Comparative Transcriptomic Analysis of the Metabolism of Betalains and Flavonoids in Red Amaranth Hypocotyl under Blue Light and Dark Conditions
Abstract
1. Introduction
2. Results
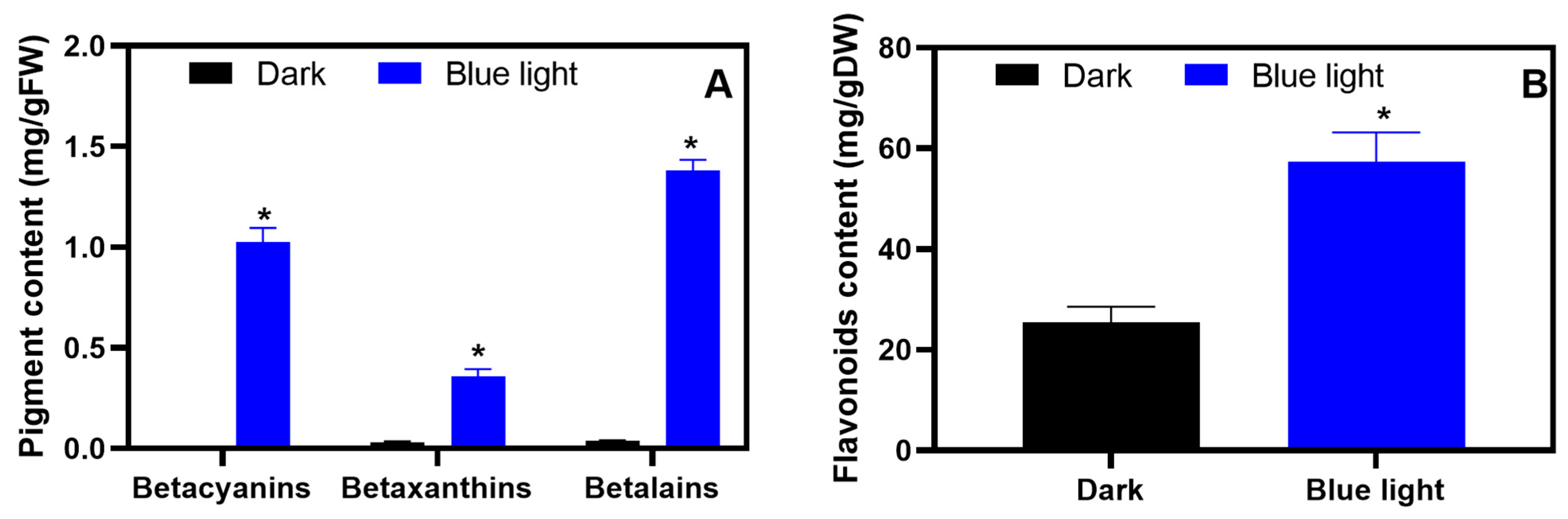
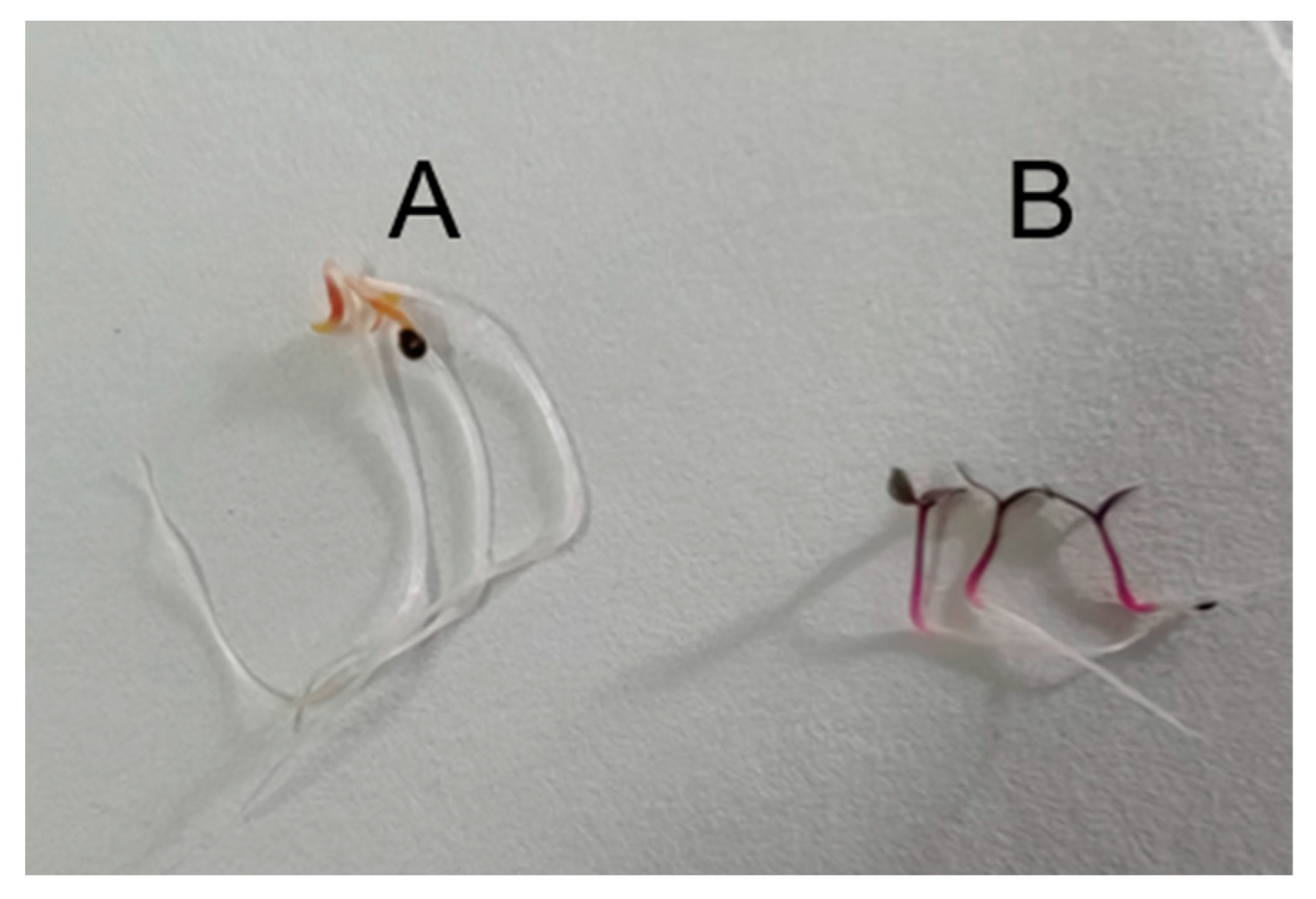
2.1. Determination of Betalain and Flavonoid Content in Amaranth Hypocotyls
2.2. Transcriptome Assembly for A. tricolor
2.3. Alternative Splicing
2.4. Prediction of Novel mRNAs
2.5. Annotation Analysis of All Protein-Coding Genes
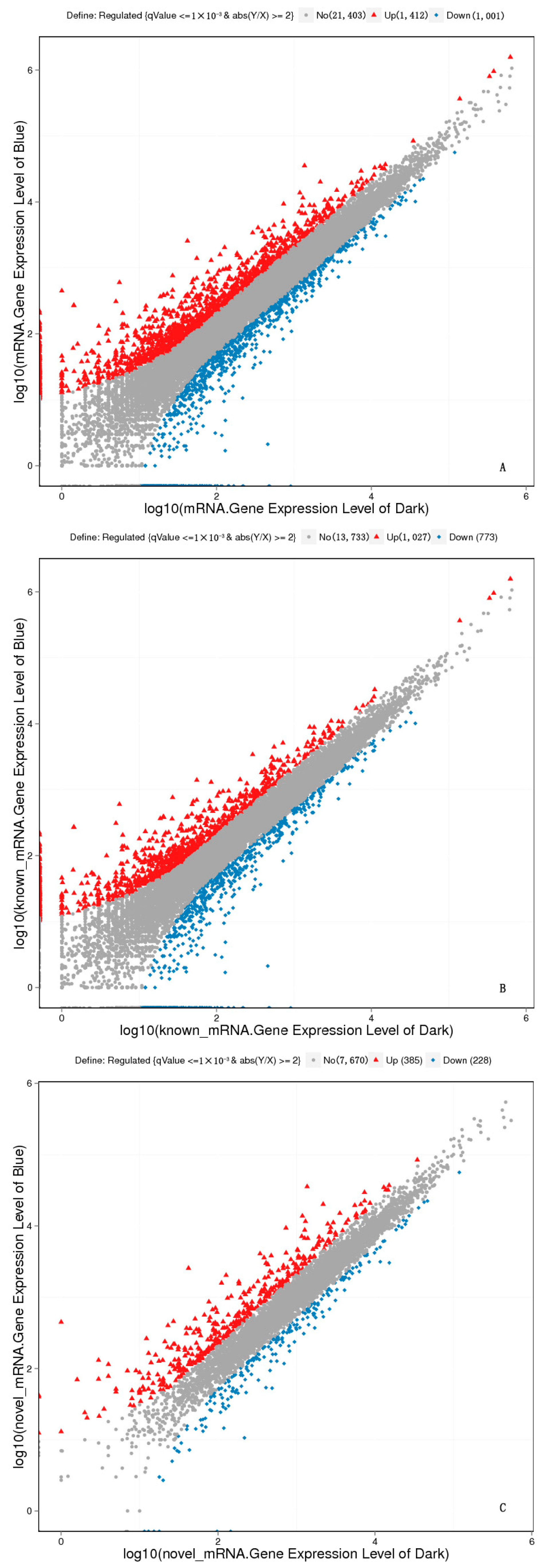
2.6. Analysis of Differently Expressed Genes Based on FPKM
2.7. Analysis of DEGs between Dark and Blue Light
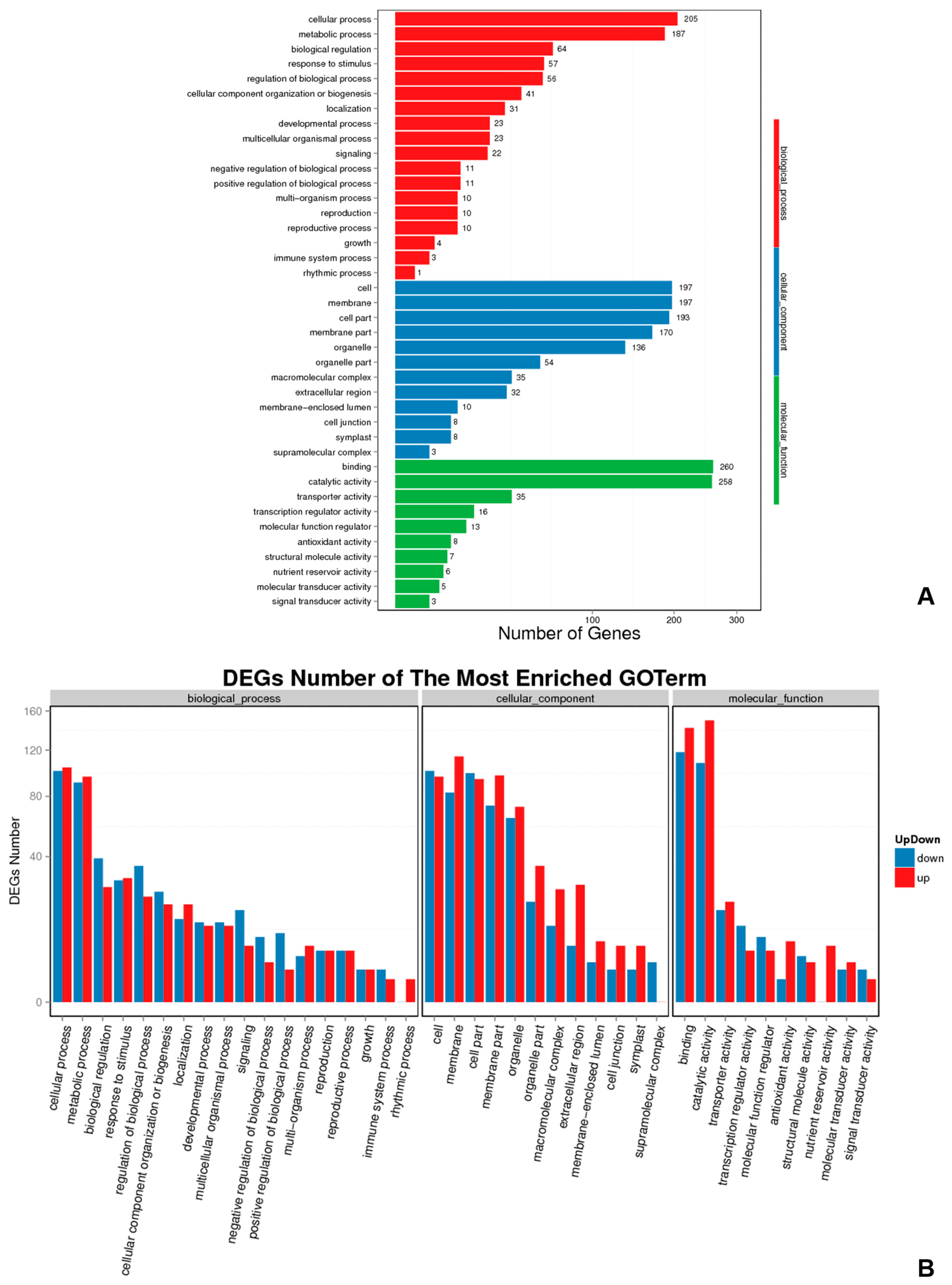
2.8. Functional Enrichment Analysis of DEGs
2.9. qRT-PCR Analysis
3. Discussion
3.1. Evaluation of the Amaranth Transcriptome
3.2. Blue Light Is Beneficial for Flavonoid and Betalain Accumulation in Amaranth
3.3. Transcriptional Downregulation of Genes Related to Anthocyanin Synthesis Might Result in Loss of Anthocyanins in A. tricolor
3.4. Blue Light Might Promote DELLA Protein Accumulation for the Regulation of Betalains and Hypocotyl Elongation in Amaranth
4. Materials and Methods
4.1. Materials and Treatment
4.2. Total RNA Extraction and Quality Detection
4.3. Construction, Detection, and Sequencing of Transcriptome Libraries
4.4. Sequencing Data Assembly
4.5. Prediction of Coding Ability and Annotation Analysis
4.6. Analysis of Expression Levels Based on FPKM
4.7. Annotation Analysis of Differentially Expressed Genes (DEGs)
4.8. Quantitative Real-Time Polymerase Chain Reaction Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| TSS | Alternative transcription start site |
| TTS | alternative transcription termination site |
| AE | alternative exon ends |
| ANS | anthocyanidin synthase |
| CHS | chalcone synthase |
| CYP76AD1/5/6 | cytochrome P450 |
| DEGs | differentially expressed genes |
| SKIP | exon skipping |
| F3H | Flavanone 3-Hydroxylase |
| GO | Gene Ontology |
| DODA | l-dihydroxyphenylalanine 4,5-dioxygenase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPO | polyphenol oxidase |
References
- Hilou, A.; Ouedraogo, I.; Sombié, P.; Guenné, S.; Paré, D.; Compaoré, M. Leafy amaranthus consumption patterns in Ouagadougou, Burkina Faso. Afr. J. Food Agric. Nutr. Dev. 2016, 16, 11248–11264. [Google Scholar] [CrossRef]
- Li, H.Y.; Deng, Z.Y.; Liu, R.H.; Zhu, H.H.; Draves, J.; Marcone, M.; Sun, Y.; Tsao, R. Characterization of phenolics, betacyanins and antioxidant activities of the seed, leaf, sprout, flower and stalk extracts of three Amaranthus species. J. Food Compos. Anal. 2015, 37, 75–81. [Google Scholar] [CrossRef]
- Rastogi, A.; Shukla, S. Amaranth: A new millennium crop of nutraceutical values. Crit. Rev. Food Sci. Nutr. 2013, 53, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, V.; Dhargalkar, S.; Vaingankar, J.; Kevat, N. Pigment Rich Amaranth by Tri-Stimulus Colorimetry and Progeny Test. Natl. Acad. Sci. Lett. 2016, 39, 411–415. [Google Scholar] [CrossRef]
- Noori, M.; Talebi, M.; Nasiri, Z. Seven Amaranthus L. (Amaranthaceae) taxa flavonoid compounds from Tehran Province, Iran. Int. J. Mod. Bot. 2015, 5, 9–17. [Google Scholar]
- Aneja, S.; Vats, M.; Aggarwal, S.; Sardana, S. Phytochemistry and hepatoprotective activity of aqueous extract of Amaranthus tricolor Linn. roots. J. Ayurveda Integr. Med. 2013, 4, 211. [Google Scholar] [CrossRef]
- Umakanta, S.; Md, T.I.; Md, G.R.; Shinya, O. Variability in total antioxidant capacity, antioxidant leaf pigments and foliage yield of vegetable amaranth. J. Integr. Agriculture. 2018, 17, 1145–1153. [Google Scholar]
- Liu, S.; Zheng, X.; Pan, J.; Peng, L.; Cheng, C.; Wang, X.; Zhao, C.; Zhang, Z.; Lin, Y.; Xuhan, X. RNA-sequencing analysis reveals betalains metabolism in the leaf of Amaranthus tricolor L. PLoS ONE 2019, 14, e0216001. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, Y.; Martin-Cabrejas, M.A.; de Mejia, E.G. Phenolic compounds in fruits and beverages consumed as part of the mediterranean diet: Their role in prevention of chronic diseases. Phytochem. Rev. 2016, 15, 405–423. [Google Scholar] [CrossRef]
- Lewandowska, H.; Kalinowska, M.; Lewandowski, W.; Stepkowski, T.M.; Brzoska, K. The role of natural polyphenols in cell signaling and cytoprotection against cancer development. J. Nutr. Biochem. 2016, 32, 1–19. [Google Scholar] [CrossRef]
- Wong, J.Y.; Matanjun, P.; Ooi, Y.B.H.; Chia, K.F. Evaluation of Antioxidant Activities in Relation to Total Phenolics and Flavonoids Content of Selected Malaysian Wild Edible Plants by Multivariate Analysis. Int. J. Food Prop. 2014, 17, 1763–1778. [Google Scholar] [CrossRef]
- Venskutonis, P.R.; Kraujalis, P. Nutritional Components of Amaranth Seeds and Vegetables: A Review on Composition, Properties, and Uses. Compr. Rev. Food Sci. Food Saf. 2013, 12, 381–412. [Google Scholar] [CrossRef]
- Adefegha, S.A.; Oboh, G. Enhancement of total phenolics and antioxidant properties of some tropical green leafy vegetables by steam cooking. J. Food Process. Preserv. 2011, 35, 615–622. [Google Scholar] [CrossRef]
- Jiménez-Aguilar, D.M.; Grusak, M.A. Minerals, vitamin C, phenolics, flavonoids and antioxidant activity of Amaranthus leafy vegetables. J. Food Compos. Anal. 2017, 58, 33–39. [Google Scholar] [CrossRef]
- Obadiah, F.; Patrick, J. Lead and cadmium concentrations in solanum lycopasicum and amaranthus indica commonly grown in plateau state, central nigeria. Int. J. Sci. Appl. Res. 2017, 2, 33–40. [Google Scholar]
- Kachiguma, N.A.; Mwase, W.; Maliro, M.; Damaliphetsa, A. Chemical and mineral composition of amaranth (Amaranthus L.) species collected from central Malawi. J. Food Res. 2015, 4, 92. [Google Scholar] [CrossRef]
- Shukla, S.; Pandey, V.; Pachauri, G.; Dixit, B.S.; Banerji, R.; Singh, S.P. Nutritional contents of different foliage cuttings of vegetable amaranth. Plant Foods Hum. Nutr. 2003, 58, 1–8. [Google Scholar] [CrossRef]
- Fernando, T.; Bean, G. Fatty acids and sterols of Amaranthus tricolor L. Food Chem. 1984, 15, 233–237. [Google Scholar] [CrossRef]
- Macdonald, H.M.; New, S.A.; Golden, M.H.; Campbell, M.K.; Reid, D.M. Nutritional associations with bone loss during the menopausal transition: Evidence of a beneficial effect of calcium, alcohol, and fruit and vegetable nutrients and of a detrimental effect of fatty acids. Am. J. Clin. Nutr. 2004, 79, 155–165. [Google Scholar] [CrossRef]
- Devaraj, V.C.; Krishna, B.G. Gastric antisecretory and cytoprotective effects of leaf extracts of Amaranthus tricolor Linn. in rats. J. Chin. Integr. Med. 2011, 9, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, V.C.; Krishna, B.G. Antiulcer activity of a polyherbal formulation (PHF) from Indian medicinal plants. Chin. J. Nat. Med. 2013, 11, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Yelisyeyeva, O.; Semen, K.; Zarkovic, N.; Kaminskyy, D.; Lutsyk, O.; Rybalchenko, V.K. Activation of aerobic metabolism by Amaranth oil improves heart rate variability both in athletes and patients with type 2 diabetes mellitus. Arch. Physiol. Biochem. 2012, 118, 47–57. [Google Scholar] [CrossRef]
- Peter, K.; Gandhi, P. Rediscovering the therapeutic potential of Amaranthus species: A review. Egypt. J. Basic Appl. Sci. 2017, 4, 196–205. [Google Scholar] [CrossRef]
- Wang, M.; Lopez-Nieves, S.; Goldman, I.L.; Maeda, H.A. Limited Tyrosine Utilization Explains Lower Betalain Contents in Yellow than Red Table Beet Genotypes. J. Agric. Food Chem. 2017, 65, 4305–4313. [Google Scholar] [CrossRef]
- Gins, M.S.; Gins, V.K.; Kononkov, P.F. Change in the biochemical composition of amaranth leaves during selection for increased amaranthine content. Appl. Biochem. Microbiol. 2002, 38, 474–479. [Google Scholar] [CrossRef]
- Schwinn, K.E. The dope on l-DOPA formation for betalain pigments. New Phytol. 2016, 210, 6–9. [Google Scholar] [CrossRef]
- Yu, Z.H.; Han, Y.N.; Xiao, X.G. A PPO Promoter from Betalain-Producing Red Swiss Chard, Directs Petiole- and Root-Preferential Expression of Foreign Gene in Anthocyanins-Producing Plants. Int. J. Mol. Sci. 2015, 16, 27032–27043. [Google Scholar] [CrossRef] [PubMed]
- Hatlestad, G.J.; Lloyd, A. The betalain secondary metabolic network. In Pigments in Fruits and Vegetables; Springer: Berlin/Heidelberg, Germany, 2015; pp. 127–140. [Google Scholar]
- Hatlestad, G.J.; Akhavan, N.A.; Sunnadeniya, R.M.; Elam, L.; Cargile, S.; Hembd, A.; Gonzalez, A.; McGrath, J.M.; Lloyd, A.M. The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 2015, 47, 92–96. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Ozeki, Y.; Sasaki, N. Biosynthesis and Regulation of Betalains in Red Beet. In Red Beet Biotechnology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 45–54. [Google Scholar]
- Gandía-Herrero, F.; García-Carmona, F. Biosynthesis of betalains: Yellow and violet plant pigments. Trends Plant Sci. 2013, 18, 334–343. [Google Scholar] [CrossRef]
- Brockington, S.F.; Yang, Y.; Gandia Herrero, F.; Covshoff, S.; Hibberd, J.M.; Sage, R.F.; Wong, G.K.; Moore, M.J.; Smith, S.A. Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytol. 2015, 207, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Sunnadeniya, R.; Bean, A.; Brown, M.; Akhavan, N.; Hatlestad, G.; Gonzalez, A.; Symonds, V.V.; Lloyd, A. Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS ONE 2016, 11, e0149417. [Google Scholar] [CrossRef]
- Xu, S.; Huang, Q.; Lin, C.; Huang, Y.; Hu, O. Comparison between betalain and flavonol accumulation in tricolor Bougainvillea peruviana ‘Thimma’ based on transcriptome. Bull. Bot. Res. 2017, 37, 249–258. [Google Scholar]
- Roberts, M.F.; Strack, D.; Wink, M. Biosynthesis of alkaloids and betalains. In Annual Plant Reviews Volume 40: Biochemistry of Plant Secondary Metabolism, 2nd ed.; Blackwell Publishing Ltd.: Oxford, UK, 2010; pp. 20–91. [Google Scholar]
- Brockington, S.F.; Walker, R.H.; Glover, B.J.; Soltis, P.S.; Soltis, D.E. Complex pigment evolution in the Caryophyllales. New Phytol. 2011, 190, 854–864. [Google Scholar] [CrossRef]
- Harris, N.N.; Javellana, J.; Davies, K.M.; Lewis, D.H.; Jameson, P.E.; Deroles, S.C.; Calcott, K.E.; Gould, K.S.; Schwinn, K.E. Betalain production is possible in anthocyanin-producing plant species given the presence of DOPA-dioxygenase and L-DOPA. BMC Plant Biol. 2012, 12, 34. [Google Scholar] [CrossRef]
- Kumar, S.S.; Manoj, P.; Shetty, N.P.; Prakash, M.; Giridhar, P. Characterization of major betalain pigments -gomphrenin, betanin and isobetanin from Basella rubra L. fruit and evaluation of efficacy as a natural colourant in product (ice cream) development. J. Food Sci. Tech. 2015, 52, 4994–5002. [Google Scholar] [CrossRef]
- Sanchez-Gonzalez, N.; Jaime-Fonseca, M.R.; San Martin-Martinez, E.; Zepeda, L.G. Extraction, Stability, and Separation of Betalains from Opuntia joconostle cv. Using Response Surface Methodology. J. Agric. Food Chem. 2013, 61, 11995–12004. [Google Scholar] [CrossRef]
- Girod, P.-A.; Zryd, J.-P. Biogenesis of betalains: Purification and partial characterization of DOPA 4, 5-dioxygenase from Amanita muscaria. Phytochemistry 1991, 30, 169–174. [Google Scholar] [CrossRef]
- da Silva, J.A.; da Silva, J.J.F.; Pombeiro, A.J. Amavadin, a vanadium natural complex: Its role and applications. Coord. Chem. Rev. 2013, 257, 2388–2400. [Google Scholar] [CrossRef]
- Li, G.; Meng, X.; Zhu, M.; Li, Z. Research Progress of Betalain in Response to Adverse Stresses and Evolutionary Relationship Compared with Anthocyanin. Molecules 2019, 24, 3078. [Google Scholar] [CrossRef] [PubMed]
- Polturak, G.; Heinig, U.; Grossman, N.; Battat, M.; Leshkowitz, D.; Malitsky, S.; Rogachev, I.; Aharoni, A. Transcriptome and Metabolic Profiling Provides Insights into Betalain Biosynthesis and Evolution in Mirabilis jalapa. Mol. Plant 2018, 11, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wen, K.; Ruan, X.; Zhao, Y.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed]
- Theis, N.; Lerdau, M.T. The Evolution of Function in Plant Secondary Metabolites. Int. J. Plant Sci. 2003, 164, S93–S102. [Google Scholar] [CrossRef]
- Coelho, G.C.; Rachwal, M.F.G.; Dedecek, R.A.; Curcio, G.R.; Nietsche, K.; Schenkel, E.P. Effect of light intensity on methylxanthine contents of Ilex paraguariensis A. St. Hil. Biochem. Syst. Ecol. 2007, 35, 75–80. [Google Scholar] [CrossRef]
- Cai, Z.Q.; Wang, W.H.; Yang, J.; Cai, C.T. Growth, photosynthesis and root reserpine concentrations of two Rauvolfia species in response to a light gradient. Ind. Crops Prod. 2009, 30, 220–226. [Google Scholar] [CrossRef]
- Becker, C.; Klaering, H.-P.; Schreiner, M.; Kroh, L.W.; Krumbein, A. Unlike Quercetin Glycosides, Cyanidin Glycoside in Red Leaf Lettuce Responds More Sensitively to Increasing Low Radiation Intensity before than after Head Formation Has Started. J. Agric. Food Chem. 2014, 62, 6911–6917. [Google Scholar] [CrossRef]
- Shi, L.; Cao, S.; Chen, W.; Yang, Z. Blue light induced anthocyanin accumulation and expression of associated genes in Chinese bayberry fruit. Entia Hortic. 2014, 179, 98–102. [Google Scholar] [CrossRef]
- Gitelson, A.; Chivkunova, O.; Zhigalova, T.; Solovchenko, A. In situ optical properties of foliar flavonoids: Implication for non-destructive estimation of flavonoid content. J. Plant Physiol. 2017, 218, 258. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.A.; Wagenmakers, P.S.; de Jager, A. Effects of light on flavonoid and chlorogenic acid levels in the skin of ‘Jonagold’ apples. Sci. Hortic. 2001, 88, 289–298. [Google Scholar] [CrossRef]
- Giliberto, L.; Perrotta, G.; Pallara, P.; Weller, J.L.; Fraser, P.D.; Bramley, P.M.; Fiore, A.; Tavazza, M.; Giuliano, G. Manipulation of the Blue Light Photoreceptor Cryptochrome 2 in Tomato Affects Vegetative Development, Flowering Time, and Fruit Antioxidant Content. Plant Physiol. 2005, 137, 199–208. [Google Scholar] [CrossRef]
- Narukawa, M.; Watanabe, K.; Inoue, Y. Light-induced root hair formation in lettuce (Lactuca sativa L. cv. Grand Rapids) roots at low pH is brought by chlorogenic acid synthesis and sugar. J. Plant Res. 2010, 123, 789–799. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhang, M.; Liang, E.; Pan, J.; Liu, S. Effects of different light qualities on callus growth and synthesis of betalain and carotenoid of Amaranthus tricolor L. J. Henan Agric. Sci. 2018, 47, 114–119. [Google Scholar]
- Yu, L.-J.; Luo, Y.-F.; Liao, B.; Xie, L.-J.; Chen, L.; Xiao, S.; Li, J.-T.; Hu, S.-N.; Shu, W.-S. Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 2012, 195, 97–112. [Google Scholar] [CrossRef]
- Suzuki, J.Y.; Amore, T.D.; Calla, B.; Palmer, N.A.; Scully, E.D.; Sattler, S.E.; Sarath, G.; Lichty, J.S.; Myers, R.Y.; Keith, L.M.; et al. Organ-specific transcriptome profiling of metabolic and pigment biosynthesis pathways in the floral ornamental progenitor species Anthurium amnicola Dressler. Sci. Rep. 2017, 7, 1596. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Liu, J.H. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. Bmc Genom. 2015, 16, 555. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, K.; Sun, L.; Qin, Q.; Sun, Y.; Pan, J.; Xue, Y. Morphological and Transcriptome Analysis of Wheat Seedlings Response to Low Nitrogen Stress. Plants 2019, 8, 98. [Google Scholar] [CrossRef]
- Deng, S.; Ma, J.; Zhang, L.; Chen, F.; Ma, L. De novo transcriptome sequencing and gene expression profiling of Magnolia wufengensis in response to cold stress. BMC Plant Biol. 2019, 19, 321. [Google Scholar] [CrossRef]
- Shengcai, L.; Huaqin, K.; Zhongxiong, L.; Junwen, W. Transcriptome Analysis by Illumina High-Throughout Paired-End Sequencing Reveals the Complexity of Differential Gene Expression during In Vitro Plantlet Growth and Flowering in Amaranthus tricolor L. PLoS ONE 2014, 9, e100919. [Google Scholar]
- Lightfoot, D.J.; Jarvis, D.E.; Ramaraj, T.; Lee, R.; Jellen, E.N.; Maughan, P.J. Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 2017, 15, 74. [Google Scholar] [CrossRef]
- Olszewski, N.E.; Sun, T.; Gubler, F. Gibberellin Signaling: Biosynthesis, Catabolism, and Response Pathways. Plant Cell 2002, 14, 561–580. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, X.; Foo, E.; Symons, G.M.; Lopez, J.; Bendehakkalu, K.T.; Xiang, J.; Weller, J.L.; Liu, X.; Reid, J.B.; et al. A Study of Gibberellin Homeostasis and Cryptochrome-Mediated Blue Light Inhibition of Hypocotyl Elongation. Plant Physiol. 2007, 145, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Junfei, P.; Liyun, P.; Chunli, Z.; Xiao, W.; Zihao, Z.; Yukun, C.; Zhongxiong, L.; Shengcai, L. Cloning of amaARF6 and its response to exogenous hormones treatment in amaranth seedlings. J. Northeast. Agric. Univ. 2018, 49, 24–32. [Google Scholar]
- Zhao, C.; Wang, X.; Pan, J.; Peng, L.; Lai, Z.; Liu, S. Cloning and Expression Analysis of AtGAI Gene in Amaranthus tricolor L. Acta Bot. Boreali-Occident. Sin. 2019, 39, 199–209. [Google Scholar]
- Sun, H.; Cui, H.; Zhang, J.; Kang, J.; Wang, Z.; Li, M.; Yi, F.; Yang, Q.; Long, R. Gibberellins Inhibit Flavonoid Biosynthesis and Promote Nitrogen Metabolism in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 9291. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiao, H.; Chen, G.; Zhao, X.; Huang, C.; Chen, C.; Wang, F. Isolation of High-Quality RNA from Reaumuria soongorica, a Desert Plant Rich in Secondary Metabolites. Mol. Biotechnol. 2011, 48, 165–172. [Google Scholar] [CrossRef]
- Vashisth, T.; Johnson, L.K.; Malladi, A. An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep. 2011, 30, 2167–2176. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 26 December 2018).
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Wang, X.; Peng, L. Comparative Transcriptomic Analysis of the Metabolism of Betalains and Flavonoids in Red Amaranth Hypocotyl under Blue Light and Dark Conditions. Molecules 2023, 28, 5627. https://doi.org/10.3390/molecules28155627
Liu S, Wang X, Peng L. Comparative Transcriptomic Analysis of the Metabolism of Betalains and Flavonoids in Red Amaranth Hypocotyl under Blue Light and Dark Conditions. Molecules. 2023; 28(15):5627. https://doi.org/10.3390/molecules28155627
Chicago/Turabian StyleLiu, Shengcai, Xiao Wang, and Liyun Peng. 2023. "Comparative Transcriptomic Analysis of the Metabolism of Betalains and Flavonoids in Red Amaranth Hypocotyl under Blue Light and Dark Conditions" Molecules 28, no. 15: 5627. https://doi.org/10.3390/molecules28155627
APA StyleLiu, S., Wang, X., & Peng, L. (2023). Comparative Transcriptomic Analysis of the Metabolism of Betalains and Flavonoids in Red Amaranth Hypocotyl under Blue Light and Dark Conditions. Molecules, 28(15), 5627. https://doi.org/10.3390/molecules28155627







