A Versatile Continuous Fluorometric Enzymatic Assay for Targeting Nicotinate Phosphoribosyltransferase
Abstract
1. Introduction
2. Results and Discussion
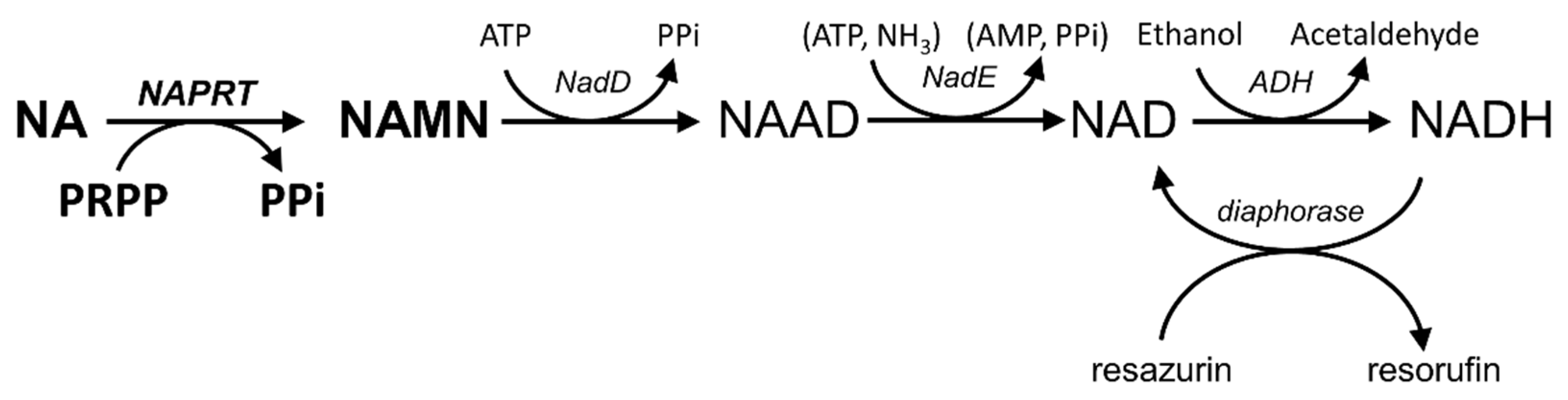
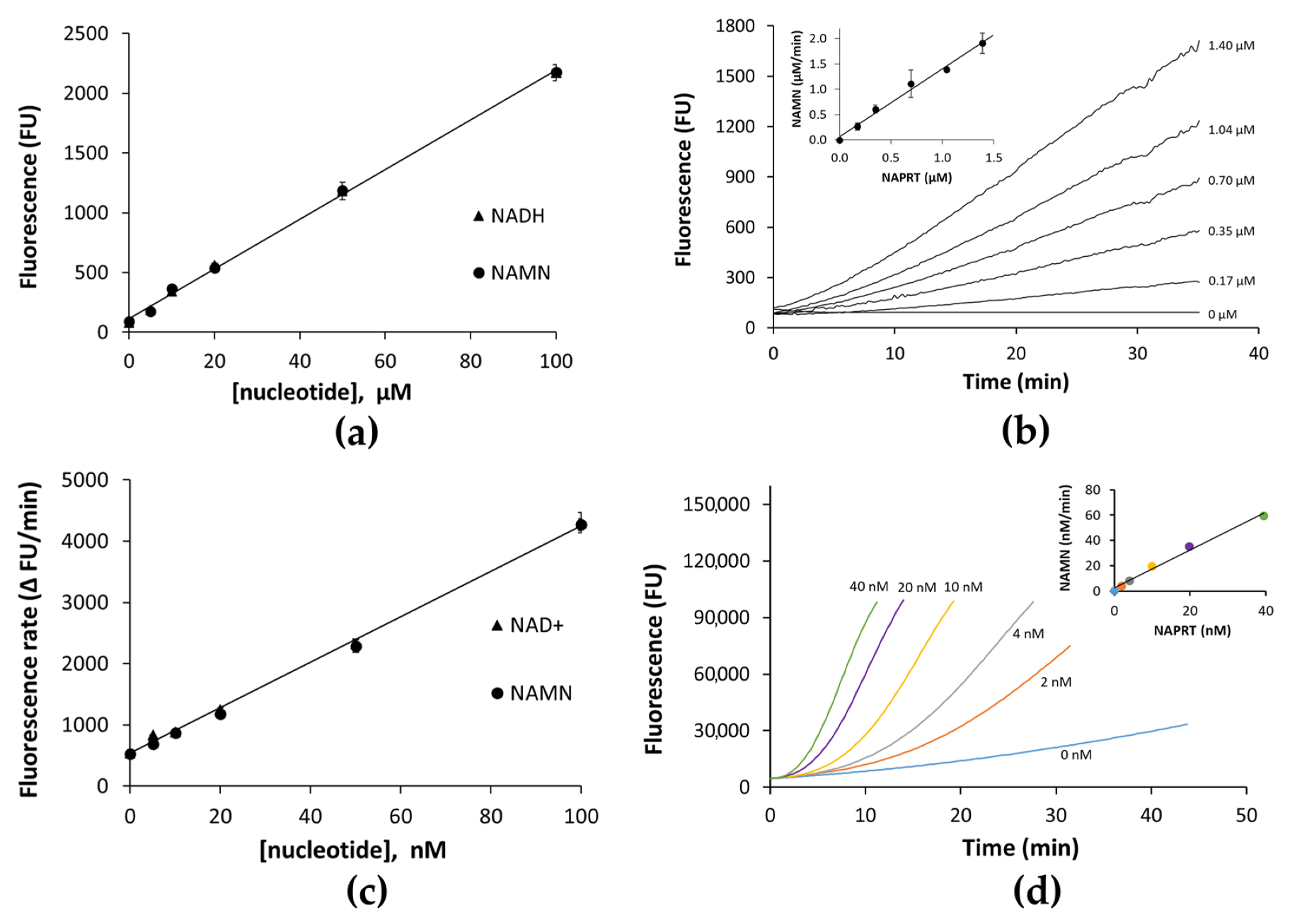
2.1. AssayDevelopment and Optimization
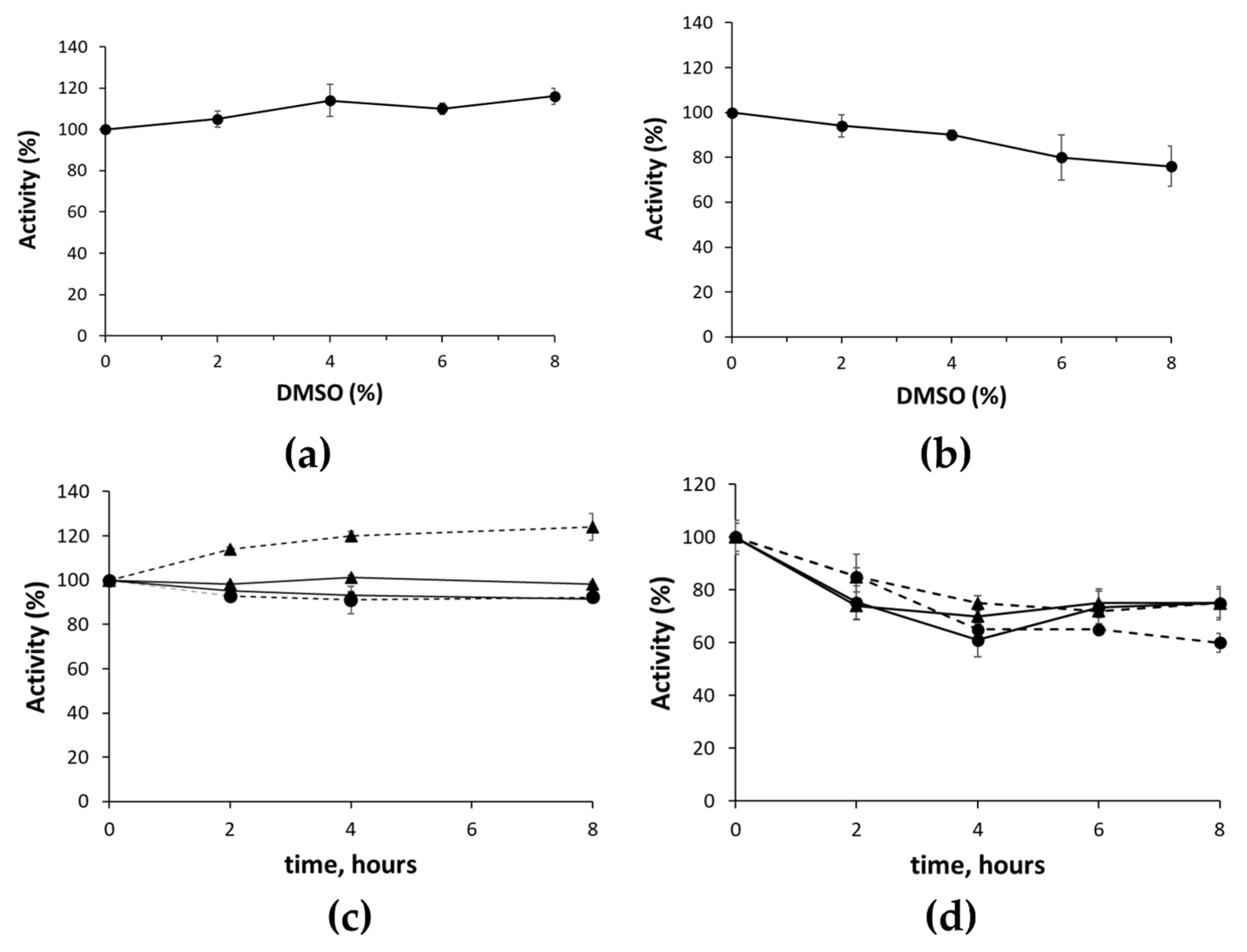
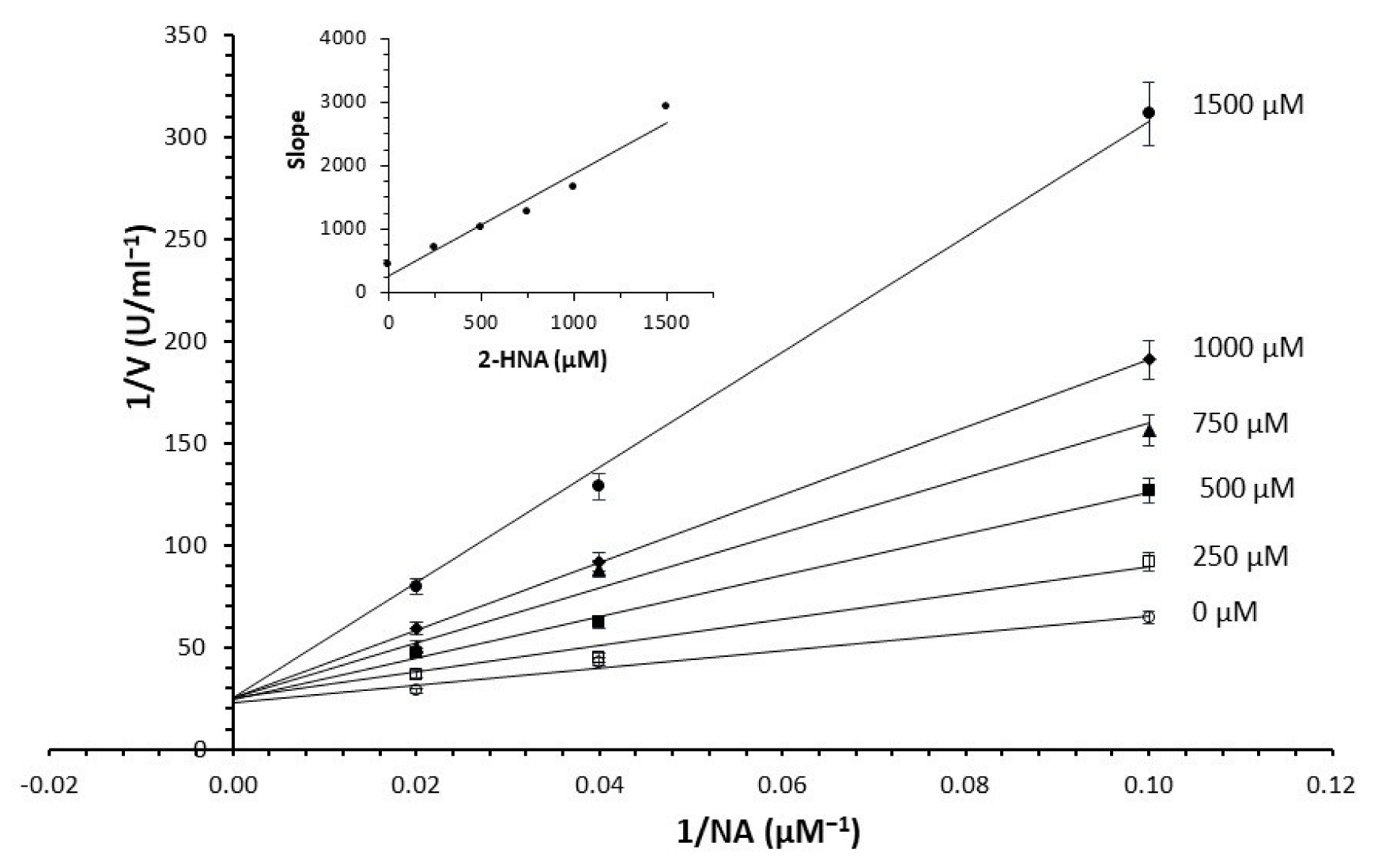
2.2. Assay’s Validation
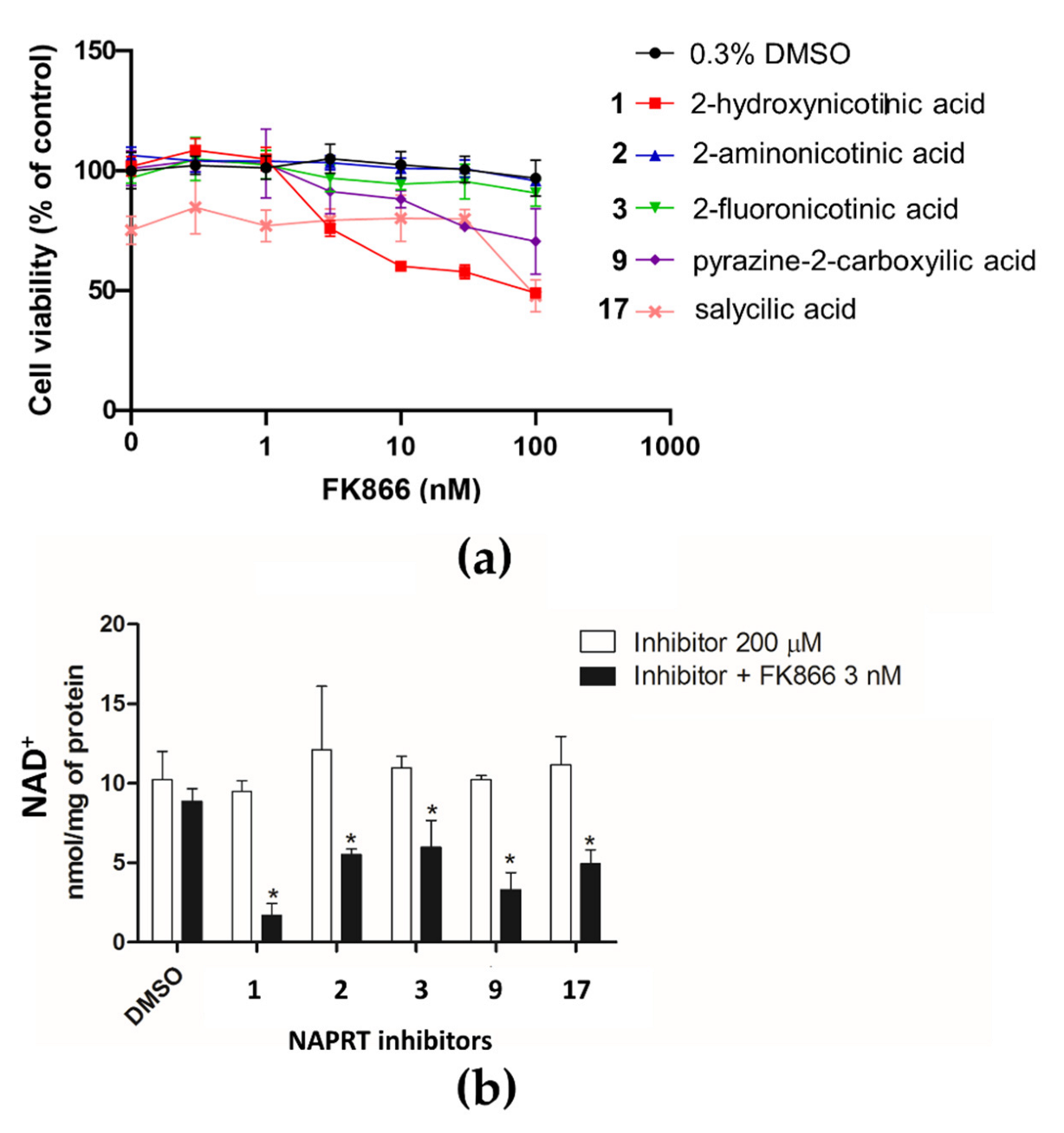
2.3. Effect of NAPRT Inhibitors on OVCAR-5 Cells
3. Materials and Methods
3.1. NAPRT Expression and Purification
3.2. NAPRT Activity Assays
3.2.1. NADH Assay
3.2.2. Resazurin/Diaphorase-Coupled Assay
3.2.3. HPLC Assay
3.3. Inhibitor Screening
3.4. Kinetic Analyses
3.5. Cell Viability Assay
3.6. NAD+ Levels Measurement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ruggieri, S.; Orsomando, G.; Sorci, L.; Raffaelli, N. Regulation of NAD biosynthetic enzymes modulates NAD-sensing processes to shape mammalian cell physiology under varying biological cues. Biochim. et Biophys. Acta (BBA) Proteins Proteom. 2015, 1854, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Piacente, F.; Caffa, I.; Ravera, S.; Sociali, G.; Passalacqua, M.; Vellone, V.G.; Becherini, P.; Reverberi, D.; Monacelli, F.; Ballestrero, A.; et al. Nicotinic Acid Phosphoribosyltransferase Regulates Cancer Cell Metabolism, Susceptibility to NAMPT Inhibitors, and DNA Repair. Cancer Res 2017, 77, 3857–3869. [Google Scholar] [CrossRef]
- Li, X.-Q.; Lei, J.; Mao, L.-H.; Wang, Q.-L.; Xu, F.; Ran, T.; Zhou, Z.-H.; He, S. NAMPT and NAPRT, Key Enzymes in NAD Salvage Synthesis Pathway, Are of Negative Prognostic Value in Colorectal Cancer. Front. Oncol. 2019, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Chowdhry, S.; Zanca, C.; Rajkumar, U.; Koga, T.; Diao, Y.; Raviram, R.; Liu, F.; Turner, K.; Yang, H.; Brunk, E.; et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 2019, 569, 570–575. [Google Scholar] [CrossRef]
- Zamporlini, F.; Ruggieri, S.; Mazzola, F.; Amici, A.; Orsomando, G.; Raffaelli, N. Novel assay for simultaneous measurement of pyridine mononucleotides synthesizing activities allows dissection of the NAD+biosynthetic machinery in mammalian cells. FEBS J. 2014, 281, 5104–5119. [Google Scholar] [CrossRef]
- Managò, A.; Audrito, V.; Mazzola, F.; Sorci, L.; Gaudino, F.; Gizzi, K.; Vitale, N.; Incarnato, D.; Minazzato, G.; Ianniello, A.; et al. Extracellular nicotinate phosphoribosyltransferase binds Toll like receptor 4 and mediates inflammation. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Gasparrini, M.; Audrito, V. NAMPT: A critical driver and therapeutic target for cancer. Int. J. Biochem. Cell Biol. 2022, 145, 106189. [Google Scholar] [CrossRef]
- Galli, U.; Colombo, G.; Travelli, C.; Tron, G.C.; Genazzani, A.A.; Grolla, A. Recent Advances in NAMPT Inhibitors: A Novel Immunotherapic Strategy. Front. Pharmacol. 2020, 11, 656. [Google Scholar] [CrossRef]
- Grozio, A.; Sociali, G.; Sturla, L.; Caffa, I.; Soncini, D.; Salis, A.; Raffaelli, N.; De Flora, A.; Nencioni, A.; Bruzzone, S. CD73 Protein as a Source of Extracellular Precursors for Sustained NAD+ Biosynthesis in FK866-treated Tumor Cells. J. Biol. Chem. 2013, 288, 25938–25949. [Google Scholar] [CrossRef]
- Cerna, D.; Li, H.; Flaherty, S.; Takebe, N.; Coleman, C.N.; Yoo, S.S. Inhibition of Nicotinamide Phosphoribosyltransferase (NAMPT) Activity by Small Molecule GMX1778 Regulates Reactive Oxygen Species (ROS)-mediated Cytotoxicity in a p53- and Nicotinic Acid Phosphoribosyltransferase1 (NAPRT1)-dependent Manner. J. Biol. Chem. 2012, 287, 22408–22417. [Google Scholar] [CrossRef]
- Fons, N.R.; Sundaram, R.K.; Breuer, G.A.; Peng, S.; McLean, R.L.; Kalathil, A.N.; Schmidt, M.S.; Carvalho, D.M.; Mackay, A.; Jones, C.; et al. PPM1D mutations silence NAPRT gene expression and confer NAMPT inhibitor sensitivity in glioma. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.; Lee, J.E.; Shin, S.-J.; Oh, S.; Kwon, S.; Kim, H.; Choi, Y.Y.; White, M.A.; Paik, S.; et al. Selective Cytotoxicity of the NAMPT Inhibitor FK866 Toward Gastric Cancer Cells With Markers of the Epithelial-Mesenchymal Transition, Due to Loss of NAPRT. Gastroenterology 2018, 155, 799–814.e13. [Google Scholar] [CrossRef]
- Shames, D.S.; Elkins, K.; Walter, K.; Holcomb, T.; Du, P.; Mohl, D.; Xiao, Y.; Pham, T.; Haverty, P.M.; Liederer, B.; et al. Loss of NAPRT1 Expression by Tumor-Specific Promoter Methylation Provides a Novel Predictive Biomarker for NAMPT Inhibitors. Clin. Cancer Res. 2013, 19, 6912–6923. [Google Scholar] [CrossRef]
- Conlon, N.; Ford, D. A systems-approach to NAD+ restoration. Biochem. Pharmacol. 2022, 198, 114946. [Google Scholar] [CrossRef]
- Gardell, S.J.; Hopf, M.; Khan, A.; Dispagna, M.; Sessions, E.H.; Falter, R.; Kapoor, N.; Brooks, J.; Culver, J.; Petucci, C.; et al. Boosting NAD+ with a small molecule that activates NAMPT. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Liu, M.; Wang, L.; Zu, Y.; Wu, C.; Li, C.; Zhang, R.; Lu, H.; Li, F.; Xi, S.; et al. Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity. Cell Res. 2022, 32, 570–584. [Google Scholar] [CrossRef]
- Hara, N.; Yamada, K.; Shibata, T.; Osago, H.; Hashimoto, T.; Tsuchiya, M. Elevation of Cellular NAD Levels by Nicotinic Acid and Involvement of Nicotinic Acid Phosphoribosyltransferase in Human Cells. J. Biol. Chem. 2007, 282, 24574–24582. [Google Scholar] [CrossRef]
- Gaut, Z.; Solomon, H. Inhibition of nicotinate phosphoribosyltransferase in human platelet lysate by nicotinic acid analogs. Biochem. Pharmacol. 1971, 20, 2903–2906. [Google Scholar] [CrossRef]
- Gaut, Z.; Solomon, H. Inhibition of Nicotinate Phosphoribosyl Transferase by Nonsteroidal Anti-Inflammatory Drugs: A Possible Mechanism of Action. J. Pharm. Sci. 1971, 60, 1887–1888. [Google Scholar] [CrossRef]
- Bruzzone, S.; De Flora, A.; Usai, C.; Graeff, R.; Lee, H.C. Cyclic ADP-ribose is a second messenger in the lipopolysaccharide-stimulated proliferation of human peripheral blood mononuclear cells. Biochem. J. 2003, 375, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Galassi, L.; Di Stefano, M.; Brunetti, L.; Orsomando, G.; Amici, A.; Ruggieri, S.; Magni, G. Characterization of human nicotinate phosphoribosyltransferase: Kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie 2012, 94, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Franco, J.; Piacente, F.; Walter, M.; Fratta, S.; Ghanem, M.; Benzi, A.; Caffa, I.; Kurkin, A.V.; Altieri, A.; Herr, P.; et al. Structure-Based Identification and Biological Characterization of New NAPRT Inhibitors. Pharmaceuticals 2022, 15, 855. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, M.S.; Caffa, I.; Del Rio, A.; Franco, J.; Parenti, M.D.; Monacelli, F.; Cea, M.; Khalifa, A.; Nahimana, A.; Duchosal, M.A.; et al. Identification of NAPRT Inhibitors with Anti-Cancer Properties by In Silico Drug Discovery. Pharmaceuticals 2022, 15, 848. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, Y.-M.; Li, G.-L. Redox-regulated synthesis of fluorescent polydopamine nanoparticles for detection of butyrylcholinesterase activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 274, 121097. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Simeonov, A.; Davis, M.I. Avoiding Fluorescence Assay Interference—The Case for Diaphorase. ASSAY Drug Dev. Technol. 2016, 14, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Niedel, J.; Dietrich, L. Nicotinate Phosphoribosyltransferase of Human Erythrocytes. J. Biol. Chem. 1973, 248, 3500–3505. [Google Scholar] [CrossRef]
- Smith, L.D.; Gholson, R.K. Allosteric Properties of Bovine Liver Nicotinate Phosphoribosyltransferase. J. Biol. Chem. 1969, 244, 68–71. [Google Scholar] [CrossRef]
- Segel, I. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; John Wiley and Sons: New York, NY, USA, 1993. [Google Scholar]
- Amici, A.; Grolla, A.A.; Del Grosso, E.; Bellini, R.; Bianchi, M.; Travelli, C.; Garavaglia, S.; Sorci, L.; Raffaelli, N.; Ruggieri, S.; et al. Synthesis and Degradation of Adenosine 5′-Tetraphosphate by Nicotinamide and Nicotinate Phosphoribosyltransferases. Cell Chem. Biol. 2017, 24, 553–564.e4. [Google Scholar] [CrossRef]
- Hasmann, M.; Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res 2003, 63, 7436–7442. [Google Scholar]
- Del Nagro, C.; Xiao, Y.; Rangell, L.; Reichelt, M.; O’Brien, T. Depletion of the Central Metabolite NAD Leads to Oncosis-mediated Cell Death. J. Biol. Chem. 2014, 289, 35182–35192. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
| ID | Structure | Compound | Inhibition (%) | IC50 (µM) | Ki (µM) Inhibition Mechanism |
|---|---|---|---|---|---|
| Nicotinic acid derivatives | |||||
| 1 |  | 2-hydroxynicotinic acid | 34 | 1214 ± 91 | 215 ± 5 competitive |
| 2 | 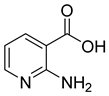 | 2-aminonicotinic acid | 38 | 609 ± 69 | 348 ± 36 mixed |
| 3 | 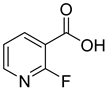 | 2-fluoronicotinic acid | 57 | 351 ± 38 | 149 ± 14 non-competitive |
| 4 |  | 6-chloronicotinic acid | 44 | 1259 ± 115 | 845 ± 69 non-competitive |
| 5 | 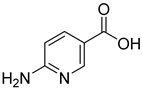 | 6-aminonicotinic acid | 19 | ||
| 6 | 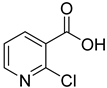 | 2-chloronicotinic acid | 12 | ||
| 7 |  | 2,3-pyridine dicarboxylic acid | 9 | ||
| 8 | 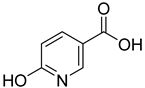 | 6-hydroxynicotinic acid | 0 | ||
| Pyrazine derivatives | |||||
| 9 |  | pyrazine-2-carboxylic acid | 68 | 365 ± 36 | 166 ± 0.14 non-competitive |
| 10 | 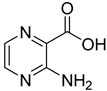 | 3-aminopyrazine-2-carboxylic acid | 6 | ||
| 11 |  | 2,3-pyrazine dicarboxylic acid | 0 | ||
| 12 |  | pyrazinamide | 0 | ||
| 13 | 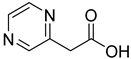 | 2-pyrazine acetic acid | 0 | ||
| 14 | 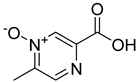 | Acipimox | 0 | ||
| Pyridazine derivatives | |||||
| 15 |  | 3-pyridazine carboxylic acid | 30 | 1370 ± 74 | 513 ± 13 mixed |
| 16 | 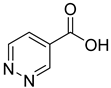 | 4-pyridazine carboxylic acid | 18 | ||
| Benzoic acid derivatives | |||||
| 17 | 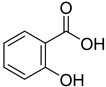 | salicylic acid | 58 | 351 ± 19 | 169 ± 15 non- competitive |
| 18 | 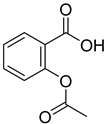 | acetyl salicylic acid | 11 | ||
| 19 | 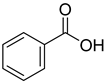 | benzoic acid | 8 | ||
| 20 | 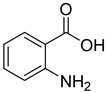 | anthranilic acid | 5 | ||
| 21 | 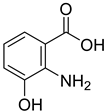 | 3-hydroxy anthranilic acid | 5 | ||
| 22 | 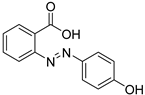 | 2-(4-hydroxyphenylazo) benzoic acid | 5 | ||
| 23 | 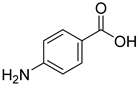 | 4-aminobenzoic acid | 0 | ||
| 24 | 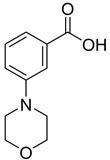 | 3-morpholinobenzoic acid | 0 | ||
| Carboxy pyridine analogues | |||||
| 25 | 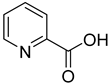 | 2-picolinic acid | 6 | ||
| 26 |  | 2,6-pyridine dicarboxylic acid | 6 | ||
| 27 | 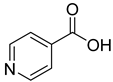 | isonicotinic acid | 0 | ||
| Pyrimidine derivatives | |||||
| 28 | 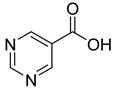 | 5-pyrimidine carboxylic acid | 20 | ||
| 29 |  | 2-pyrimidine carboxylic acid | 14 | ||
| 30 | 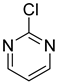 | 2-chloropyrimidine | 12 | ||
| 31 | 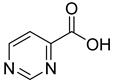 | 4-pyrimidine carboxylic acid | 8 | ||
| 32 |  | 2-hydroxypyrimidine | 0 | ||
| 33 | 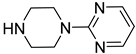 | 2-(1-piperazinyl)pyrimidine | 0 | ||
| Aliphatic carboxylic acids | |||||
| 34 | 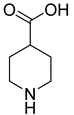 | piperidine-4-carboxylic acid | 0 | ||
| 35 | 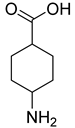 | 4-aminocyclohexane carboxylic acid | 0 | ||
| Other compounds | |||||
| 36 |  | pyrrole-2-carboxylic acid | 13 | ||
| 37 |  | acetanilide | 0 | ||
| Pyridine derivatives | |||||
| 38 |  | nicotinuric acid | 0 | ||
| 39 | 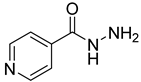 | isonicotinic acid hydrazide | 0 | ||
| 40 | 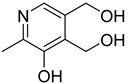 | pyridoxine | 0 | ||
| 41 | 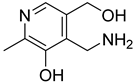 | pyridoxamine | 0 | ||
| 42 | 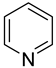 | pyridine | 0 | ||
| 43 | 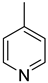 | 4-methylpyridine | 0 | ||
| 44 | 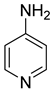 | 4-aminopyridine | 0 | ||
| 45 | 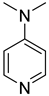 | 4-dimethylaminopyridine | 0 | ||
| 46 | 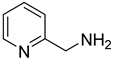 | 2-pyridinemethanamine | 0 | ||
| 47 | 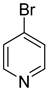 | 4-bromopyridine | 0 | ||
| 48 | 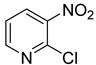 | 2-chloro-3-nitropyridine | 0 | ||
| 49 | 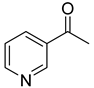 | 3-acetylpyridine | 0 | ||
| 50 | 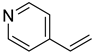 | 4-vinylpyridine | 0 | ||
| 51 | 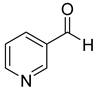 | 3-pyridinecarboxaldehyde | 0 | ||
| 52 | 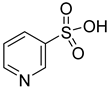 | 3-pyridinesulfonic acid | 0 | ||
| 53 | 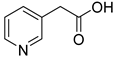 | 3-pyridineacetic acid | 0 | ||
| 54 | 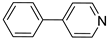 | 4-phenylpyridine | 0 | ||
| 55 | 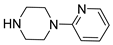 | 1-(2-pyridyl)piperazine | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minazzato, G.; Marangoni, E.; Fortunato, C.; Petrelli, R.; Cappellacci, L.; Del Bello, F.; Sorci, L.; Gasparrini, M.; Piacente, F.; Bruzzone, S.; et al. A Versatile Continuous Fluorometric Enzymatic Assay for Targeting Nicotinate Phosphoribosyltransferase. Molecules 2023, 28, 961. https://doi.org/10.3390/molecules28030961
Minazzato G, Marangoni E, Fortunato C, Petrelli R, Cappellacci L, Del Bello F, Sorci L, Gasparrini M, Piacente F, Bruzzone S, et al. A Versatile Continuous Fluorometric Enzymatic Assay for Targeting Nicotinate Phosphoribosyltransferase. Molecules. 2023; 28(3):961. https://doi.org/10.3390/molecules28030961
Chicago/Turabian StyleMinazzato, Gabriele, Elisa Marangoni, Carlo Fortunato, Riccardo Petrelli, Loredana Cappellacci, Fabio Del Bello, Leonardo Sorci, Massimiliano Gasparrini, Francesco Piacente, Santina Bruzzone, and et al. 2023. "A Versatile Continuous Fluorometric Enzymatic Assay for Targeting Nicotinate Phosphoribosyltransferase" Molecules 28, no. 3: 961. https://doi.org/10.3390/molecules28030961
APA StyleMinazzato, G., Marangoni, E., Fortunato, C., Petrelli, R., Cappellacci, L., Del Bello, F., Sorci, L., Gasparrini, M., Piacente, F., Bruzzone, S., & Raffaelli, N. (2023). A Versatile Continuous Fluorometric Enzymatic Assay for Targeting Nicotinate Phosphoribosyltransferase. Molecules, 28(3), 961. https://doi.org/10.3390/molecules28030961











