Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners
Abstract
1. Introduction
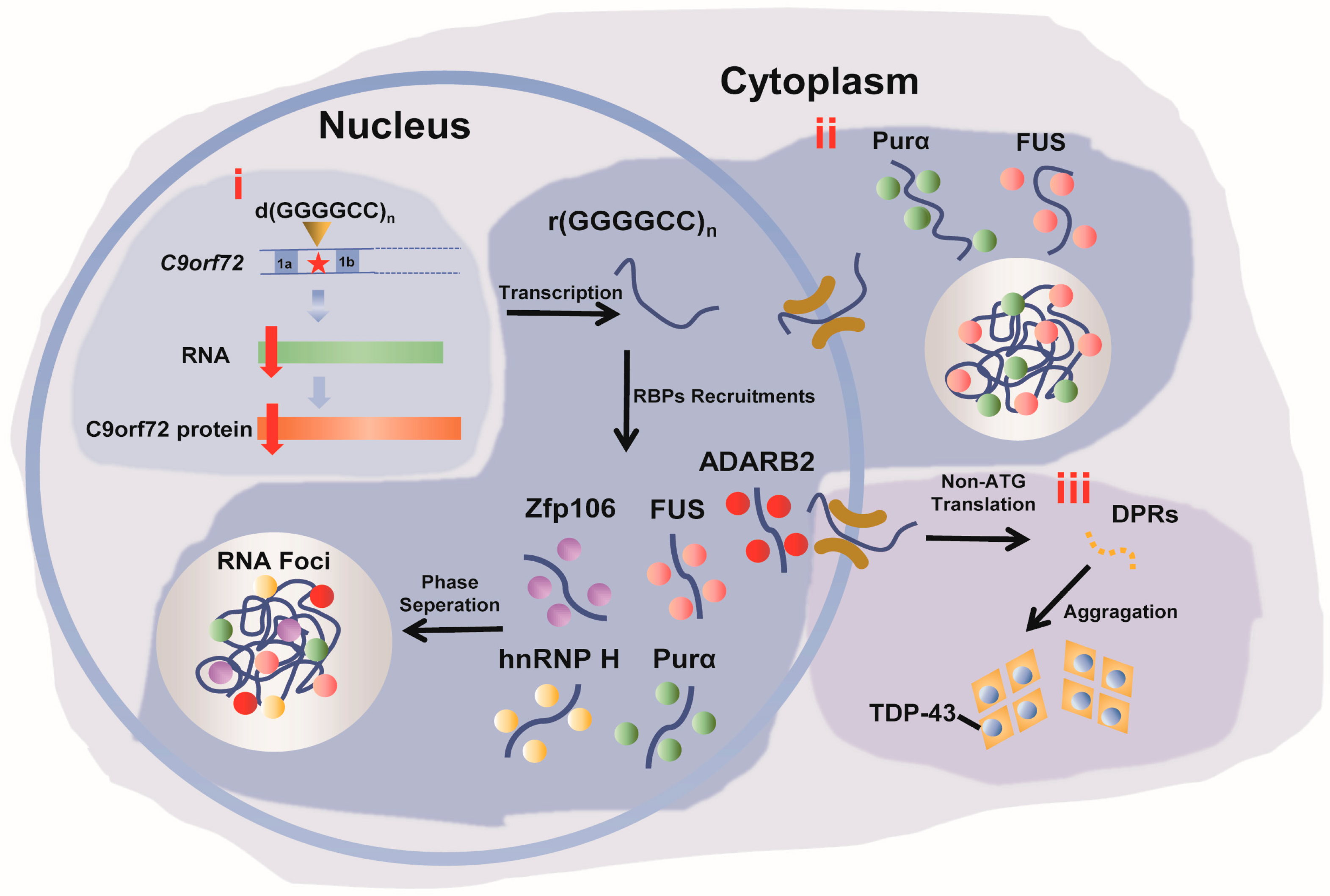
2. The Structure of r(GGGGCC)n RNA Repeats and the RNA within the RNA Foci of r(GGGGCC)n
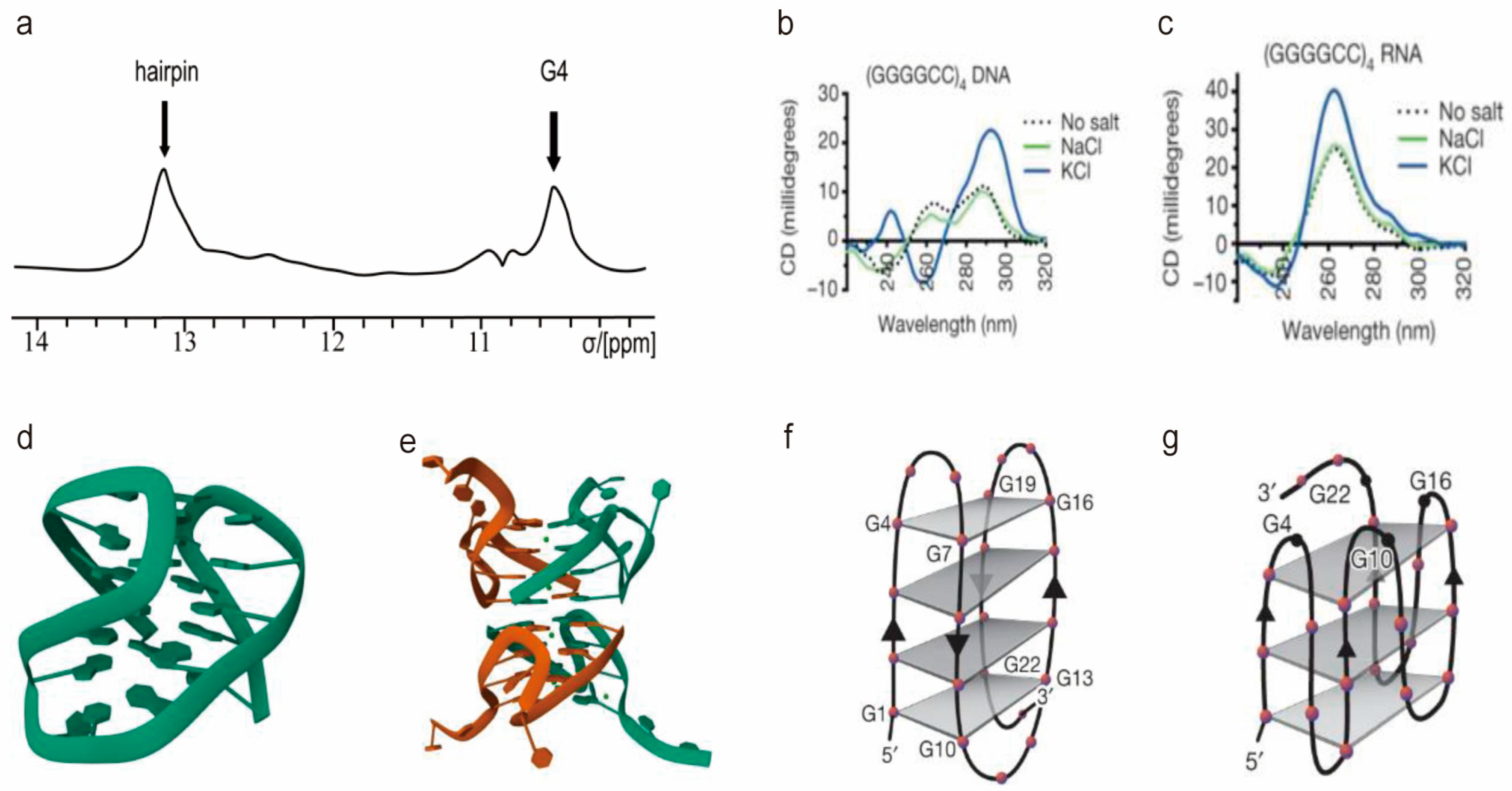
2.1. The Solution Structures of r(GGGGCC)n RNA
2.2. Structure of d(GGGGCC)n DNA
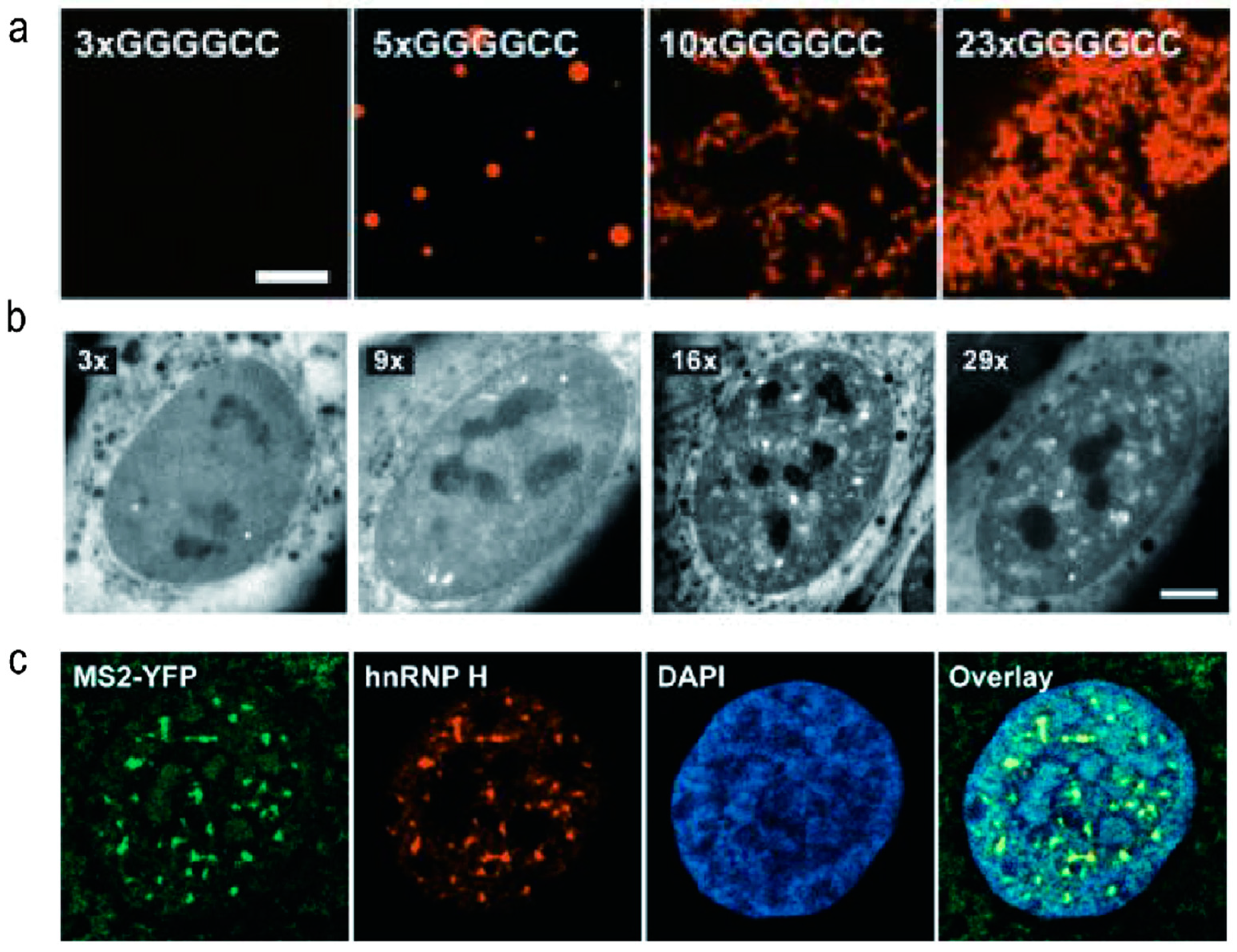
2.3. Biological Phase Separation and Transition of r(GGGGCC)n
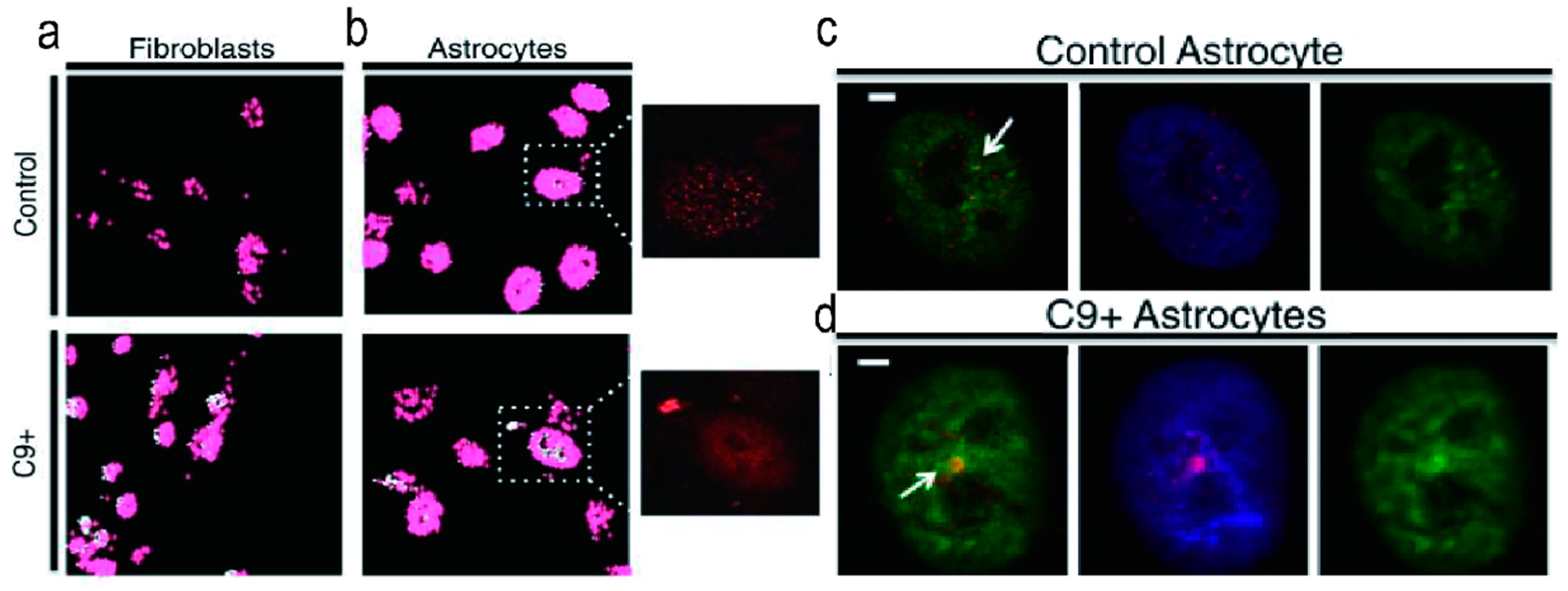
3. Disease Related RBPs That Bind to r(GGGGCC)n
3.1. hnRNP H and TDP-43
3.2. FUS
3.3. Zfp106
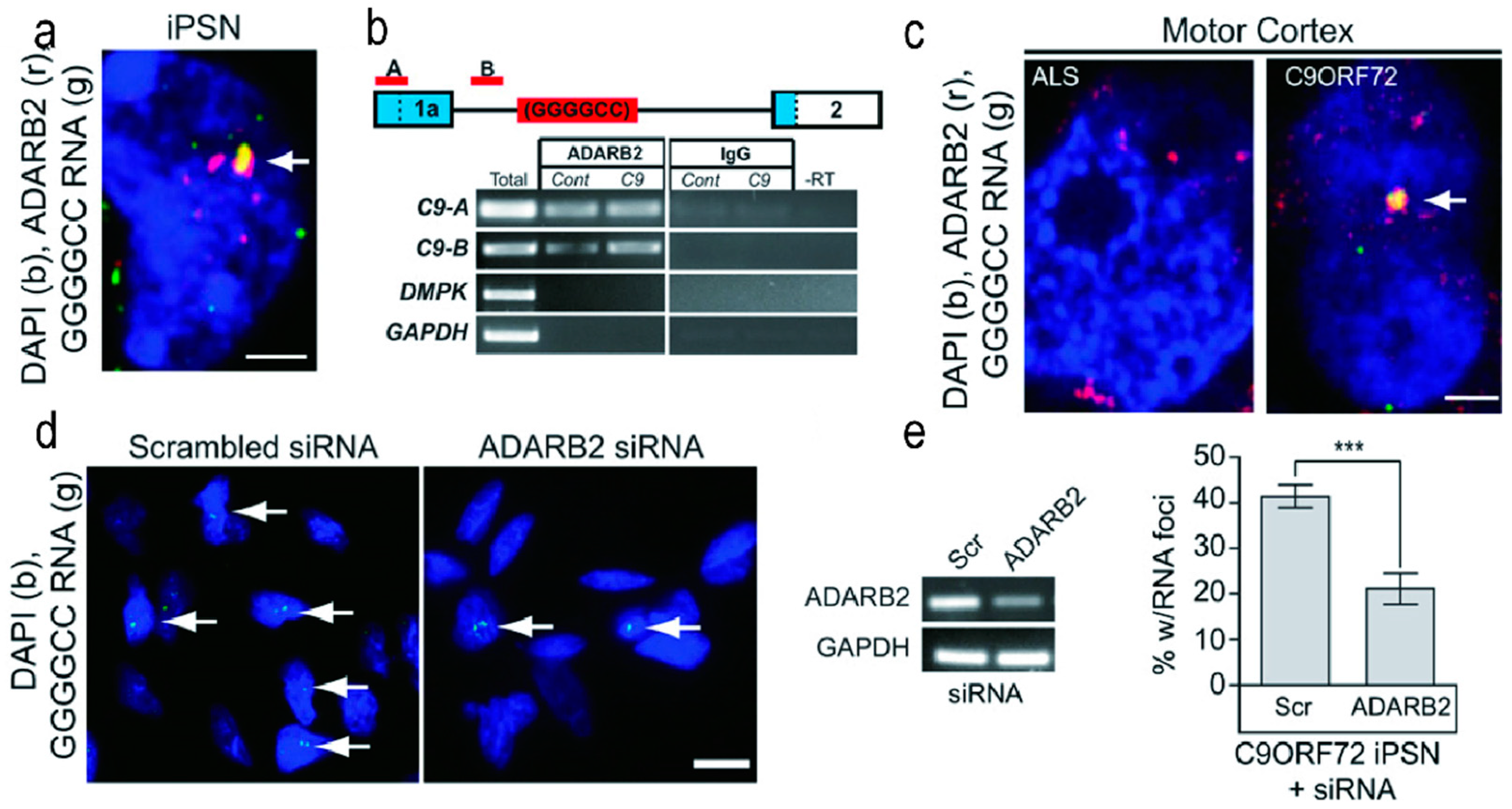
3.4. ADARB2
3.5. Purα
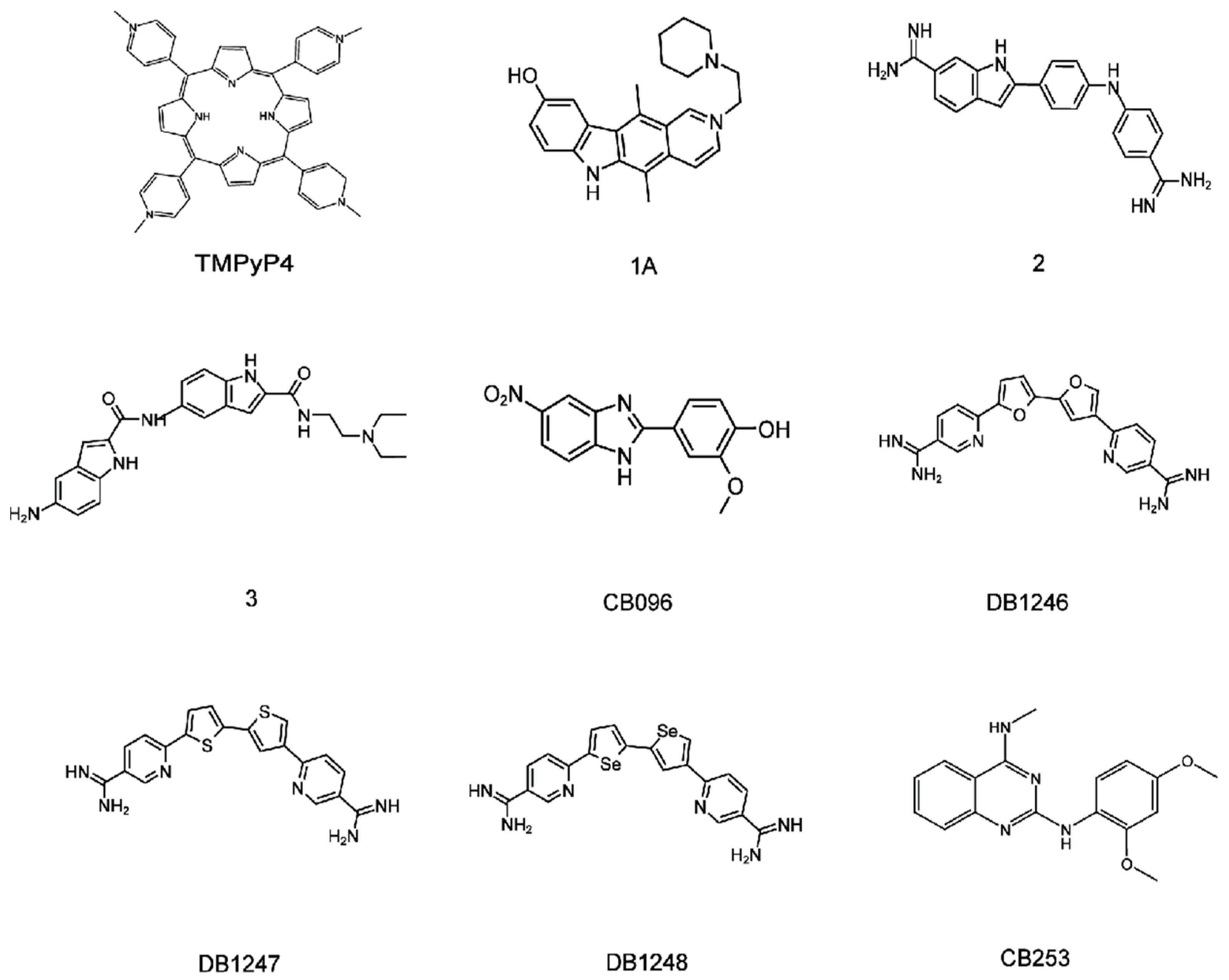
4. Lead Small Molecules Binds to r(GGGGCC)n
4.1. Binding of r(GGGGCC)8 with the TMPyP4
4.2. Binding of r(GGGGCC)8 with Other Liands
4.3. Binding of r(GGGGCC)8 with CB096
4.4. Binding of r(GGGGCC)n with DB1246, DB1247, and DB1273
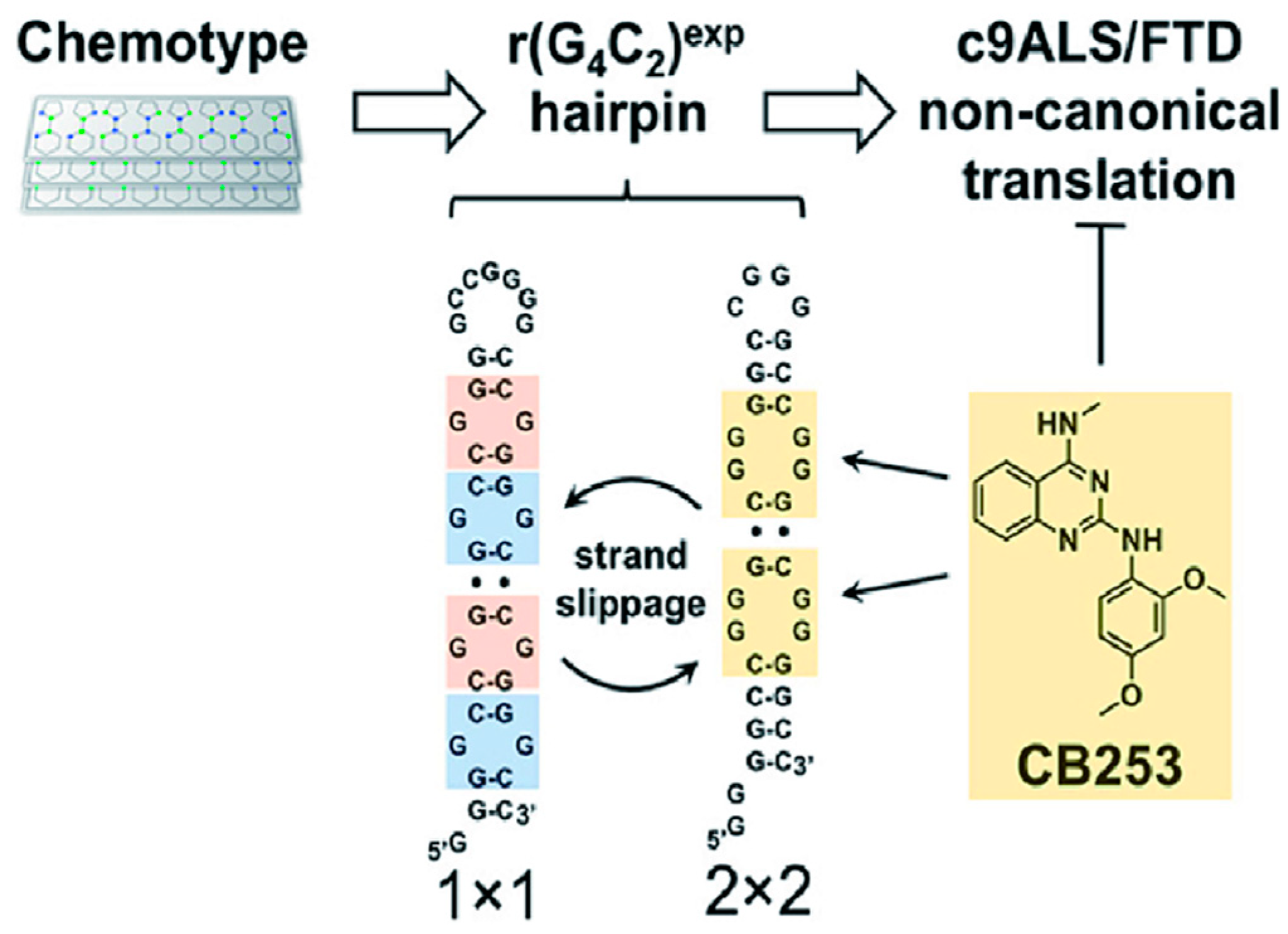
4.5. Binding of r(GGGGCC)n with CB253
5. Summary and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ling, S.-C.; Polymenidou, M.; Cleveland, D.W. Converging Mechanisms in ALS and FTD: Disrupted RNA and Protein Homeostasis. Neuron 2013, 79, 416–438. [Google Scholar] [CrossRef]
- Sagui, C. Structure and Dynamics of DNA and RNA Double Helices Obtained from the GGGGCC and CCCCGG Hexanucleotide Repeats That Are the Hallmark of C9FTD/ALS Diseases. ACS Chem. Neurosci. 2017, 8, 578–591. [Google Scholar]
- Ash, P.E.A.; Bieniek, K.F.; Gendron, T.F.; Caulfield, T.; Lin, W.-L.; DeJesus-Hernandez, M.; van Blitterswijk, M.M.; Jansen-West, K.; Paul, J.W., III; Rademakers, R.; et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 2013, 77, 639–646. [Google Scholar] [CrossRef] [PubMed]
- van der Ende, E.L.; Jackson, J.L.; White, A.; Seelaar, H.; van Blitterswijk, M.; Van Swieten, J.C. Unravelling the clinical spectrum and the role of repeat length in C9ORF72 repeat expansions. J. Neurol. Neurosurg. Psychiatry 2021, 92, 502–509. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Z.; Yang, F.; Jiao, B.; Peng, H.; Shi, Y.; Wang, Y.; Huang, F.; Wang, J.; Shen, L.; et al. Analysis of the GGGGCC Repeat Expansions of the C9orf72 Gene in SCA3/MJD Patients from China. PLoS ONE 2015, 10, e0130336. [Google Scholar] [CrossRef]
- Lee, Y.-B.; Chen, H.-J.; Peres, J.N.; Gomez-Deza, J.; Attig, J.; Štalekar, M.; Troakes, C.; Nishimura, A.L.; Scotter, E.L.; Vance, C.; et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013, 5, 1178–1186. [Google Scholar] [CrossRef]
- Balendra, R.; Isaacs, A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018, 14, 544–558. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Dodd, K.C.; Power, R.; Ealing, J.; Hamdalla, H. FUS-ALS presenting with myoclonic jerks in a 17-year-old man. Amyotroph. Lateral Scler. Front. Degener. 2019, 20, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, C.; Geng, Y.; Zhu, G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015, 5, 16673. [Google Scholar] [CrossRef]
- Calcoen, D.; Elias, L.; Yu, X. What does it take to produce a breakthrough drug? Nat. Rev. Drug Discov. 2015, 14, 161–162. [Google Scholar]
- Tamaki, Y.; Urushitani, M. Molecular Dissection of TDP-43 as a Leading Cause of ALS/FTLD. Int. J. Mol. Sci. 2022, 23, 12508. [Google Scholar] [CrossRef]
- van Zundert, B.; Brown, R.H., Jr. Silencing strategies for therapy of SOD1-mediated ALS. Neurosci. Lett. 2017, 636, 32–39. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Saánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed]
- O’rourke, J.G.; Bogdanik, L.; Muhammad, A.; Gendron, T.F.; Kim, K.J.; Austin, A.; Cady, J.; Liu, E.Y.; Zarrow, J.; Grant, S.; et al. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron 2015, 88, 892–901. [Google Scholar]
- Jiang, J.; Zhu, Q.; Gendron, T.F.; Saberi, S.; McAlonis-Downes, M.; Seelman, A.; Stauffer, J.E.; Jafar-Nejad, P.; Drenner, K.; Schulte, D.; et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron 2016, 90, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Jiang, J.; Gendron, T.F.; McAlonis-Downes, M.; Jiang, L.; Taylor, A.; Garcia, S.D.; Dastidar, S.G.; Rodriguez, M.J.; King, P.; et al. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat. Neurosci. 2020, 23, 615–624. [Google Scholar] [CrossRef]
- Rodriguez, C.; Todd, P. New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol. Dis. 2019, 130, 104515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Daigle, J.G.; Cunningham, K.M.; Coyne, A.N.; Ruan, K.; Grima, J.C.; Bowen, K.E.; Wadhwa, H.; Yang, P.; Rigo, F.; et al. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 2018, 173, 958–971. [Google Scholar] [PubMed]
- Zaepfel, B.L.; Zhang, Z.; Maulding, K.; Coyne, A.N.; Cheng, W.; Hayes, L.R.; Lloyd, T.E.; Sun, S.; Rothstein, J.D. UPF1 reduces C9orf72 HRE-induced neurotoxicity in the absence of nonsense-mediated decay dysfunction. Cell Rep. 2021, 34, 108925. [Google Scholar] [CrossRef]
- Wen, X.; Westergard, T.; Pasinelli, P.; Trotti, D. Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene. Neurosci. Lett. 2017, 636, 16–26. [Google Scholar] [CrossRef]
- Mizielinska, S.; Grönke, S.; Niccoli, T.; Ridler, C.E.; Clayton, E.L.; Devoy, A.; Moens, T.; Norona, F.E.; Woollacott, I.O.C.; Pietrzyk, J.; et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 2014, 345, 1192–1194. [Google Scholar] [CrossRef]
- Zamiri, B.; Reddy, K.; Macgregor, R.B., Jr.; Pearson, C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014, 289, 4653–4659. [Google Scholar] [CrossRef]
- West, R.J.; Sharpe, J.L.; Voelzmann, A.; Munro, A.L.; Hahn, I.; Baines, R.A.; Pickering-Brown, S. Co-expression of C9orf72 related dipeptide-repeats over 1000 repeat units reveals age- and combination-specific phenotypic profiles in Drosophila. Acta Neuropathol. Commun. 2020, 8, 158. [Google Scholar] [CrossRef]
- Nordin, A.; Akimoto, C.; Wuolikainen, A.; Alstermark, H.; Jonsson, P.; Birve, A.; Marklund, S.L.; Graffmo, K.S.; Forsberg, K.; Brännström, T.; et al. Extensive size variability of the GGGGCC expansion in C9orf72 in both neuronal and non-neuronal tissues in 18 patients with ALS or FTD. Hum. Mol. Genet. 2015, 24, 3133–3142. [Google Scholar] [CrossRef]
- Goodman, L.D.; Bonini, N.M. New Roles for Canonical Transcription Factors in Repeat Expansion Diseases. Trends Genet. 2020, 36, 81–92. [Google Scholar] [CrossRef]
- Wang, J.; Wang, B.; Zhou, T. The Advance on Frontotemporal Dementia (FTD)’s Neuropathology and Molecular Genetics. Mediat. Inflamm. 2022, 2022, 5003902. [Google Scholar] [CrossRef]
- Abramzon, Y.A.; Fratta, P.; Traynor, B.J.; Chia, R. The Overlapping Genetics of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Front. Neurosci. 2020, 14, 42. [Google Scholar] [CrossRef]
- Echeverria, G.V.; Cooper, T.A. RNA-binding proteins in microsatellite expansion disorders: Mediators of RNA toxicity. Brain Res. 2012, 1462, 100–111. [Google Scholar] [CrossRef]
- Gitler, A.D.; Tsuiji, H. There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 2016, 1647, 19–29. [Google Scholar] [CrossRef]
- Gijselinck, I.; Van Langenhove, T.; van der Zee, J.; Sleegers, K.; Philtjens, S.; Kleinberger, G.; Janssens, J.; Bettens, K.; Van Cauwenberghe, C.; Pereson, S.; et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012, 11, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Anderson, P.J.; Ivanov, P. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep. 2017, 21, 3573–3584. [Google Scholar] [CrossRef]
- Mehta, A.R.; Selvaraj, B.T.; Barton, S.K.; McDade, K.; Abrahams, S.; Chandran, S.; Smith, C.; Gregory, J.M. Improved detection of RNA foci in C9orf72 amyotrophic lateral sclerosis post-mortem tissue using BaseScope™ shows a lack of association with cognitive dysfunction. Brain Commun. 2020, 2, fcaa009. [Google Scholar] [CrossRef]
- Malnar, M.; Rogelj, B. SFPQ regulates the accumulation of RNA foci and dipeptide repeat proteins from the expanded repeat mutation in C9orf72. J. Cell Sci. 2021, 134, jcs256602. [Google Scholar] [CrossRef]
- Nedelsky, N.B.; Taylor, J.P. Bridging biophysics and neurology: Aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 272–286. [Google Scholar] [CrossRef]
- Wang, X.; Goodrich, K.J.; Conlon, E.G.; Gao, J.; Erbse, A.; Manley, J.L.; Cech, T.R. C9orf72 and triplet repeat disorder RNAs: G-quadruplex formation, binding to PRC2 and implications for disease mechanisms. RNA 2019, 25, 935–947. [Google Scholar] [CrossRef]
- Swinnen, B.; Bento-Abreu, A.; Gendron, T.F.; Boeynaems, S.; Bogaert, E.; Nuyts, R.; Timmers, M.; Scheveneels, W.; Hersmus, N.; Wang, J.; et al. A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. 2018, 135, 427–443. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, H.; Xia, Q.; Li, K.; Li, K.; Jiang, X.; Xu, G.; Wang, G.; Ying, Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015, 24, 2426–2441. [Google Scholar] [CrossRef]
- Mori, K.; Weng, S.-M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzschmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Y.-N.; Paul, T.; Periz, G.; Banco, M.T.; Ferré-D’amaré, A.R.; Rothstein, J.D.; Hayes, L.R.; Myong, S.; Wang, J. A Helicase Unwinds Hexanucleotide Repeat RNA G-Quadruplexes and Facilitates Repeat-Associated Non-AUG Translation. J. Am. Chem. Soc. 2021, 143, 7368–7379. [Google Scholar] [CrossRef]
- Flores, B.N.; Dulchavsky, M.E.; Krans, A.; Sawaya, M.R.; Paulson, H.L.; Todd, P.K.; Barmada, S.J.; Ivanova, M.I. Distinct C9orf72-Associated Dipeptide Repeat Structures Correlate with Neuronal Toxicity. PLoS ONE 2016, 11, e0165084. [Google Scholar] [CrossRef]
- Cheng, W.; Wang, S.; Mestre, A.A.; Fu, C.; Makarem, A.; Xian, F.; Hayes, L.R.; Lopez-Gonzalez, R.; Drenner, K.; Jiang, J.; et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun. 2018, 9, 51. [Google Scholar] [CrossRef]
- Jain, A.; Vale, R.D. RNA phase transitions in repeat expansion disorders. Nature 2017, 546, 243–247. [Google Scholar] [CrossRef]
- Penumutchu, S.R.; Chiu, L.-Y.; Meagher, J.L.; Hansen, A.L.; Stuckey, J.A.; Tolbert, B.S. Differential Conformational Dynamics Encoded by the Linker between Quasi RNA Recognition Motifs of Heterogeneous Nuclear Ribonucleoprotein H. J. Am. Chem. Soc. 2018, 140, 11661–11673. [Google Scholar] [CrossRef]
- Celona, B.; Dollen, J.V.; Vatsavayai, S.C.; Kashima, R.; Johnson, J.R.; Tang, A.A.; Hata, A.; Miller, B.L.; Huang, E.J.; Krogan, N.J.; et al. Suppression of C9orf72 RNA repeat-induced neurotoxicity by the ALS-associated RNA-binding protein Zfp106. eLife 2017, 6, e19032. [Google Scholar] [CrossRef]
- Donnelly, C.J.; Zhang, P.W.; Pham, J.T.; Haeusler, A.R.; Mistry, N.A.; Vidensky, S.; Daley, E.L.; Poth, E.M.; Hoover, B.; Fines, D.M.; et al. RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron 2013, 80, 415–428. [Google Scholar] [CrossRef]
- Xu, Z.; Poidevin, M.; Li, X.; Li, Y.; Shu, L.; Nelson, D.L.; Li, H.; Hales, C.M.; Gearing, M.; Wingo, T.S.; et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 7778–7783. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, Y.; Zhao, S.; Mao, H.; Wang, Z.; Li, H.; Xu, Z. Purα Repaired Expanded Hexanucleotide GGGGCC Repeat Noncoding RNA-Caused Neuronal Toxicity in Neuro-2a Cells. Neurotox. Res. 2018, 33, 693–701. [Google Scholar] [CrossRef]
- Ishiguro, A.; Lu, J.; Ozawa, D.; Nagai, Y.; Ishihama, A. ALS-linked FUS mutations dysregulate G-quadruplex-dependent liquid-liquid phase separation and liquid-to-solid transition. J. Biol. Chem. 2021, 297, 101284. [Google Scholar] [CrossRef]
- Murakami, T.; Qamar, S.; Lin, J.Q.; Schierle, G.S.K.; Rees, E.; Miyashita, A.; Costa, A.R.; Dodd, R.B.; Chan, F.T.S.; Michel, C.H.; et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 2015, 88, 678–690. [Google Scholar] [CrossRef]
- Cooper-Knock, J.; Walsh, M.J.; Higginbottom, A.; Robin Highley, J.; Dickman, M.J.; Edbauer, D.; Ince, P.G.; Wharton, S.B.; Wilson, S.A.; Kirby, J.; et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 2014, 137, 2040–2051. [Google Scholar] [CrossRef]
- Wang, Q.; Conlon, E.G.; Manley, J.L.; Rio, D.C. Widespread intron retention impairs protein homeostasis in C9orf72 ALS brains. Genome Res. 2020, 30, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Simone, R.; Balendra, R.; Moens, T.G.; Preza, E.; Wilson, K.M.; Heslegrave, A.; Woodling, N.S.; Niccoli, T.; Gilbert-Jaramillo, J.; Abdelkarim, S.; et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018, 10, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Millevoi, S.; Moine, H.; Vagner, S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA 2012, 3, 495–507. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, Y.; Gendron, T.F.; Bauer, P.O.; Chew, J.; Yang, W.-Y.; Fostvedt, E.; Jansen-West, K.; Belzil, V.V.; Desaro, P.; et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 2014, 83, 1043–1050. [Google Scholar] [CrossRef]
- Jaiswal, M.K. Riluzole and edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Meijboom, K.E.; Abdallah, A.; Fordham, N.P.; Nagase, H.; Rodriguez, T.; Kraus, C.; Gendron, T.F.; Krishnan, G.; Esanov, R.; Andrade, N.S.; et al. CRISPR/Cas9-mediated excision of ALS/FTD-causing hexanucleotide repeat expansion in C9ORF72 rescues major disease mechanisms in vivo and in vitro. Nat. Commun. 2022, 13, 6286. [Google Scholar] [CrossRef]
- Maity, A.; Winnerdy, F.R.; Chen, G.; Phan, A.T. Duplexes Formed by G4C2 Repeats Contain Alternate Slow- and Fast-Flipping G.G Base Pairs. Biochemistry 2021, 60, 1097–1107. [Google Scholar] [CrossRef]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.; Shaw, P.G.; Kim, M.-S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef]
- Brčić, J.; Plavec, J. G-quadruplex formation of oligonucleotides containing ALS and FTD related GGGGCC repeat. Front. Chem. Sci. Eng. 2016, 10, 222–237. [Google Scholar] [CrossRef]
- Fratta, P.; Mizielinska, S.; Nicoll, A.J.; Zloh, M.; Fisher, E.M.C.; Parkinson, G.; Isaacs, A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012, 2, srep01016. [Google Scholar] [CrossRef]
- Reddy, K.; Zamiri, B.; Stanley, S.Y.; Macgregor, R.B.; Pearson, C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013, 288, 9860–9866. [Google Scholar] [CrossRef]
- Božič, T.; Zalar, M.; Rogelj, B.; Plavec, J.; Šket, P. Structural Diversity of Sense and Antisense RNA Hexanucleotide Repeats Associated with ALS and FTLD. Molecules 2020, 25, 525. [Google Scholar] [CrossRef]
- Brčić, J.; Plavec, J. ALS and FTD linked GGGGCC-repeat containing DNA oligonucleotide folds into two distinct G-quadruplexes. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 1237–1245. [Google Scholar] [CrossRef]
- Thys, R.G.; Wang, Y.-H. DNA Replication Dynamics of the GGGGCC Repeat of the C9orf72 Gene. J. Biol. Chem. 2015, 290, 28953–28962. [Google Scholar] [CrossRef]
- Brčić, J.; Plavec, J. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015, 43, 8590–8600. [Google Scholar] [CrossRef]
- Brčić, J.; Plavec, J. NMR structure of a G-quadruplex formed by four d(G4C2) repeats: Insights into structural polymorphism. Nucleic Acids Res. 2018, 46, 11605–11617. [Google Scholar] [CrossRef]
- Geng, Y.; Liu, C.; Cai, Q.; Luo, Z.; Miao, H.; Shi, X.; Xu, N.; Fung, C.P.; Choy, T.T.; Yan, B.; et al. Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence. Nucleic Acids Res. 2021, 49, 5881–5890. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, W.; Chang, R.; Zhang, S.; Yang, G.; Zhao, G. Liquid-Liquid Phase Separation: Unraveling the Enigma of Biomolecular Condensates in Microbial Cells. Front. Microbiol. 2021, 12, 751880. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; van den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Bertrand, E.; Demongin, C.; Dobra, I.; Rengifo-Gonzalez, J.C.; Singatulina, A.S.; Sukhanova, M.V.; Lavrik, O.I.; Pastré, D.; Hamon, L. FUS fibrillation occurs through a nucleation-based process below the critical concentration required for liquid–liquid phase separation. Sci. Rep. 2023, 13, 7772. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Inami, H.; Oiwa, K.; Murata, Y.; Sakai, S.; Komine, O.; Sobue, A.; Iguchi, Y.; Katsuno, M.; Yamanaka, K. Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis. 2020, 11, 909. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J. Phase separation of RNA-binding proteins in physiology and disease: An introduction to the JBC Reviews thematic series. J. Biol. Chem. 2019, 294, 7113–7114. [Google Scholar] [CrossRef]
- Portz, B.; Lee, B.L.; Shorter, J. FUS and TDP-43 Phases in Health and Disease. Trends Biochem. Sci. 2021, 46, 550–563. [Google Scholar] [CrossRef]
- Maharana, S.; Wang, J.; Papadopoulos, D.K.; Richter, D.; Pozniakovsky, A.; Poser, I.; Bickle, M.; Rizk, S.; Guillén-Boixet, J.; Franzmann, T.M.; et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Markovtsov, V.; Nikolic, J.M.; Goldman, J.A.; Turck, C.W.; Chou, M.Y.; Black, D.L. Cooperative Assembly of an hnRNP Complex Induced by a Tissue-Specific Homolog of Polypyrimidine Tract Binding Protein. Mol. Cell. Biol. 2000, 20, 7463–7479. [Google Scholar] [CrossRef]
- Wang, E.; Cambi, F. Heterogeneous Nuclear Ribonucleoproteins H and F Regulate the Proteolipid Protein/DM20 Ratio by Recruiting U1 Small Nuclear Ribonucleoprotein through a Complex Array of G Runs. J. Biol. Chem. 2009, 284, 11194–11204. [Google Scholar] [CrossRef]
- Van Dusen, C.M.; Yee, L.; McNally, L.M.; McNally, M.T. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell. Biol. 2010, 30, 2552–2562. [Google Scholar] [CrossRef]
- Conlon, E.G.; Lu, L.; Sharma, A.; Yamazaki, T.; Tang, T.; Shneider, N.A.; Manley, J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife 2016, 5, e17820. [Google Scholar] [CrossRef]
- Liao, Y.-Z.; Ma, J.; Dou, J.-Z. The Role of TDP-43 in Neurodegenerative Disease. Mol. Neurobiol. 2022, 59, 4223–4241. [Google Scholar] [CrossRef]
- Lee, E.B.; Lee, V.M.-Y.; Trojanowski, J.Q. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2011, 13, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Conicella, A.E.; Zerze, G.H.; Mittal, J.; Fawzi, N.L. ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 2016, 24, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Chew, J.; Cook, C.; Gendron, T.F.; Jansen-West, K.; del Rosso, G.; Daughrity, L.M.; Castanedes-Casey, M.; Kurti, A.; Stankowski, J.N.; Disney, M.D.; et al. Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 2019, 14, 9. [Google Scholar] [PubMed]
- Solomon, D.A.; Stepto, A.; Au, W.H.; Adachi, Y.; Diaper, D.C.; Hall, R.; Rekhi, A.; Boudi, A.; Tziortzouda, P.; Lee, Y.B.; et al. A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain 2018, 141, 2908–2924. [Google Scholar] [CrossRef]
- Mori, K.; Lammich, S.; Mackenzie, I.R.A.; Forné, I.; Zilow, S.; Kretzschmar, H.; Edbauer, D.; Janssens, J.; Kleinberger, G.; Cruts, M.; et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013, 125, 413–423. [Google Scholar]
- Peters, O.M.; Cabrera, G.T.; Tran, H.; Gendron, T.F.; McKeon, J.E.; Metterville, J.; Weiss, A.; Wightman, N.; Salameh, J.; Kim, J.; et al. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron 2015, 88, 902–909. [Google Scholar]
- Lagier-Tourenne, C.; Polymenidou, M.; Cleveland, D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010, 19, R46–R64. [Google Scholar] [CrossRef]
- Kanai, Y.; Dohmae, N.; Hirokawa, N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 2004, 43, 513–525. [Google Scholar] [CrossRef]
- Yoshimura, A.; Fujii, R.; Watanabe, Y.; Okabe, S.; Fukui, K.; Takumi, T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 2006, 16, 2345–2351. [Google Scholar]
- Vance, C.; Rogelj, B.; Hortobágyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA Processing Protein, Cause Familial Amyotrophic Lateral Sclerosis Type 6. Science 2009, 323, 1208–1211. [Google Scholar]
- Niaki, A.G.; Sarkar, J.; Cai, X.; Rhine, K.; Vidaurre, V.; Guy, B.; Hurst, M.; Lee, J.C.; Koh, H.R.; Guo, L.; et al. Loss of Dynamic RNA Interaction and Aberrant Phase Separation Induced by Two Distinct Types of ALS/FTD-Linked FUS Mutations. Mol. Cell 2020, 77, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, H.; Bell, G.I. Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int. J. Biochem. Cell Biol. 2005, 37, 1421–1437. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Cannavino, J.; Li, H.; Anderson, K.M.; Nelson, B.R.; McAnally, J.; Bezprozvannaya, S.; Liu, Y.; Lin, W.; Liu, N.; et al. Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106. Proc. Natl. Acad. Sci. USA 2016, 113, E4494–E4503. [Google Scholar] [CrossRef] [PubMed]
- reibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.-H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015, 525, 129–133. [Google Scholar]
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2012, 45, 1121–1128. [Google Scholar] [CrossRef]
- Chen, C.-X.; Cho, D.-S.C.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar]
- White, M.K.; Johnson, E.M.; Khalili, K. Multiple roles for Pur-α in cellular and viral regulation. Cell Cycle 2009, 8, 414–420. [Google Scholar] [CrossRef]
- Johnson, E.M.; Kinoshita, Y.; Weinreb, D.B.; Wortman, M.J.; Simon, R.; Khalili, K.; Winckler, B.; Gordon, J. Role of Purα in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 2006, 83, 929–943. [Google Scholar] [CrossRef]
- Daniel, D.C.; Wortman, M.J.; Schiller, R.J.; Liu, H.; Gan, L.; Mellen, J.S.; Chang, C.F.; Gallia, G.L.; Rappaport, J.; Khalili, K.; et al. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Purα on DNA replication initiated at the JC virus origin. J. Gen. Virol. 2001, 82, 1543–1553. [Google Scholar] [CrossRef][Green Version]
- Daniel, D.C.; Johnson, E.M. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene 2018, 643, 133–143. [Google Scholar] [CrossRef]
- Khalili, K.; Del Valle, L.; Muralidharan, V.; Gault, W.J.; Darbinian, N.; Otte, J.; Meier, E.; Johnson, E.M.; Daniel, D.C.; Kinoshita, Y.; et al. Purα is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Biol. 2003, 23, 6857–6875. [Google Scholar] [CrossRef]
- Barbe, M.F.; Krueger, J.J.; Loomis, R.; Otte, J.; Gordon, J. Memory Deficits, Gait Ataxia and Neuronal Loss in the Hippocampus and Cerebellum in Mice That Are Heterozygous for Pur-Alpha. Neuroscience 2016, 337, 177–190. [Google Scholar] [CrossRef]
- Rossi, S.; Serrano, A.; Gerbino, V.; Giorgi, A.; Di Francesco, L.; Nencini, M.; Bozzo, F.; Schininà, M.E.; Bagni, C.; Cestra, G.; et al. Nuclear accumulation of mRNAs underlies G4C2 repeat-induced translational repression in a cellular model of C9orf72 ALS. J. Cell Sci. 2015, 128, 1787–1799. [Google Scholar] [CrossRef]
- Daigle, J.G.; Krishnamurthy, K.; Ramesh, N.; Casci, I.; Monaghan, J.; McAvoy, K.; Godfrey, E.W.; Daniel, D.C.; Johnson, E.M.; Monahan, Z.; et al. Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity. Acta Neuropathol. 2016, 131, 605–620. [Google Scholar] [CrossRef]
- Di Salvio, M.; Piccinni, V.; Gerbino, V.; Mantoni, F.; Camerini, S.; Lenzi, J.; Rosa, A.; Chellini, L.; Loreni, F.; Carrì, M.T.; et al. Pur-alpha functionally interacts with FUS carrying ALS-associated mutations. Cell Death Dis. 2015, 6, e1943. [Google Scholar] [CrossRef]
- Muralidharan, V.; Sweet, T.; Nadraga, Y.; Amini, S.; Khalili, K. Regulation of Purα gene transcription: Evidence for autoregulation of Purα promoter. J. Cell. Physiol. 2001, 186, 406–413. [Google Scholar] [CrossRef]
- Martino, L.; Pagano, B.; Fotticchia, I.; Neidle, S.; Giancola, C. Shedding Light on the Interaction between TMPyP4 and Human Telomeric Quadruplexes. J. Phys. Chem. B 2009, 113, 14779–14786. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.J.; Wingate, K.L.; Silwal, J.; Leeper, T.C.; Basu, S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012, 40, 4137–4145. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Wheelhouse, R.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644. [Google Scholar]
- Mulholland, K.; Sullivan, H.J.; Garner, J.; Cai, J.; Chen, B.; Wu, C. Three-Dimensional Structure of RNA Monomeric G-Quadruplex Containing ALS and FTD Related G4C2 Repeat and Its Binding with TMPyP4 Probed by Homology Modeling based on Experimental Constraints and Molecular Dynamics Simulations. ACS Chem. Neurosci. 2020, 11, 57–75. [Google Scholar] [CrossRef]
- Disney, M.D.; Liu, B.; Yang, W.-Y.; Sellier, C.; Tran, T.; Charlet-Berguerand, N.; Childs-Disney, J.L. A small molecule that targets r(CGG)exp and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem. Biol. 2012, 7, 1711–1718. [Google Scholar] [CrossRef]
- Ursu, A.; Wang, K.W.; Bush, J.A.; Choudhary, S.; Chen, J.L.; Baisden, J.T.; Zhang, Y.J.; Gendron, T.F.; Petrucelli, L.; Yildirim, I.; et al. Structural Features of Small Molecules Targeting the RNA Repeat Expansion That Causes Genetically Defined ALS/FTD. ACS Chem. Biol. 2020, 15, 3112–3123. [Google Scholar] [CrossRef] [PubMed]
- Ursu, A.; Baisden, J.T.; Bush, J.A.; Taghavi, A.; Choudhary, S.; Zhang, Y.J.; Gendron, T.F.; Petrucelli, L.; Yildirim, I.; Disney, M.D. A Small Molecule Exploits Hidden Structural Features within the RNA Repeat Expansion That Causes c9ALS/FTD and Rescues Pathological Hallmarks. ACS Chem. Neurosci. 2021, 12, 4076–4089. [Google Scholar] [CrossRef]
- Guo, Q.; Lehmer, C.; Martínez-Sánchez, A.; Rudack, T.; Beck, F.; Hartmann, H.; Pérez-Berlanga, M.; Frottin, F.; Hipp, M.S.; Hartl, F.U.; et al. In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 2018, 172, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yang, Y.; Zhao, Y.; Li, X.; Xue, Y.; Wang, S. High-resolution solid-state NMR spectroscopy of hydrated non-crystallized RNA. Chem. Commun. 2019, 55, 13991–13994. [Google Scholar] [CrossRef] [PubMed]
- Marchanka, A.; Simon, B.; Althoff-Ospelt, G.; Carlomagno, T. RNA structure determination by solid-state NMR spectroscopy. Nat. Commun. 2015, 6, 7024. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, X.; Wen, Z.; Zou, M.; Yu, G.; Liu, X.; Mao, J.; Zhang, L.; Xue, Y.; Fu, R.; et al. Dynamics of base pairs with low stability in RNA by solid-state nuclear magnetic resonance exchange spectroscopy. iScience 2022, 25, 105322. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, S. RNA Characterization by Solid-State NMR Spectroscopy. Chemistry 2018, 24, 8698–8707. [Google Scholar] [CrossRef]
- Yang, Y.; Xiang, S.; Liu, X.; Pei, X.; Wu, P.; Gong, Q.; Li, N.; Baldus, M.; Wang, S. Proton-detected solid-state NMR detects the inter-nucleotide correlations and architecture of dimeric RNA in microcrystals. Chem. Commun. 2017, 53, 12886–12889. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhao, X.; He, J.; Wang, S.; Shen, X.; Liu, Q.; Wang, S. Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners. Molecules 2023, 28, 5801. https://doi.org/10.3390/molecules28155801
Liu X, Zhao X, He J, Wang S, Shen X, Liu Q, Wang S. Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners. Molecules. 2023; 28(15):5801. https://doi.org/10.3390/molecules28155801
Chicago/Turabian StyleLiu, Xiaole, Xinyue Zhao, Jinhan He, Sishi Wang, Xinfei Shen, Qingfeng Liu, and Shenlin Wang. 2023. "Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners" Molecules 28, no. 15: 5801. https://doi.org/10.3390/molecules28155801
APA StyleLiu, X., Zhao, X., He, J., Wang, S., Shen, X., Liu, Q., & Wang, S. (2023). Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners. Molecules, 28(15), 5801. https://doi.org/10.3390/molecules28155801