Two Antimicrobial Peptides Derived from Bacillus and Their Properties
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening of Bacteriostatic Bacillus spp. Strains
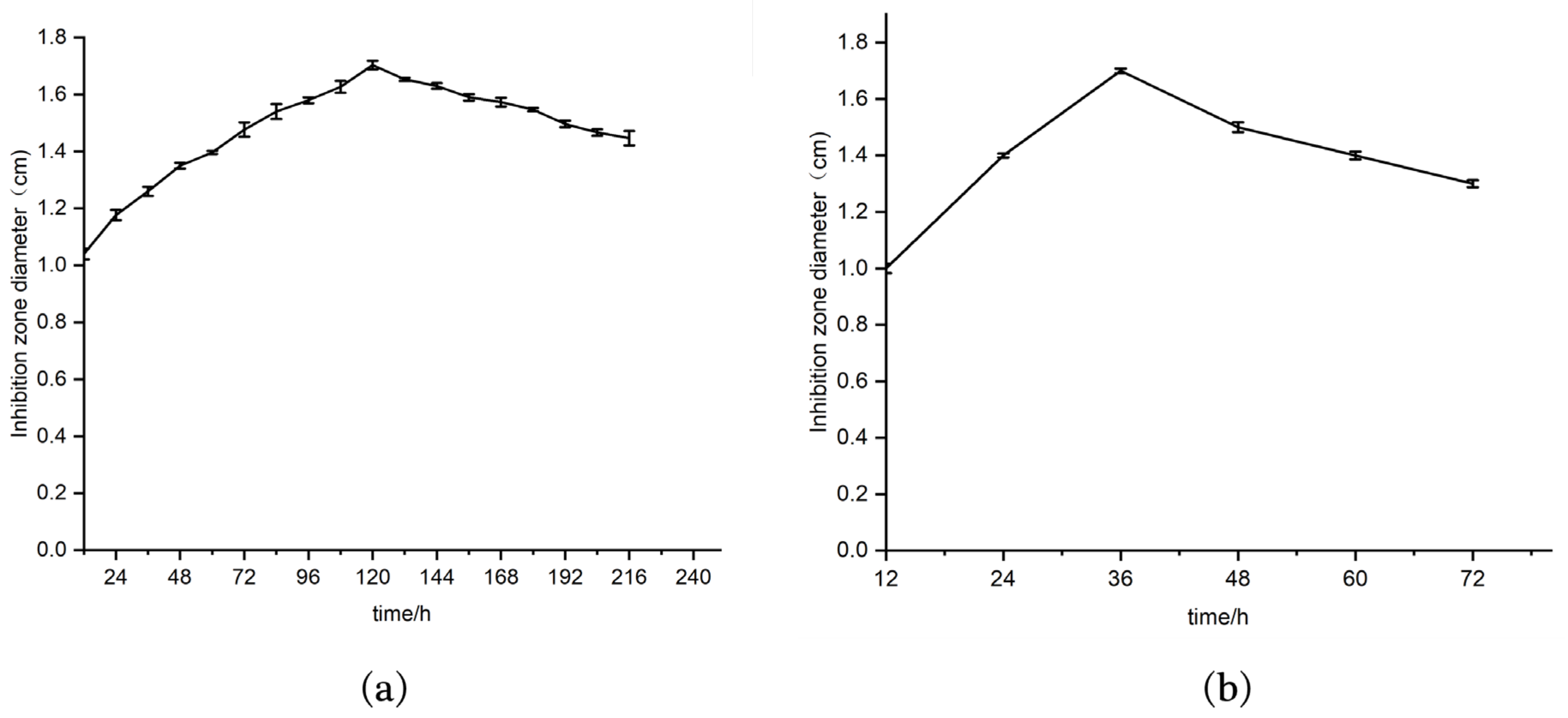
2.2. Determination of the Optimal Fermentation Time
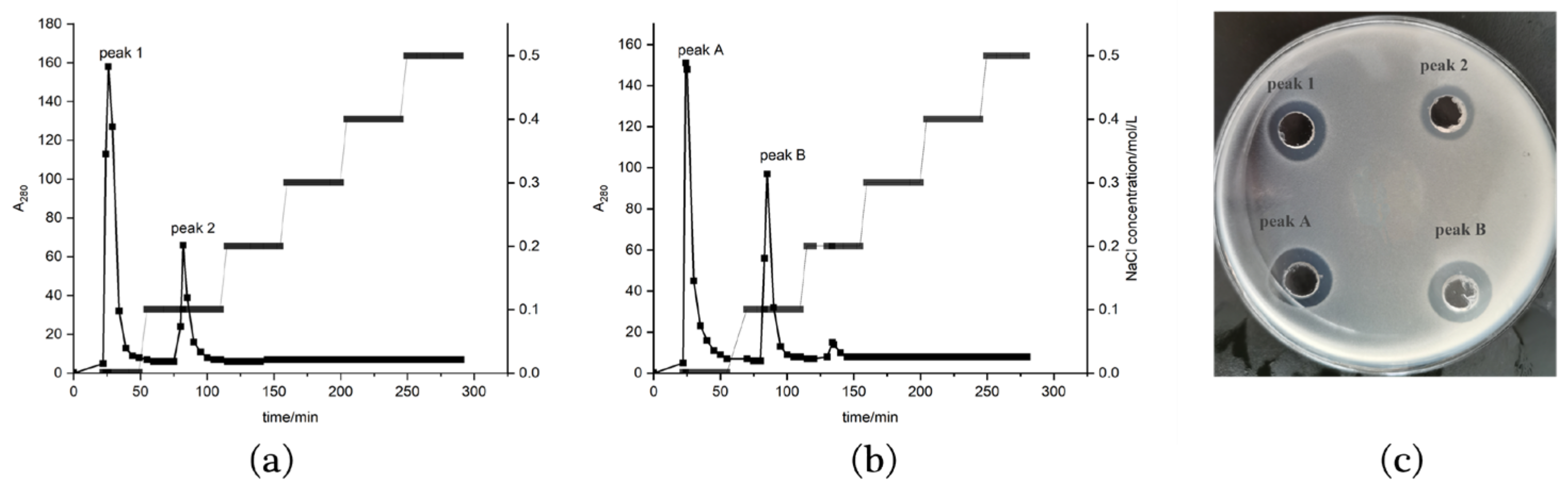
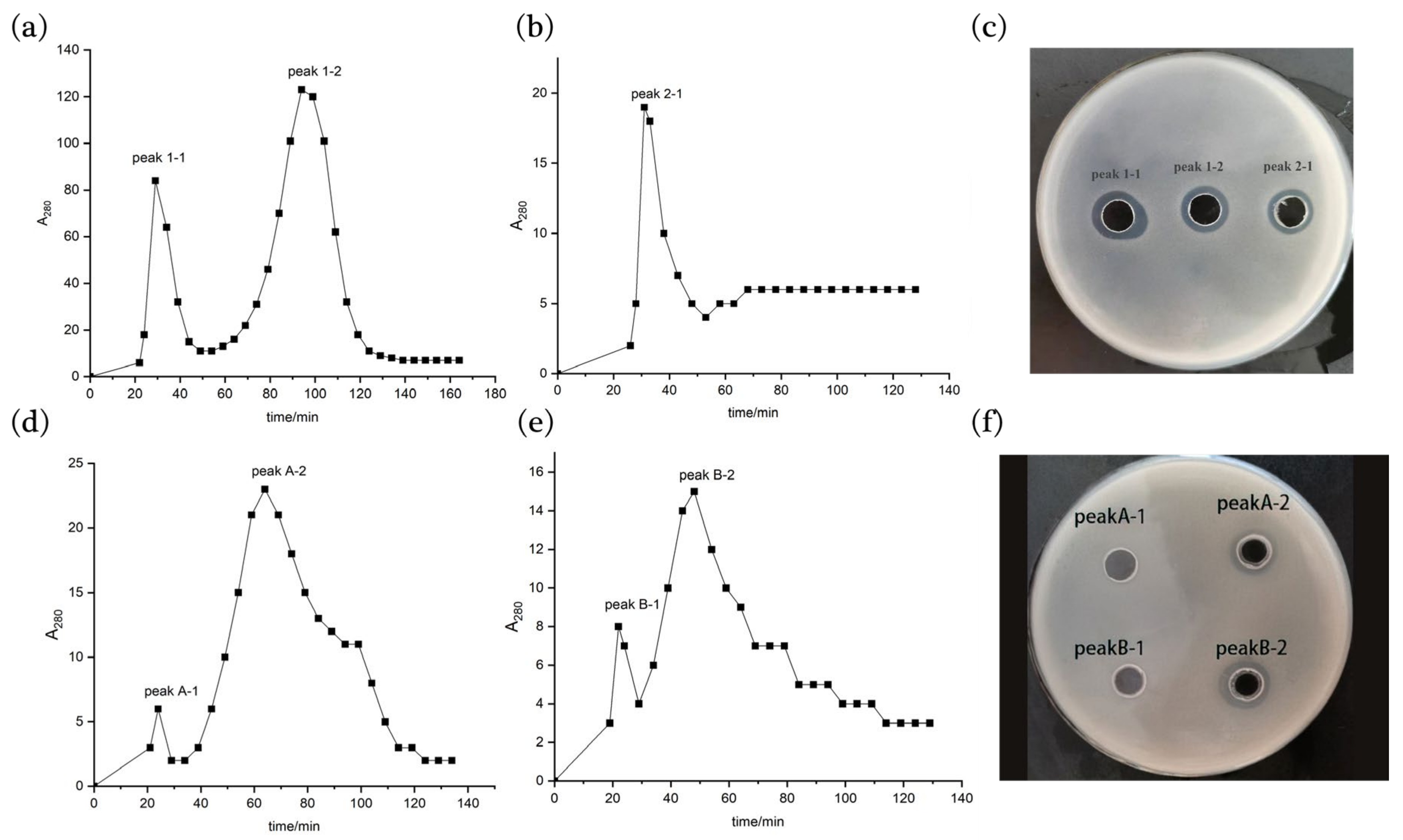
2.3. Isolation and Purification of the Antimicrobial Peptides
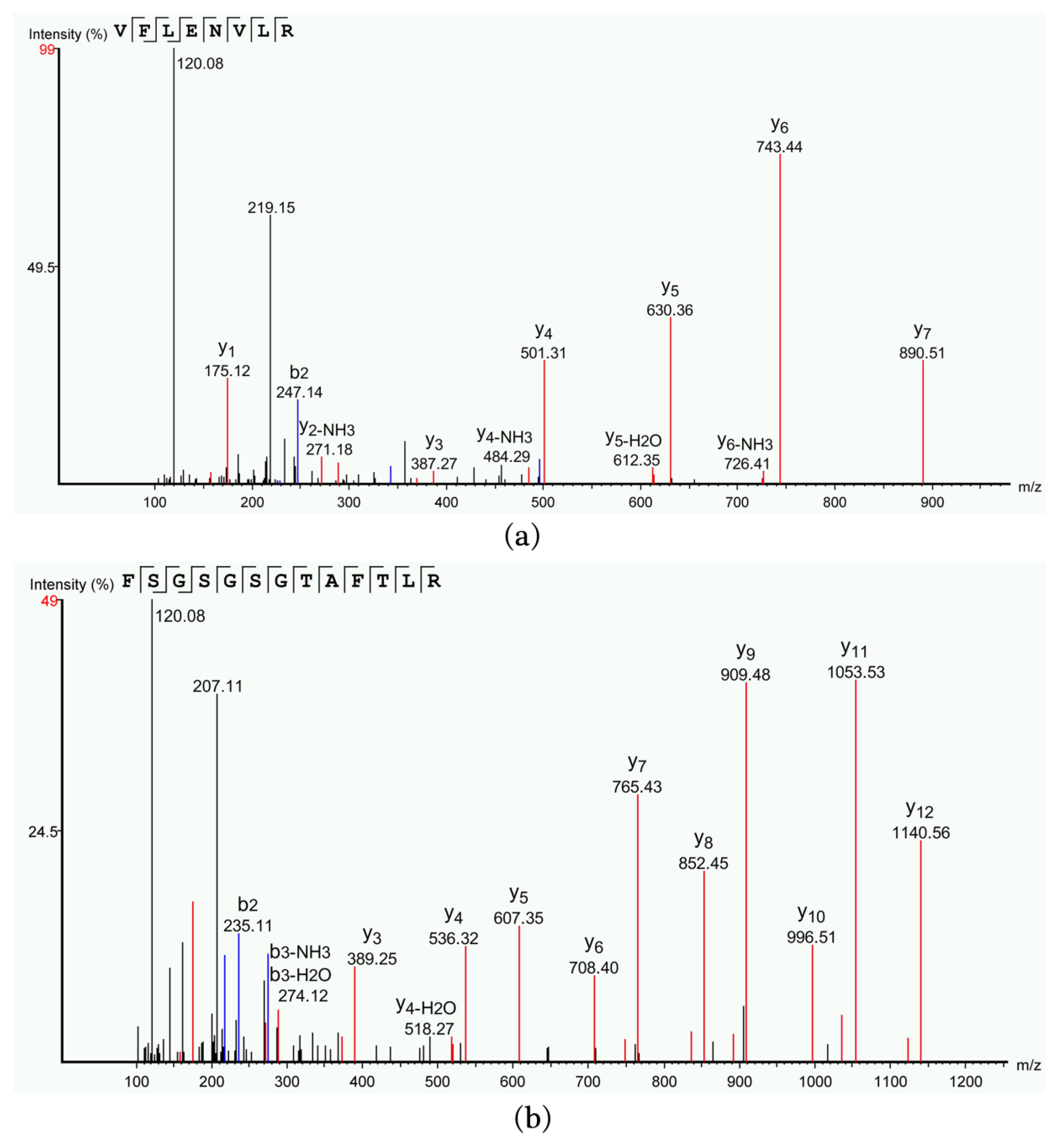
2.4. Identification and Property Analysis of Antimicrobial Peptides
3. Materials and Methods
3.1. Screening of the Antimicrobial Active Bacillus sp.
3.2. Optimization of the Production Conditions for Antimicrobial Peptides
3.2.1. Determination of the Optimal Fermentation Time
3.2.2. Identification of the Optimal Ammonium Sulfate Saturation
3.3. Antibacterial Peptide Purification
3.3.1. Dialysis Desalting
3.3.2. DEAE Cellulose-52 ion-Exchange Chromatography
3.3.3. Sephadex G50 Gel Chromatography
3.3.4. Anti-Phase High-Performance Liquid Chromatography (RP-HPLC) Purification and Preparation
3.4. Identification and Property Analysis of Antimicrobial Peptides
3.4.1. LC–MS/MS Analysis
3.4.2. Determination of Antimicrobial Peptide Stability
3.4.3. The Minimum Inhibitory Concentration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dibner, J.J.; Richards, J.D. Antibiotic growth promoters in agriculture: History and mode of action. Poult. Sci. 2005, 84, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Ashayerizadeh, A.; Dabiri, N.; Ashayerizadeh, O.; Mirzadeh, K.H.; Roshanfekr, H.; Mamooee, M. Effect of dietary antibiotic, probiotic and prebiotic as growth promoters, on growth performance, carcass characteristics and hematological indices of broiler chickens. Pak. J. Biol. Sci. 2009, 12, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. Antibiotic growth promoters enhance animal production by targeting intestinal bile salt hydrolase and its producers. Front. Microbiol. 2014, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, G.L. Why and how antibiotics are used in swine production. Anim. Biotechnol. 2002, 13, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Wierup, M. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 2001, 7, 183–190. [Google Scholar] [CrossRef]
- Li, Z.; Hu, Y.; Yang, Y.; Lu, Z.; Wang, Y. Antimicrobial resistance in livestock: Antimicrobial peptides provide a new solution for a growing challenge. Anim. Front. 2018, 8, 21–29. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K. Use of antibiotics as feed additives: A burning question. Front. Microbiol. 2014, 5, 334. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Puan, S.L.; Erriah, P.; Baharudin, M.M.A.A.; Yahaya, N.M.; Kamil, W.N.I.W.A.; Ali, M.S.M.; Ahmad, S.A.; Oslan, S.N.; Lim, S.; Sabri, S. Antimicrobial peptides from Bacillus spp. and strategies to enhance their yield. Appl. Microbiol. Biotechnol. 2023, 107, 5569–5593. [Google Scholar] [CrossRef] [PubMed]
- Arvidson, S.; Dornbusch, H.; Ericsson, H. Interpretation of the agar diffusion method for bacterial susceptibility testing. J. Antimicrob. Chemoth. 1981, 7, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Hou, Z.; Liu, L.; Xuan, Y.; Chen, X.; Fan, L.; Li, Z.; Xu, B. (1)Progress, applications, challenges and prospects of protein purification technology. Front. Bioeng. Biotechnol. 2022, 10, 1028691. [Google Scholar] [CrossRef] [PubMed]
- Mercado, V.; Olmos, J. Bacteriocin Production by Bacillus Species: Isolation, Characterization, and Application. Probiotics Antimicro. 2022, 14, 1151–1169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Zhu, J.; Xie, W.; Hu, X.; Song, L.; Zi, J.; Yu, R. Purification and characterization of an antibacterial and anti-inflammatory polypeptide from Arca subcrenata. Int. J. Biol. Macromol. 2017, 96, 177–184. [Google Scholar] [CrossRef]
- D’Hondt, M.; Gevaert, B.; Stalmans, S.; Van Dorpe, S.; Wynendaele, E.; Peremans, K.; Burvenich, C.; De Spiegeleer, B. Reversed-phase fused-core HPLC modeling of peptides. J. Pharm. Anal. 2013, 3, 93–101. [Google Scholar] [CrossRef]
- Josic, D.; Kovac, S. Reversed-phase High Performance Liquid Chromatography of proteins. Curr. Protoc. Protein Sci. 2010, 61, 7–8. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, S.; Zhang, J.; Wu, W. Antibacterial activity composition of the fermentation broth of Streptomyces djakartensis NW35. Molecules 2013, 18, 2763–2768. [Google Scholar] [CrossRef]
- Zhou, X.L.; Xiao, C.J.; Wu, L.B.; Huang, B.; Dong, X.; Jiang, B. Five new terpenoids from the rhizomes of Isodon adenantha. J. Asian Nat. Prod. Res. 2014, 16, 555–564. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Mota-Meira, M.; LaPointe, G.; Lacroix, C.; Lavoie, M.C. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 2000, 44, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhou, Q.; Li, P.; Gu, Q. Purification, characterization, and mode of action of Paracin 54, a novel bacteriocin against Staphylococci. Appl. Microbiol. Biotechnol. 2021, 105, 6735–6748. [Google Scholar] [CrossRef] [PubMed]


| Indicator Bacteria | MIC (μg/mL) | |
|---|---|---|
| Peptide-I | Peptide-II | |
| St. aureus ACCC 10499 | 64 | 16 |
| B. cereus ACCC 04315 | 32 | 64 |
| Sa. enterica ACCC 01996 | 8 | 16 |
| Time (min) | Strain 9-1 | Time (min) | Strain 76-1 | ||
|---|---|---|---|---|---|
| Mobile Phase A Acetonitrile | Mobile Phase B Ultrapure Water | Mobile Phase A Acetonitrile | Mobile Phase B Ultrapure Water | ||
| 0–30 | 30~70% | 70~30% | 0–60 | 40~100% | 60~0 |
| 30–50 | 70~80% | 30~20% | - | - | - |
| 50–70 | 80~100% | 20~0 | - | - | - |
| 70–80 | 100% | 0 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Meng, Z.; Li, S.; Liu, T.; Song, J.; Li, J.; Zhang, X. Two Antimicrobial Peptides Derived from Bacillus and Their Properties. Molecules 2023, 28, 7899. https://doi.org/10.3390/molecules28237899
Zhang Y, Meng Z, Li S, Liu T, Song J, Li J, Zhang X. Two Antimicrobial Peptides Derived from Bacillus and Their Properties. Molecules. 2023; 28(23):7899. https://doi.org/10.3390/molecules28237899
Chicago/Turabian StyleZhang, Yujia, Zinuo Meng, Shilong Li, Ting Liu, Juan Song, Jia Li, and Xiumin Zhang. 2023. "Two Antimicrobial Peptides Derived from Bacillus and Their Properties" Molecules 28, no. 23: 7899. https://doi.org/10.3390/molecules28237899
APA StyleZhang, Y., Meng, Z., Li, S., Liu, T., Song, J., Li, J., & Zhang, X. (2023). Two Antimicrobial Peptides Derived from Bacillus and Their Properties. Molecules, 28(23), 7899. https://doi.org/10.3390/molecules28237899








