, F.M. Alternative algorithm for the search of an optimal set of descriptors in QSAR-QSPR studies. MATCH Commun. Math. Comput. Chem. 2006, 55, 179–192. [Google Scholar]
- Duchowicz, P.R.; Bennardi, D.O.; Bacelo, D.E.; Bonifazi, E.L.; Rios-Luci, C.; Padrón, J.M.; Burton, G.; Misico, R.I. QSAR on antiproliferative naphthoquinones based on a conformation-independent approach. Eur. J. Med. Chem. 2014, 77, 176–184. [Google Scholar] [CrossRef]
- Goodarzi, M.; Bacelo, D.E.; Fioressi, S.E.; Duchowicz, P.R. Replacement Orthogonal Wavelengths Selection as a new method for multivariate calibration in spectroscopy. Microchem. J. 2019, 145, 872–882. [Google Scholar] [CrossRef]
- Leardi, R.; Lupiáñez González, A. Genetic algorithms applied to feature selection in PLS regression: How and when to use them. Chemom. Intell. Lab. Syst. 1998, 41, 195–207. [Google Scholar] [CrossRef]

| Training Set | Predicted Class | |||||
| PLS-DA on All | PLS-DA on Selected | SIMCA | ||||
| True Class | Canadian | Non-Canadian | Canadian | Non-Canadian | Canadian | Non-Canadian |
| Canadian | 67 88.2% | 9 11.8% | 59 86.8% | 9 13.2% | 60 88.2% | 8 11.8% |
| Non-Canadian | 11 5.0% | 213 95.0% | 11 4.7% | 221 95.3% | 33 14.2% | 199 85.8% |
| Validation Set | Predicted Class | |||||
| PLS-DA on All | PLS-DA on Selected | SIMCA | ||||
| True Class | Canadian | Non-Canadian | Canadian | Non-Canadian | Canadian | Non-Canadian |
| Canadian | 21 72.4% | 8 27.6% | 29 78.4% | 8 21.6% | 31 83.8% | 6 16.2% |
| Non-Canadian | 7 7.4% | 88 92.6% | 4 4.6% | 83 95.4% | 8 9.2% | 79 90.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burton, I.W.; Kompany-Zareh, M.; Haverstock, S.; Haché, J.; Martinez-Farina, C.F.; Wentzell, P.D.; Berrué, F. Analysis and Discrimination of Canadian Honey Using Quantitative NMR and Multivariate Statistical Methods. Molecules 2023, 28, 1656. https://doi.org/10.3390/molecules28041656
Burton IW, Kompany-Zareh M, Haverstock S, Haché J, Martinez-Farina CF, Wentzell PD, Berrué F. Analysis and Discrimination of Canadian Honey Using Quantitative NMR and Multivariate Statistical Methods. Molecules. 2023; 28(4):1656. https://doi.org/10.3390/molecules28041656
Chicago/Turabian StyleBurton, Ian W., Mohsen Kompany-Zareh, Sophie Haverstock, Jonathan Haché, Camilo F. Martinez-Farina, Peter D. Wentzell, and Fabrice Berrué. 2023. "Analysis and Discrimination of Canadian Honey Using Quantitative NMR and Multivariate Statistical Methods" Molecules 28, no. 4: 1656. https://doi.org/10.3390/molecules28041656
APA StyleBurton, I. W., Kompany-Zareh, M., Haverstock, S., Haché, J., Martinez-Farina, C. F., Wentzell, P. D., & Berrué, F. (2023). Analysis and Discrimination of Canadian Honey Using Quantitative NMR and Multivariate Statistical Methods. Molecules, 28(4), 1656. https://doi.org/10.3390/molecules28041656







