Design, Synthesis, Docking Study, and Antiproliferative Evaluation of Novel Schiff Base–Benzimidazole Hybrids with VEGFR-2 Inhibitory Activity
Abstract
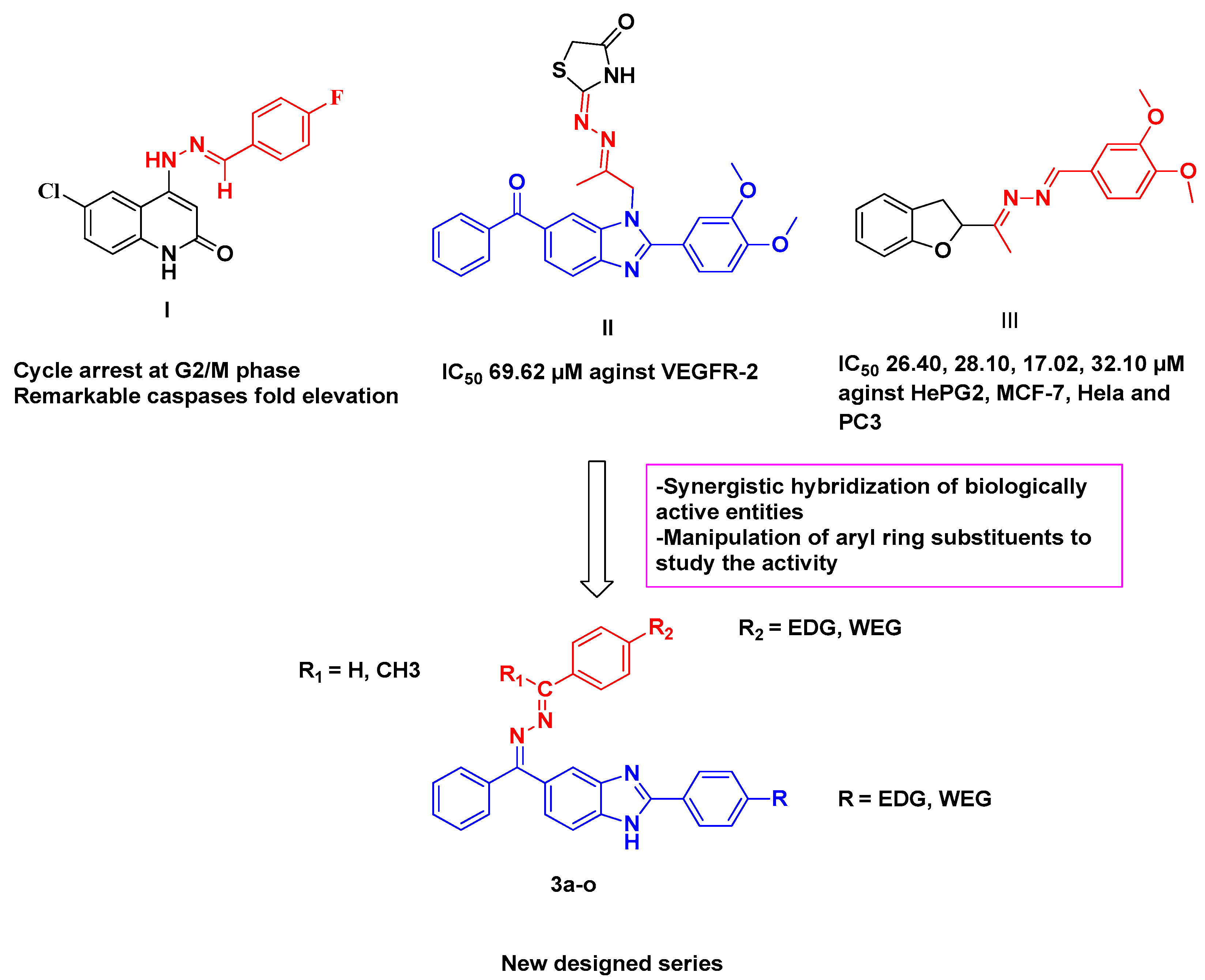
1. Introduction
2. Results
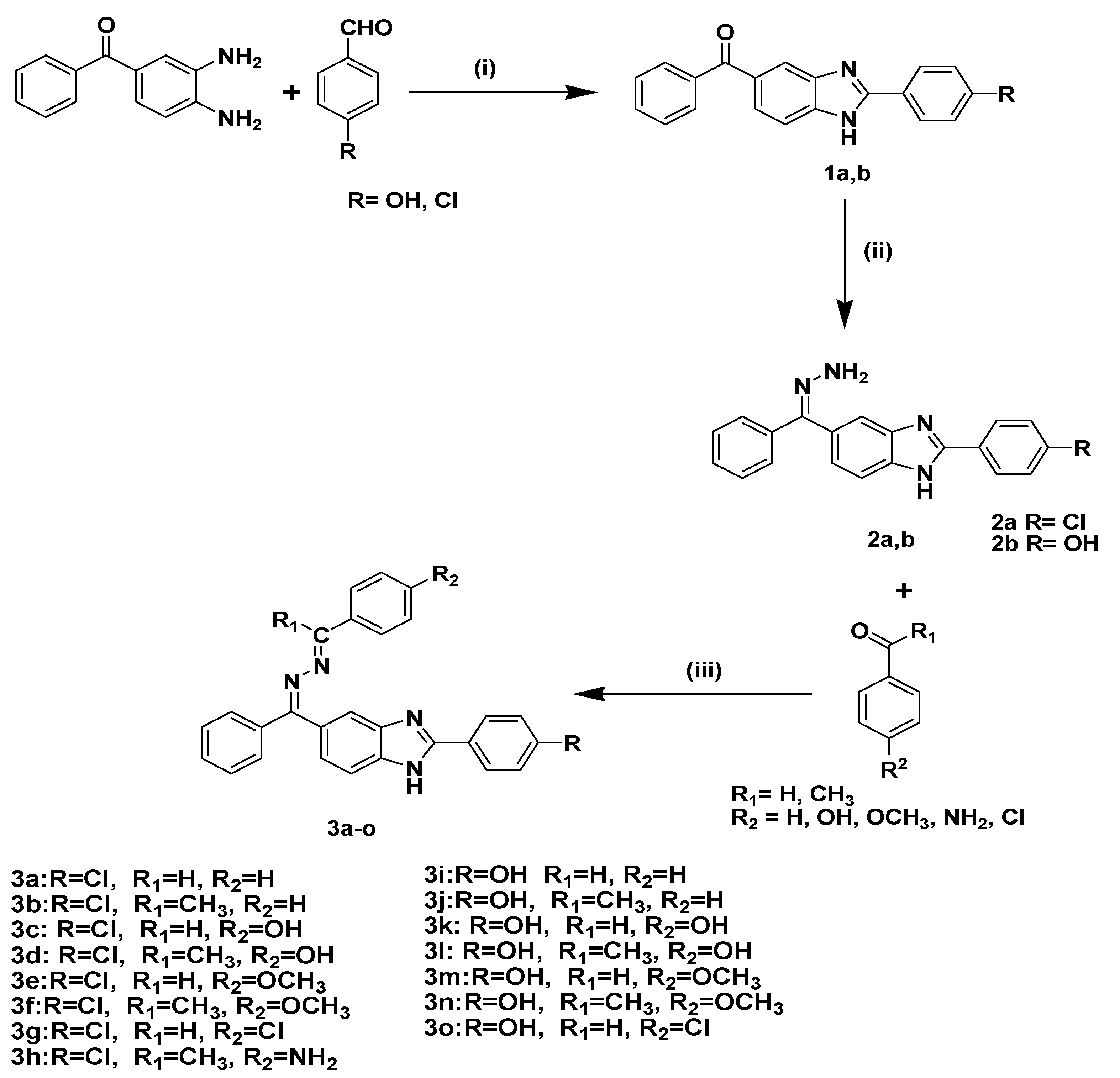
2.1. Chemistry
2.2. Biology
2.2.1. Primary In Vitro One-Dose Anticancer Assay
2.2.2. Full In Vitro Five-Dose Anticancer Assay
2.2.3. MTT Assay against Lung Cancer Cell Lines
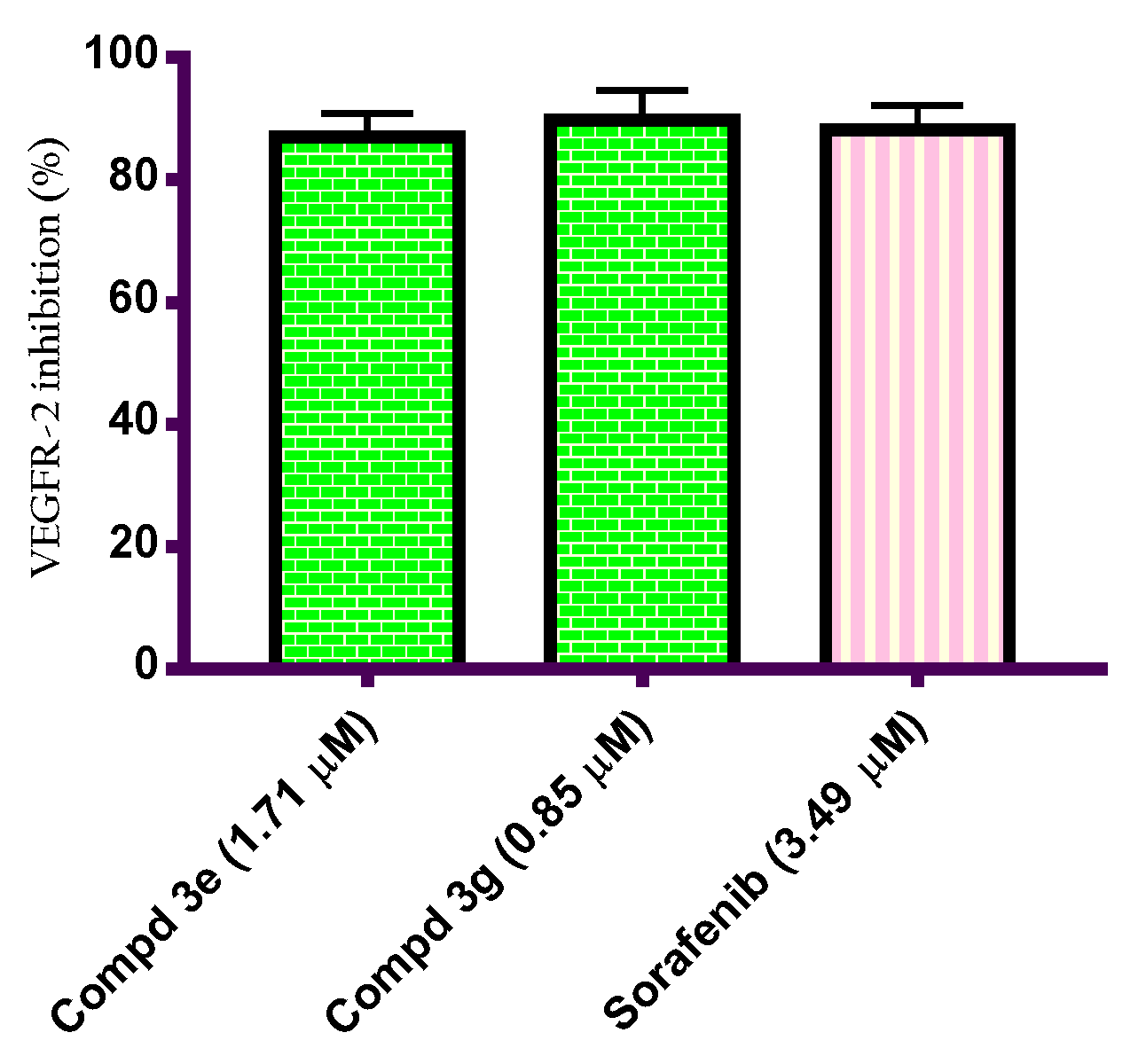
2.2.4. In Vitro VEGFR-2 Inhibition Assay
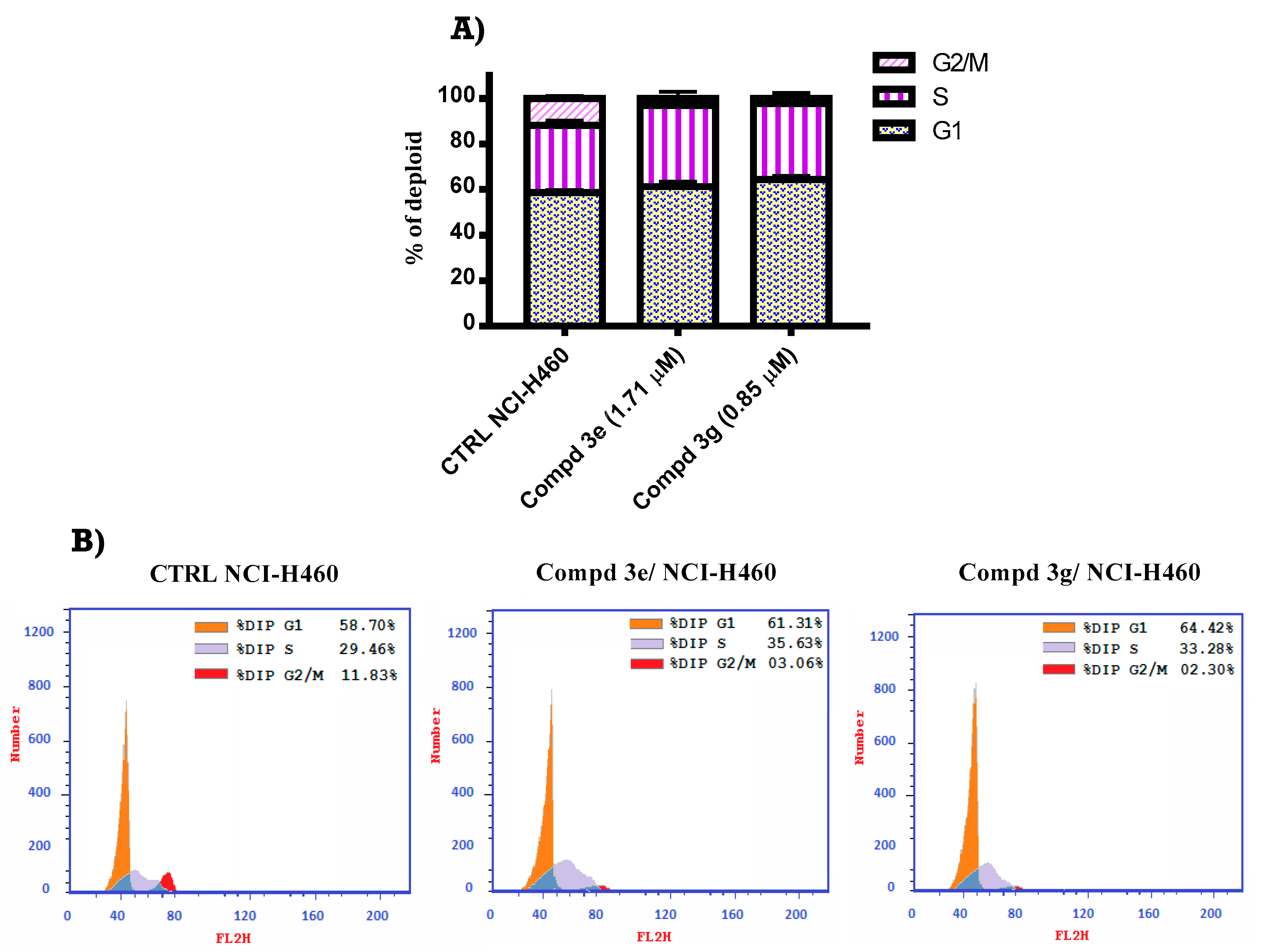
2.2.5. Cell Cycle Analysis of Benzimidazole Hybrids 3e and 3g
2.2.6. Annexin V/ FITC Apoptosis Staining Assay
2.2.7. Caspase Assay
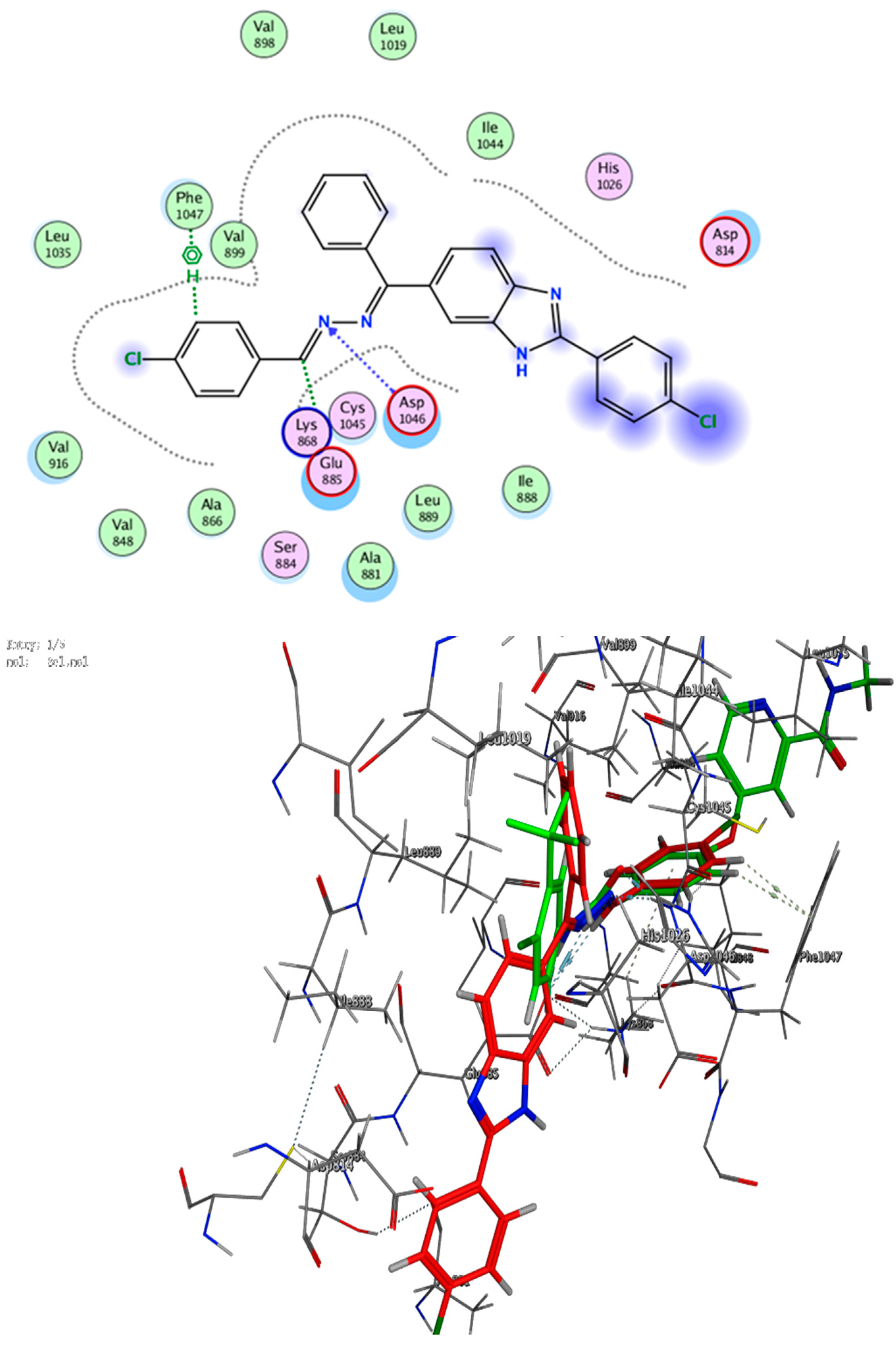
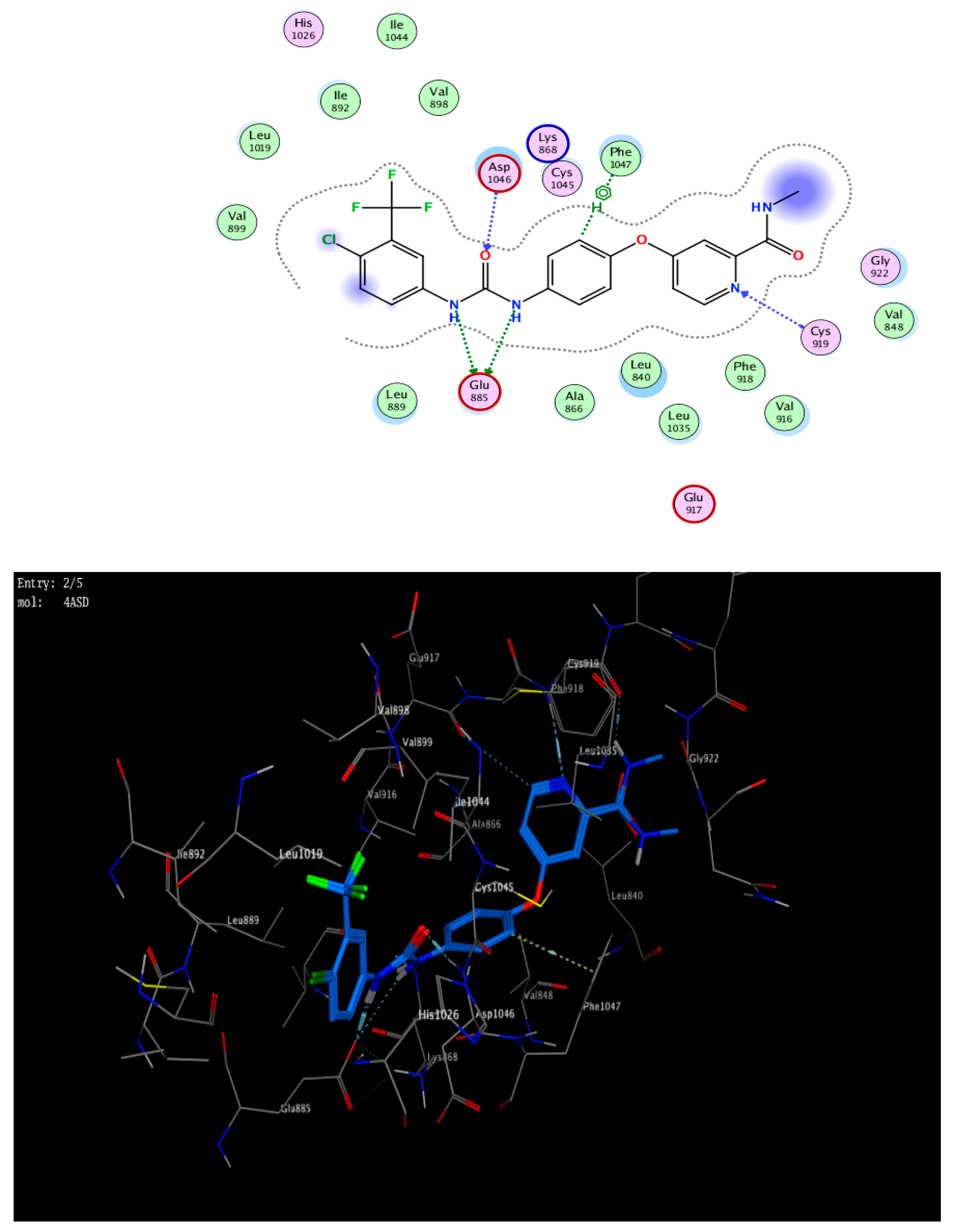
3. Docking Study
4. Conclusions
5. Experimental Section
5.1. Chemistry
5.1.1. General Synthesis of Compounds 3a–o
6-((Z)-((E)-Benzylidenehydrazono)(phenyl)methyl)-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3a)
2-(4-Chlorophenyl)-5-((Z)-phenyl((E)-(1-phenylethylidene)hydrazono)methyl)-1H-benzo[d]imidazole (3b)
4-((E)-((Z)-((2-(4-Chlorophenyl)-1H-benzo[d]imidazol-6-yl)(phenyl) methylene)hydrazono)methyl)phenol (3c)
4-((E)-1-((Z)-((2-(4-Chlorophenyl)-1H-benzo[d]imidazol-5-yl)(phenyl)methylene)hydrazono)ethyl)phenol (3d)
2-(4-Chlorophenyl)-6-((Z)-((E)-(4-methoxybenzylidene)hydrazono)(phenyl)methyl)-1H-benzo[d]imidazole (3e)
2-(4-Chlorophenyl)-5-((Z)-((E)-(1-(4-methoxyphenyl)ethylidene)hydrazono) (phenyl)methyl)-1H-benzo[d]imidazole (3f)
6-((Z)-((E)-(4-Chlorobenzylidene)hydrazono)(phenyl)methyl)-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3g)
4-((E)-1-((Z)-((2-(4-Chlorophenyl)-1H-benzo[d]imidazol-5-yl)(phenyl)methylene)hydrazono)ethyl)aniline (3h)
4-(6-((Z)-((E)-benzylidenehydrazono)(phenyl)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3i)
4-(5-((Z)-Phenyl((E)-(1-phenylethylidene)hydrazono)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3j)
4-(6-((Z)-((E)-(4-hydroxybenzylidene)hydrazono)(phenyl)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3k)
4-((E)-1-((Z)-((2-(4-Hydroxyphenyl)-1H-benzo[d]imidazol-5-yl)(phenyl)methylene)hydrazono)ethyl)phenol (3l)
4-(6-((Z)-((E)-(4-Methoxybenzylidene)hydrazono)(phenyl)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3m)
4-(5-((Z)-((E)-(1-(4-Methoxyphenyl)ethylidene)hydrazono)(phenyl)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3n)
4-(6-((Z)-((E)-(4-chlorobenzylidene)hydrazono)(phenyl)methyl)-1H-benzo[d]imidazol-2-yl)phenol (3o)
5.2. Biological Evaluation
5.2.1. NCI Screening Assay
5.2.2. MTT Assay for Cell Viability
5.2.3. VEGFR Inhibitory Assay
5.2.4. Cell Cycle Analysis and Apoptotic Assay
Cell Apoptosis and Apoptotic Detection
5.2.5. Activation of Caspases
5.2.6. Docking Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Appendix A
Appendix A.1. Biological Studies
Appendix A.1.1. Cytotoxic Activity Evaluation against Lung Cancer Cell Lines
Appendix A.1.2. VEGFR-2 Inhibition Assay
Appendix A.1.3. Cell Cycle Analysis of Compounds 3e and 3g
Appendix A.1.4. Apoptosis Assay for Compounds 3e and 3g
Appendix A.1.5. Caspase 9 Assay for Compounds 3e and 3g
Appendix A.1.6. Molecular Modeling Study
References
- Aly, A.A.; Ramadan, M.; Abuo-Rahma, G.E.-D.A.; Elshaier, Y.A.; Elbastawesy, M.A.; Brown, A.B.; Bräse, S. Quinolones as prospective drugs: Their syntheses and biological applications. In Advances in Heterocyclic Chemistry; Academic Press: Cambridge, MA, USA, 2020; pp. 147–196. [Google Scholar]
- Tiglani, D.; Salahuddin; Mazumder, A.; Yar, M.S.; Kumar, R.; Ahsan, M.J. Benzimidazole-quinoline hybrid scaffold as promising pharmacological Agents: A review. Polycycl. Aromat. Compd. 2022, 42, 5044–5066. [Google Scholar] [CrossRef]
- Brishty, S.R.; Hossain, M.; Khandaker, M.U.; Faruque, M.R.I.; Osman, H.; Rahman, S. A comprehensive account on recent progress in pharmacological activities of benzimidazole derivatives. Front. Pharmacol. 2021, 1, 2863. [Google Scholar] [CrossRef] [PubMed]
- Faheem, M.; Rathaur, A.; Pandey, A.; Kumar Singh, V.; Tiwari, A.K. A review on the modern synthetic approach of benzimidazole candidate. ChemistrySelect 2020, 5, 3981–3994. [Google Scholar] [CrossRef]
- Choudhary, S.; Arora, M.; Verma, H.; Kumar, M.; Silakari, O. Benzimidazole based hybrids against complex diseases: A catalogue of the SAR profile. Eur. J. Pharmacol. 2021, 899, 174027. [Google Scholar] [CrossRef]
- Satija, G.; Sharma, B.; Madan, A.; Iqubal, A.; Shaquiquzzaman, M.; Akhter, M.; Parvez, S.; Khan, M.A.; Alam, M.M. Benzimidazole based derivatives as anticancer agents: Structure activity relationship analysis for various targets. J. Heterocycl. Chem. 2022, 59, 22–66. [Google Scholar] [CrossRef]
- Khan, S.; Alhumaydhi, F.A.; Ibrahim, M.M.; Alqahtani, A.; Alshamrani, M.; Alruwaili, A.S.; Hassanian, A.A.; Khan, S. Recent advances and therapeutic journey of Schiff base complexes with selected metals (Pt, Pd, Ag, Au) as potent anticancer agents: A review. Anti-Cancer Agents Med. Chem. 2022, 22, 3086–3096. [Google Scholar] [CrossRef]
- Korkmaz, A.; Bursal, E. Benzothiazole sulfonate derivatives bearing azomethine: Synthesis, characterization, enzyme inhibition, and molecular docking study. J. Mol. Struct. 2022, 1257, 132641. [Google Scholar] [CrossRef]
- Matela, G. Schiff bases and complexes: A review on anti-cancer activity. Anti-Cancer Agents Med. Chem. 2020, 20, 1908–1917. [Google Scholar] [CrossRef]
- Elbastawesy, M.A.; Ramadan, M.; El-Shaier, Y.A.; Aly, A.A.; Abuo-Rahma, G.E.-D.A. Arylidenes of Quinolin-2-one scaffold as Erlotinib analogues with activities against leukemia through inhibition of EGFR TK/STAT-3 pathways. Bioorg. Chem. 2020, 96, 103628. [Google Scholar] [CrossRef]
- El-Khouly, O.A.; Henen, M.A.; El-Sayed, M.A.-A.; El-Messery, S.M. Design, synthesis and computational study of new benzofuran hybrids as dual PI3K/VEGFR2 inhibitors targeting cancer. Sci. Rep. 2022, 12, 17104. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Wang, Y. Therapeutic angiogenesis: Controlled delivery of angiogenic factors. Ther. Deliv. 2012, 3, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Teleanu, R.I.; Chircov, C.; Grumezescu, A.M.; Teleanu, D.M. Tumor angiogenesis and anti-angiogenic strategies for cancer treatment. J. Clin. Med. 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Tyrosine kinase receptor Flt/VEGFR family: Its characterization related to angiogenesis and cancer. Genes Cancer 2010, 1, 1119–1123. [Google Scholar] [CrossRef]
- Elbastawesy, M.A.; Aly, A.A.; Ramadan, M.; Elshaier, Y.A.; Youssif, B.G.; Brown, A.B.; Abuo-Rahma, G.E.-D.A. Novel pyrazoloquinolin-2-ones: Design, synthesis, docking studies, and biological evaluation as antiproliferative EGFR-TK inhibitors. Bioorg. Chem. 2019, 90, 103045. [Google Scholar] [CrossRef]
- S Sharma, P.; Sharma, R.; Tyagi, T. VEGF/VEGFR pathway inhibitors as anti-angiogenic agents: Present and future. Curr. Cancer Drug Targets 2011, 11, 624–653. [Google Scholar] [CrossRef]
- Poliaková, M.; Aebersold, D.M.; Zimmer, Y.; Medová, M. The relevance of tyrosine kinase inhibitors for global metabolic pathways in cancer. Mol. Cancer 2018, 17, 27. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Wang, Y.; Lin, C.; Zhang, D.; Chen, J.; Ouyang, L.; Wu, F.; Zhang, J.; Chen, L. Recent progress on vascular endothelial growth factor receptor inhibitors with dual targeting capabilities for tumor therapy. J. Hematol. Oncol. 2022, 15, 89. [Google Scholar] [CrossRef]
- Soltan, O.M.; Shoman, M.E.; Abdel-Aziz, S.A.; Narumi, A.; Konno, H.; Abdel-Aziz, M. Molecular hybrids: A five-year survey on structures of multiple targeted hybrids of protein kinase inhibitors for cancer therapy. Eur. J. Med. Chem. 2021, 225, 113768. [Google Scholar] [CrossRef]
- Obeng, E. Apoptosis (programmed cell death) and its signals—A review. Braz. J. Biol. 2020, 81, 1133–1143. [Google Scholar] [CrossRef]
- Jan, R. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv. Pharm. Bull. 2019, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Meguid, E.A.; El-Deen, E.M.M.; Nael, M.A.; Anwar, M.M. Novel benzimidazole derivatives as anti-cervical cancer agents of potential multi-targeting kinase inhibitory activity. Arab. J. Chem. 2020, 13, 9179–9195. [Google Scholar] [CrossRef]
- Zaki, I.; Moustafa, A.M.; Beshay, B.Y.; Masoud, R.E.; Elbastawesy, M.A.; Abourehab, M.A.; Zakaria, M.Y. Design and synthesis of new trimethoxylphenyl-linked combretastatin analogues loaded on diamond nanoparticles as a panel for ameliorated solubility and antiproliferative activity. J. Enzym. Inhib. Med. Chem. 2022, 37, 2679–2701. [Google Scholar] [CrossRef]
- El-Sheref, E.M.; Elbastawesy, M.A.; Brown, A.B.; Shawky, A.M.; Gomaa, H.A.; Bräse, S.; Youssif, B.G. Design and Synthesis of (2-oxo-1,2-Dihydroquinolin-4-yl)-1,2,3-triazole Derivatives via Click Reaction: Potential Apoptotic Antiproliferative Agents. Molecules 2021, 26, 6798. [Google Scholar] [CrossRef]
- Morcoss, M.M.; Abdelhafez, E.S.; Abdel-Rahman, H.M.; Abdel-Aziz, M.; El-Ella, A.; Dalal, A. Novel Benzimidazole/Hydrazone Derivatives as Promising Anticancer Lead Compounds: Design, Synthesis, and Molecular Docking Study. J. Adv. Biomed. Pharm. Sci. 2020, 3, 45–52. [Google Scholar] [CrossRef]
- Zha, G.-F.; Qin, H.-L.; Youssif, B.G.; Amjad, M.W.; Raja, M.A.G.; Abdelazeem, A.H.; Bukhari, S.N.A. Discovery of potential anticancer multi-targeted ligustrazine based cyclohexanone and oxime analogs overcoming the cancer multidrug resistance. Eur. J. Med. Chem. 2017, 135, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Ceci, C.; Atzori, M.G.; Lacal, P.M.; Graziani, G. Role of VEGFs/VEGFR-1 signaling and its inhibition in modulating tumor invasion: Experimental evidence in different metastatic cancer models. Int. J. Mol. Sci. 2020, 21, 1388. [Google Scholar] [CrossRef]
- Ahmed, M.F.; Santali, E.Y.; El-Haggar, R. Novel piperazine–chalcone hybrids and related pyrazoline analogues targeting VEGFR-2 kinase; design, synthesis, molecular docking studies, and anticancer evaluation. J. Enzym. Inhib. Med. Chem. 2021, 36, 308–319. [Google Scholar] [CrossRef]
- Avrutsky, M.I.; Troy, C.M. Caspase-9: A multimodal therapeutic target with diverse cellular expression in human disease. Front. Pharmacol. 2021, 12, 1728. [Google Scholar] [CrossRef]
- Morcoss, M.M.; El Shimaa, M.; Ibrahem, R.A.; Abdel-Rahman, H.M.; Abdel-Aziz, M.; Abou El-Ella, D.A. Design, synthesis, mechanistic studies and in silico ADME predictions of benzimidazole derivatives as novel antifungal agents. Bioorg. Chem. 2020, 101, 103956. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.S.; Gomaa, R.M.; Elmorsy, M.A. Design and synthesis of 2-phenyl benzimidazole derivatives as VEGFR-2 inhibitors with anti-breast cancer activity. Chem. Biol. Drug Des. 2019, 93, 454–463. [Google Scholar] [CrossRef] [PubMed]


| Comp No | IC50 Value (μM) | |
|---|---|---|
| A549 | NCI-H460 | |
| 3c | 4.95 ± 0.61 | 3.07 ± 0.42 |
| 3e | 3.58 ± 0.53 | 1.71 ± 0.17 |
| 3g | 1.88 ± 0.35 | 0.85 ± 0.24 |
| 3o | 5.98 ± 0.55 | 5.22 ± 0.42 |
| Sorafenib | 4.84 ± 0.23 | 3.49 ± 0.18 |
| Comp. No. | VEGFR-2 | ||||
|---|---|---|---|---|---|
| Affinity kcal/mol | Distance (in Ao) from Main Residue | Amino Acids Residue | Functional Group | Type of Interactions | |
| 3g | −8.59 | 3.38 | Glu 885 | C24 | H-donor |
| 3.09 | ASP 1046 | N23 | H-acceptor | ||
| 3.72 | PHE 1047 | 6-ring | pi-H | ||
| Sorafenib | −8.74 | 2.91 | Glu 885 | N12 | H-donor |
| 3.45 | Glu 885 | N14 | H-donor | ||
| 2.90 | Asp 1046 | O15 | H-acceptor | ||
| 3.25 | Cys 919 | N26 | H-acceptor | ||
| 3.67 | PHE 1047 | 5-ring | pi-H | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd El-Lateef, H.M.; Elbastawesy, M.A.I.; Abdelghani Ibrahim, T.M.; Khalaf, M.M.; Gouda, M.; Wahba, M.G.F.; Zaki, I.; Morcoss, M.M. Design, Synthesis, Docking Study, and Antiproliferative Evaluation of Novel Schiff Base–Benzimidazole Hybrids with VEGFR-2 Inhibitory Activity. Molecules 2023, 28, 481. https://doi.org/10.3390/molecules28020481
Abd El-Lateef HM, Elbastawesy MAI, Abdelghani Ibrahim TM, Khalaf MM, Gouda M, Wahba MGF, Zaki I, Morcoss MM. Design, Synthesis, Docking Study, and Antiproliferative Evaluation of Novel Schiff Base–Benzimidazole Hybrids with VEGFR-2 Inhibitory Activity. Molecules. 2023; 28(2):481. https://doi.org/10.3390/molecules28020481
Chicago/Turabian StyleAbd El-Lateef, Hany M., Mohammed A. I. Elbastawesy, Tamer Mohamed Abdelghani Ibrahim, Mai M. Khalaf, Mohamed Gouda, Mariam G. F. Wahba, Islam Zaki, and Martha M. Morcoss. 2023. "Design, Synthesis, Docking Study, and Antiproliferative Evaluation of Novel Schiff Base–Benzimidazole Hybrids with VEGFR-2 Inhibitory Activity" Molecules 28, no. 2: 481. https://doi.org/10.3390/molecules28020481
APA StyleAbd El-Lateef, H. M., Elbastawesy, M. A. I., Abdelghani Ibrahim, T. M., Khalaf, M. M., Gouda, M., Wahba, M. G. F., Zaki, I., & Morcoss, M. M. (2023). Design, Synthesis, Docking Study, and Antiproliferative Evaluation of Novel Schiff Base–Benzimidazole Hybrids with VEGFR-2 Inhibitory Activity. Molecules, 28(2), 481. https://doi.org/10.3390/molecules28020481








