Structure-Based Design of Novel MAO-B Inhibitors: A Review
Abstract
1. Introduction
1.1. Types of Molecular Docking
1.2. Docking Software
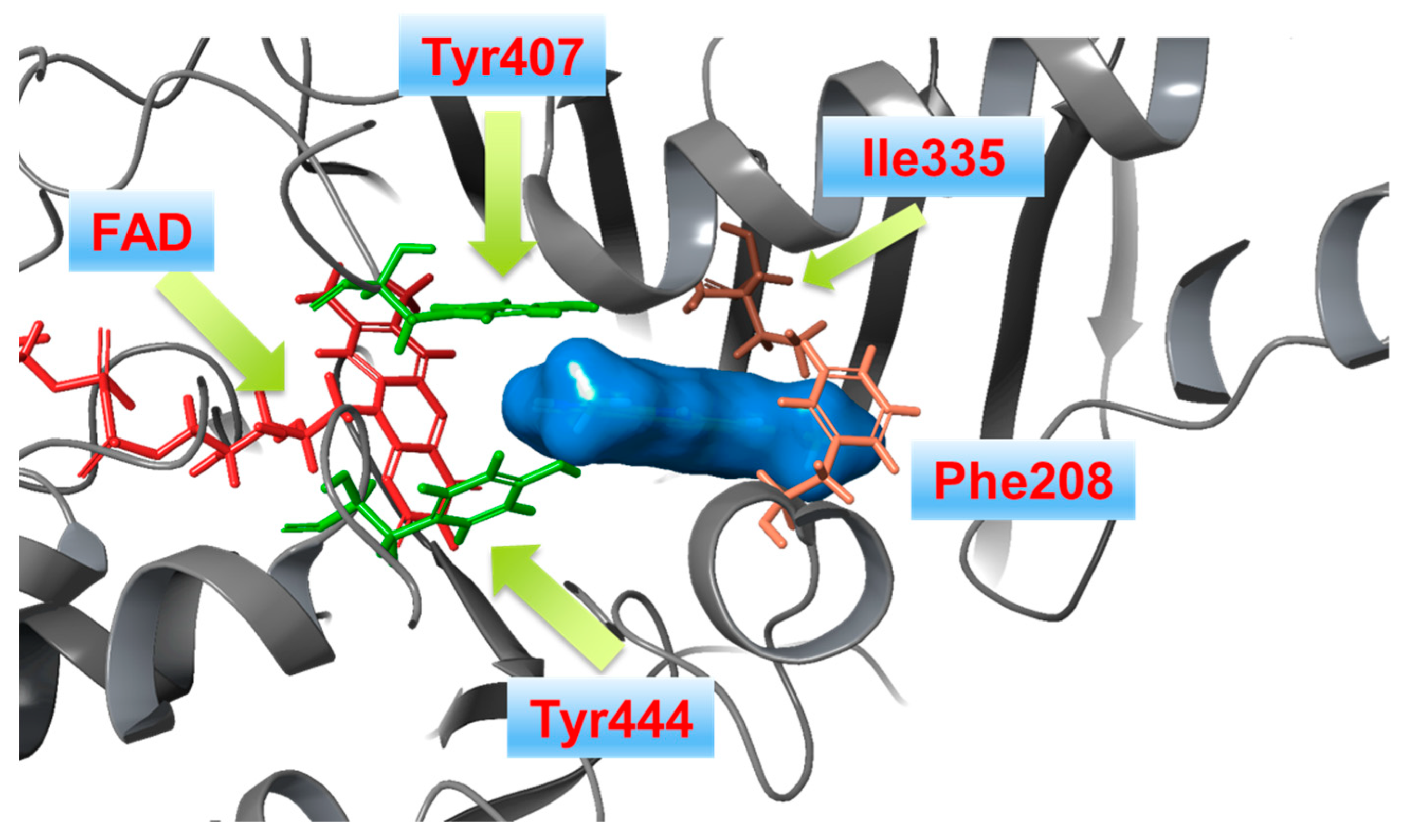
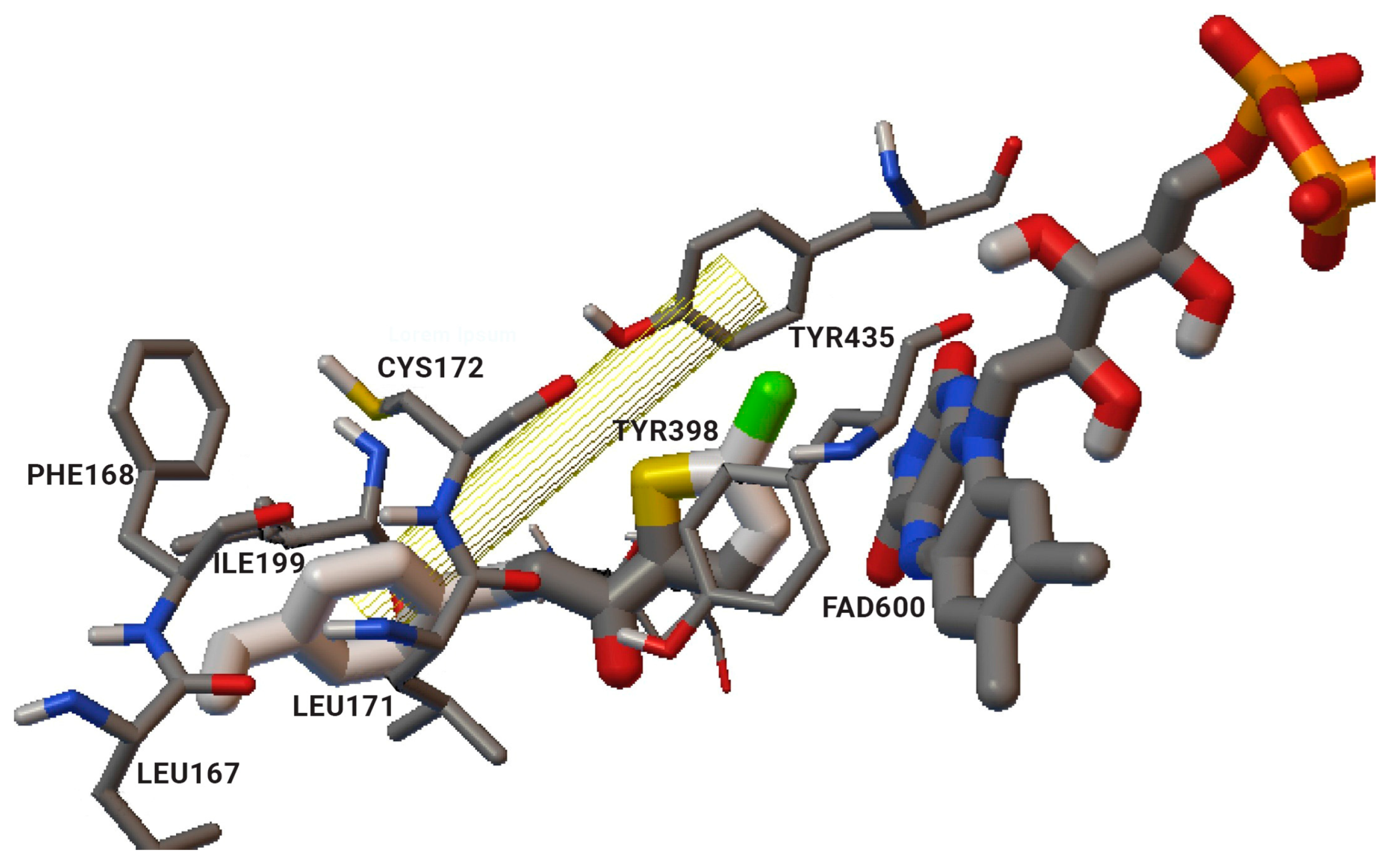
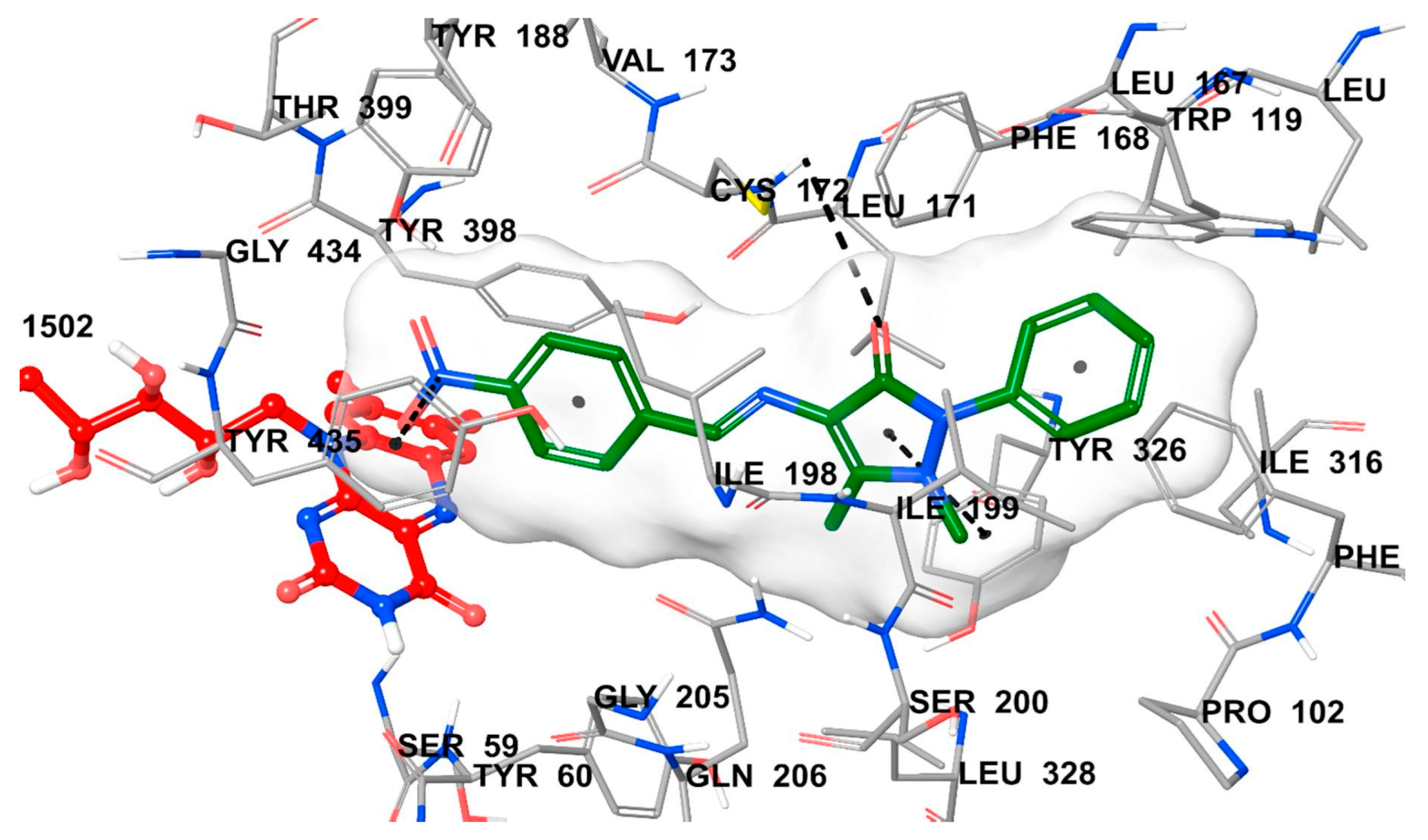
1.3. Structure and Active Site of MAO-B
2. Docking Studies of Recently Synthesized MAO-B Inhibitors
2.1. Chalcones
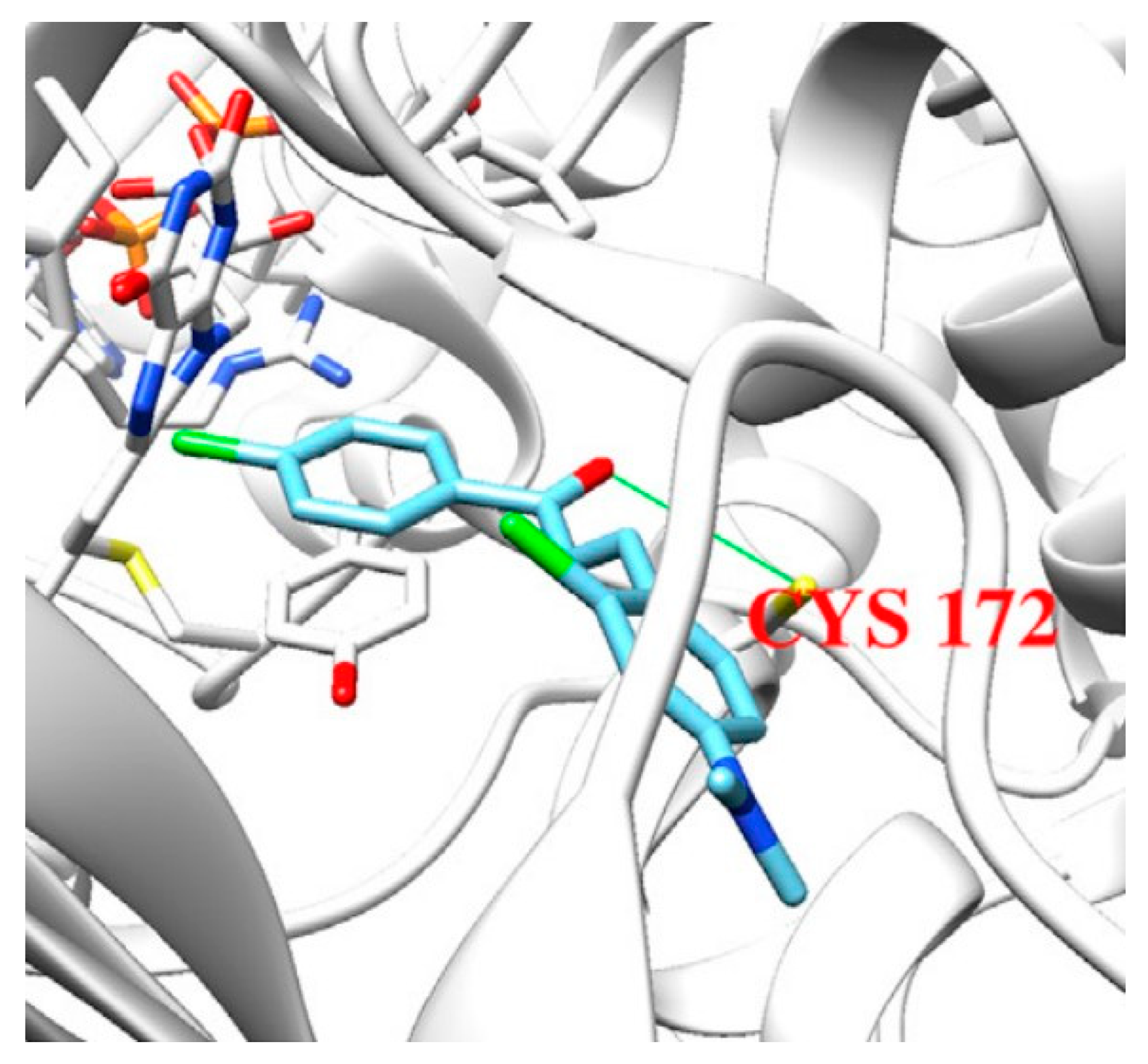
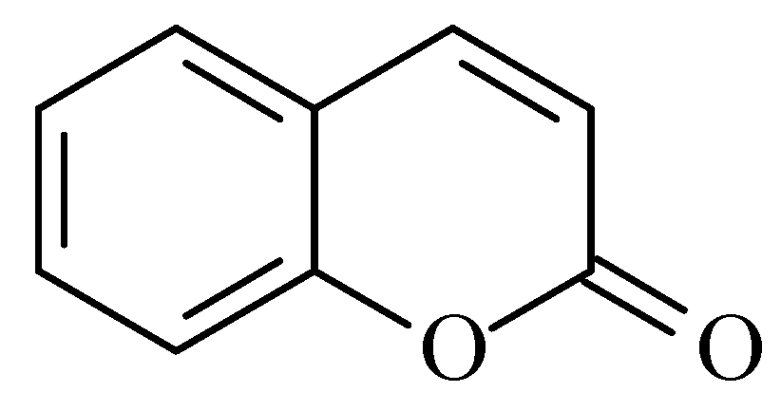
2.2. Coumarins
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 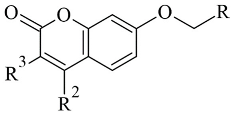 | IC50 0.0005—28.5 (μM) | Molecular Operating Environment (MOE) | 2V61 | [81] |
| 2. | 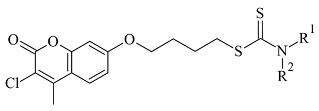 | IC50 0.101—47.16 (μM) | Molecular Operating Environment (MOE) | 2V61 | [82] |
| 3. | 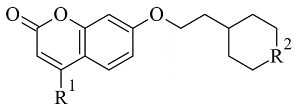 | IC50 0.372—94.32 (μM) | Discovery Studio (DC) | 2V61 | [84] |
| 4. |  | IC50 4.62—>200 (μM) | Discovery Studio (DC) | 4A79 | [86] |
| 5. | 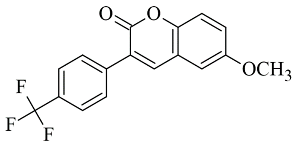 | IC50 56—8476 (nM) | Glide | 2V60 | [87] |
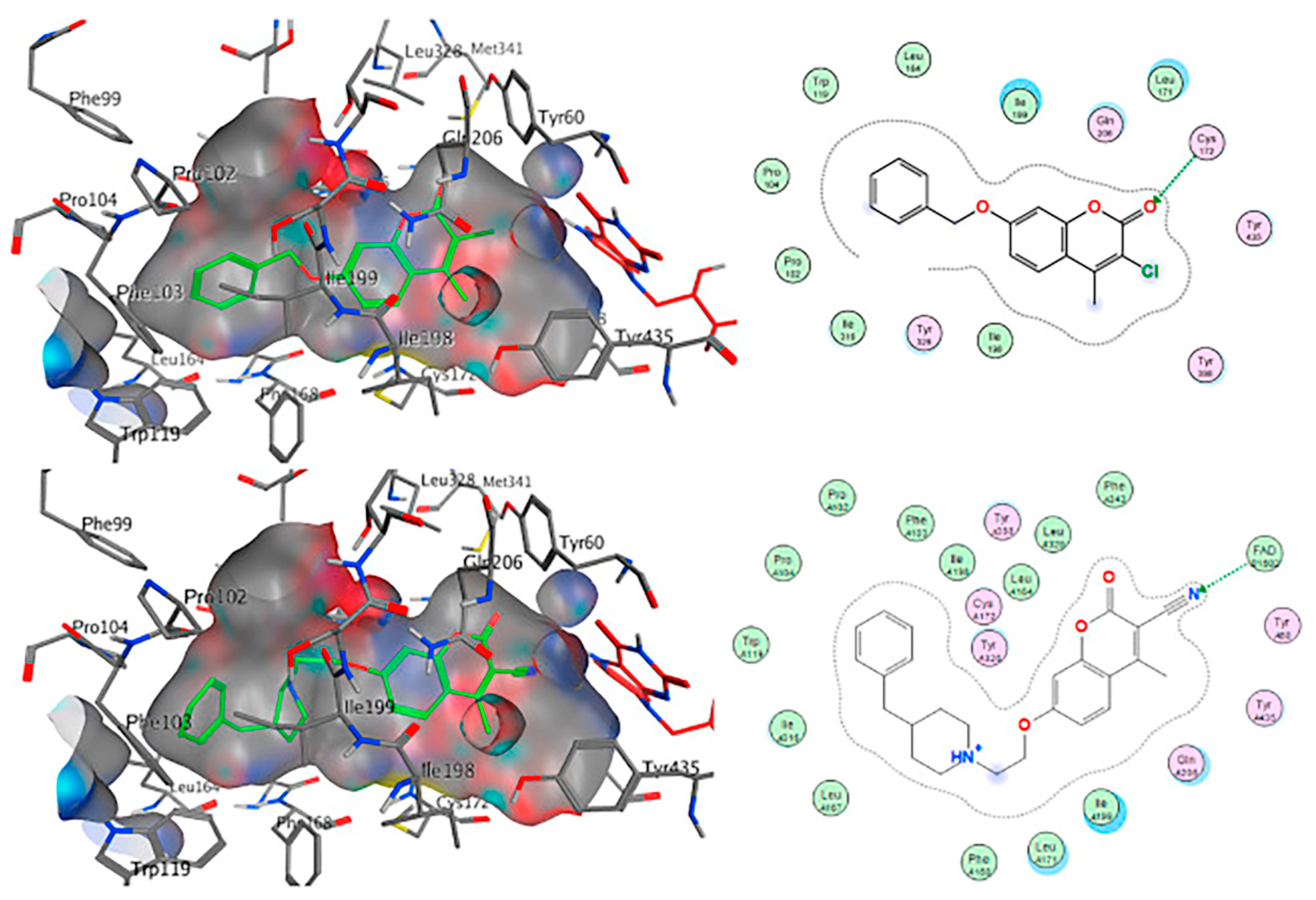
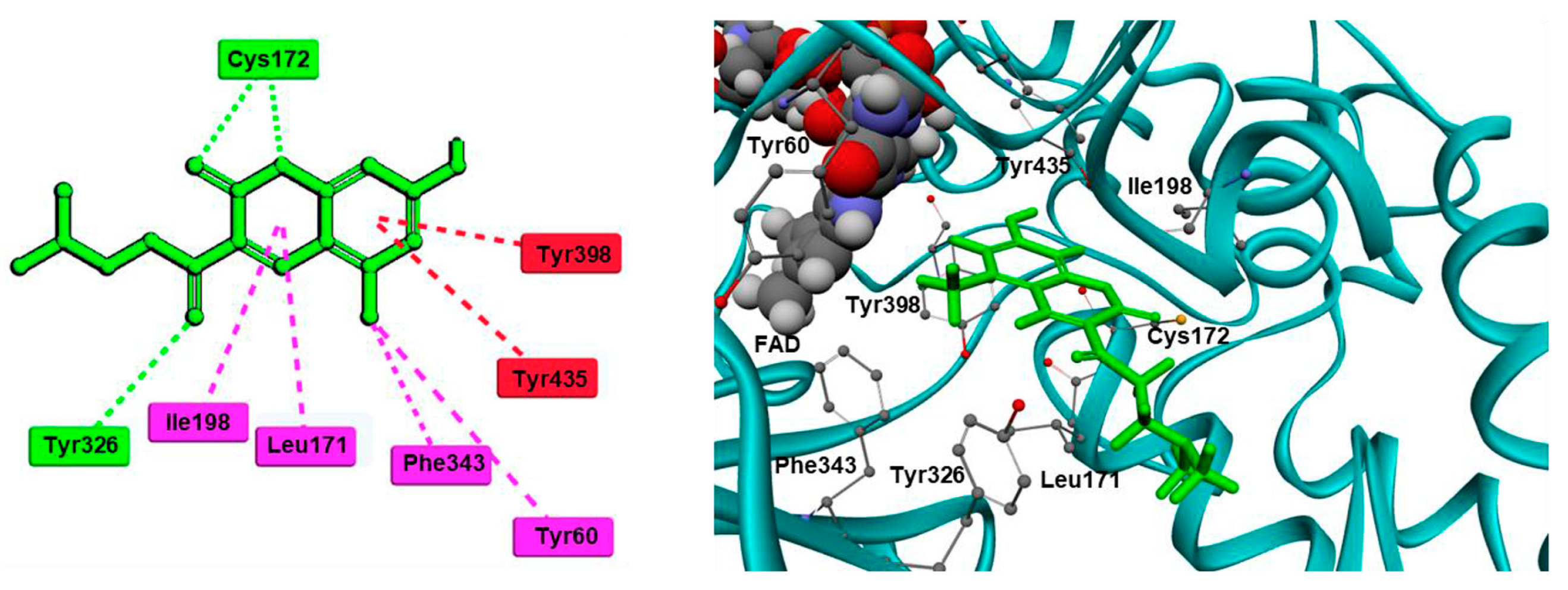
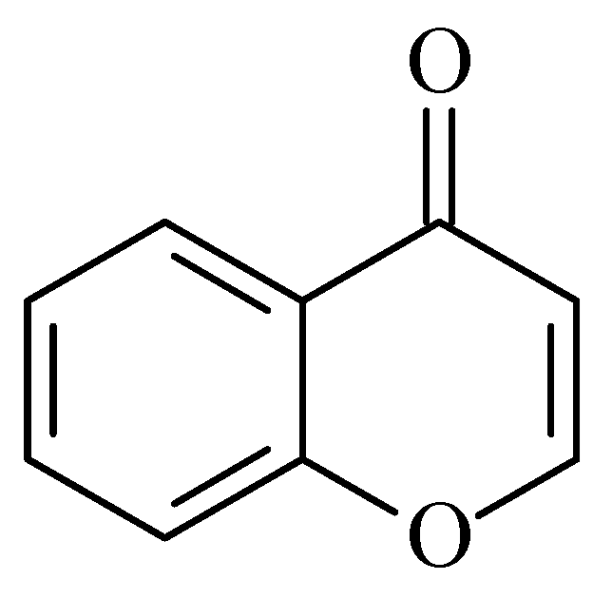
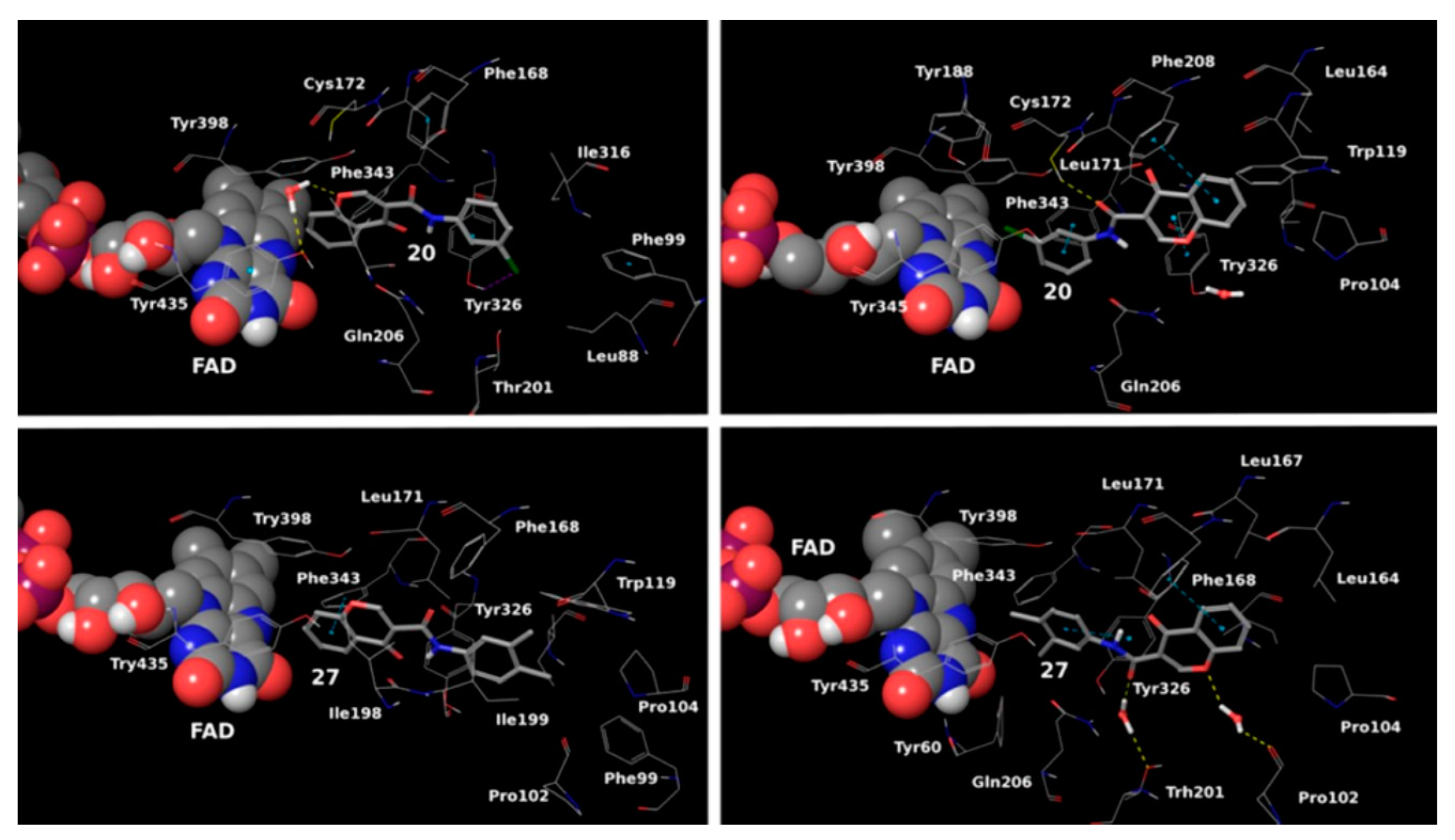
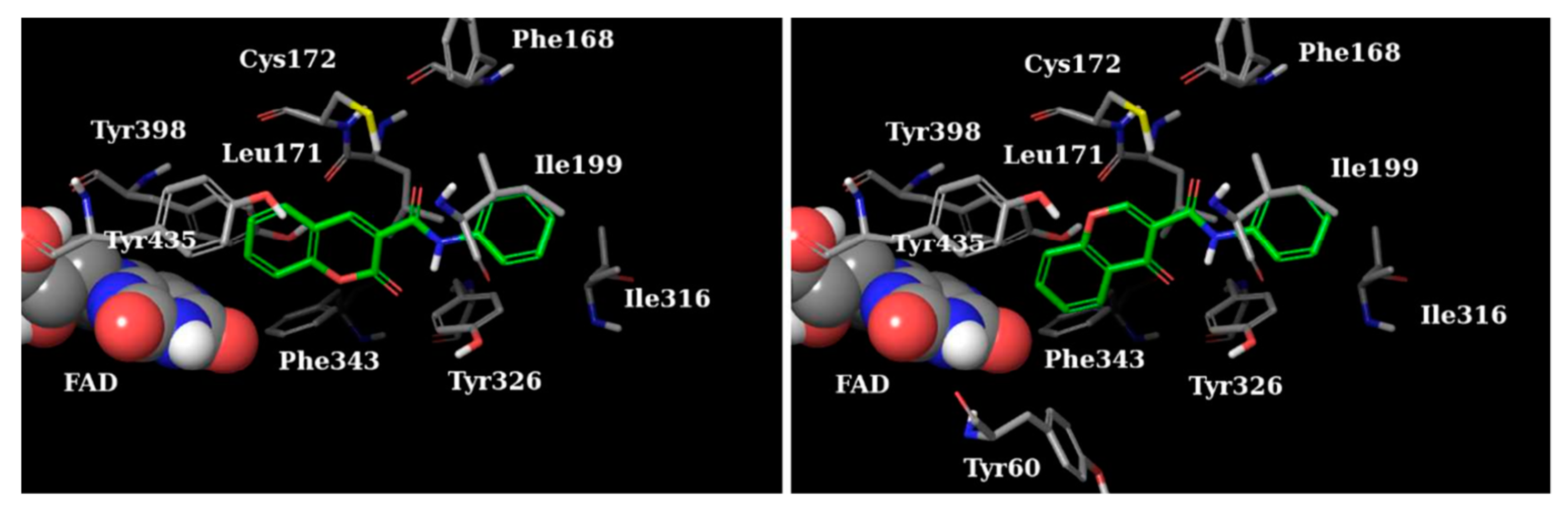
2.3. Chromones
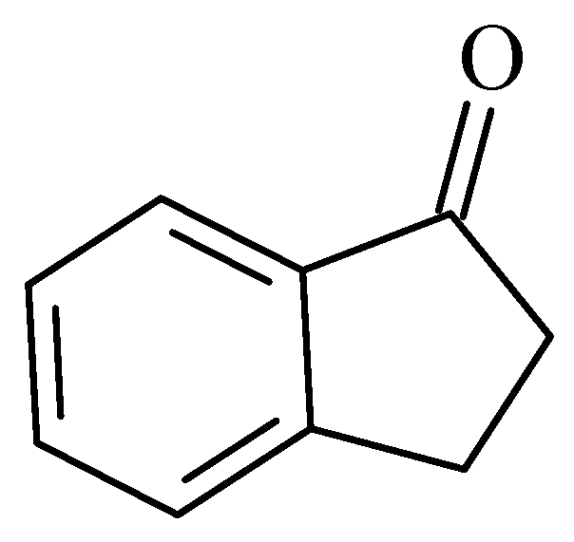
2.4. Indanones
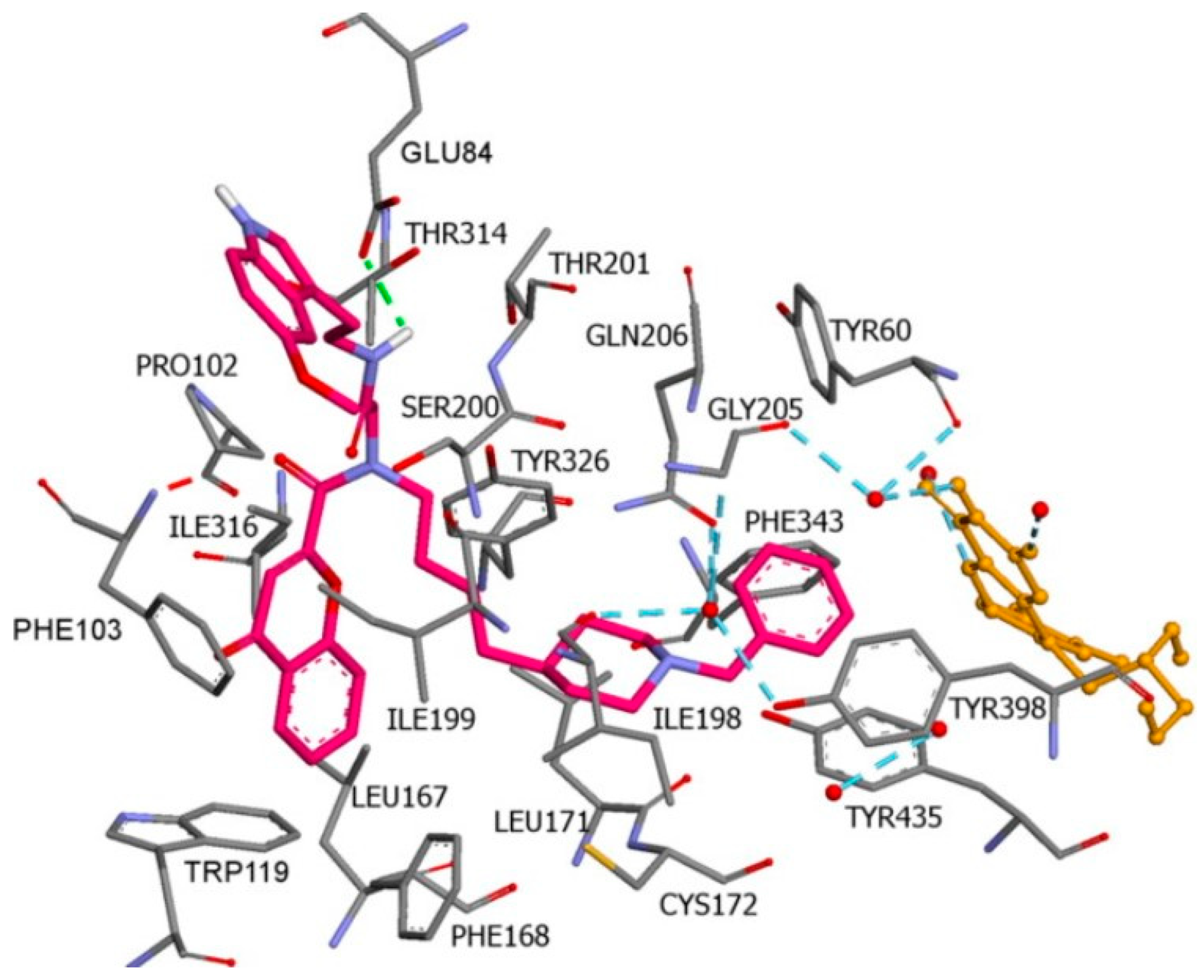
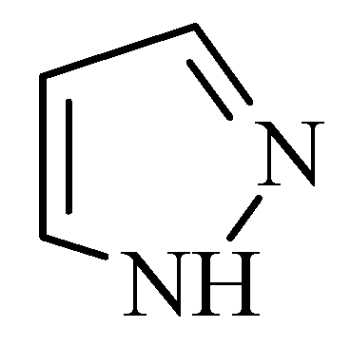
2.5. Pyrazoles
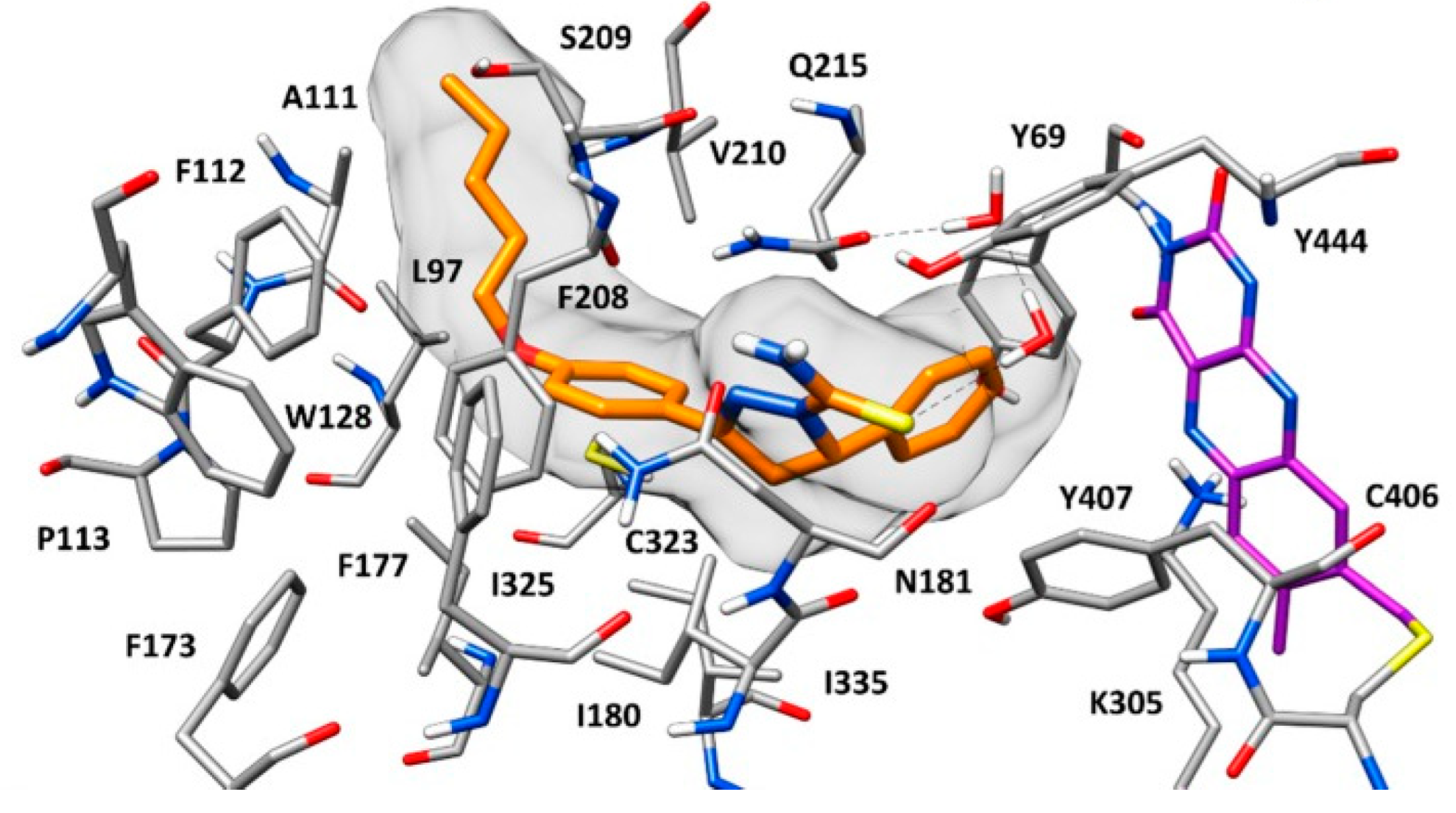
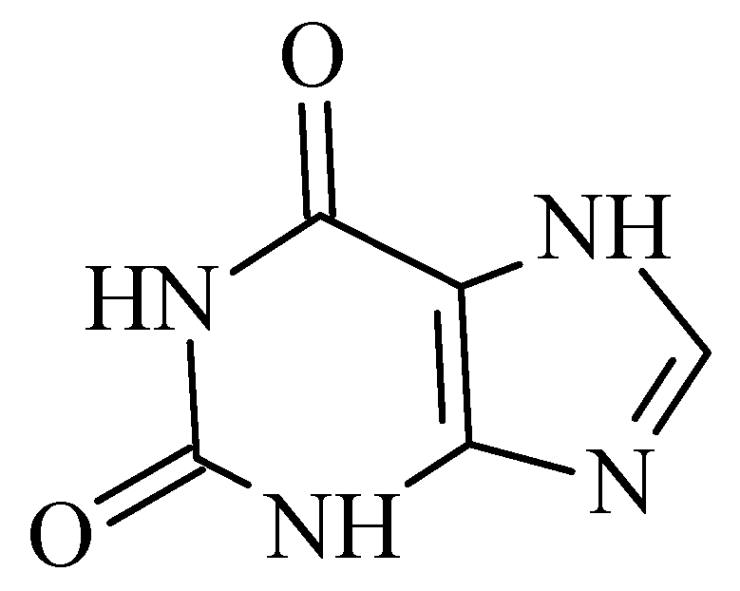
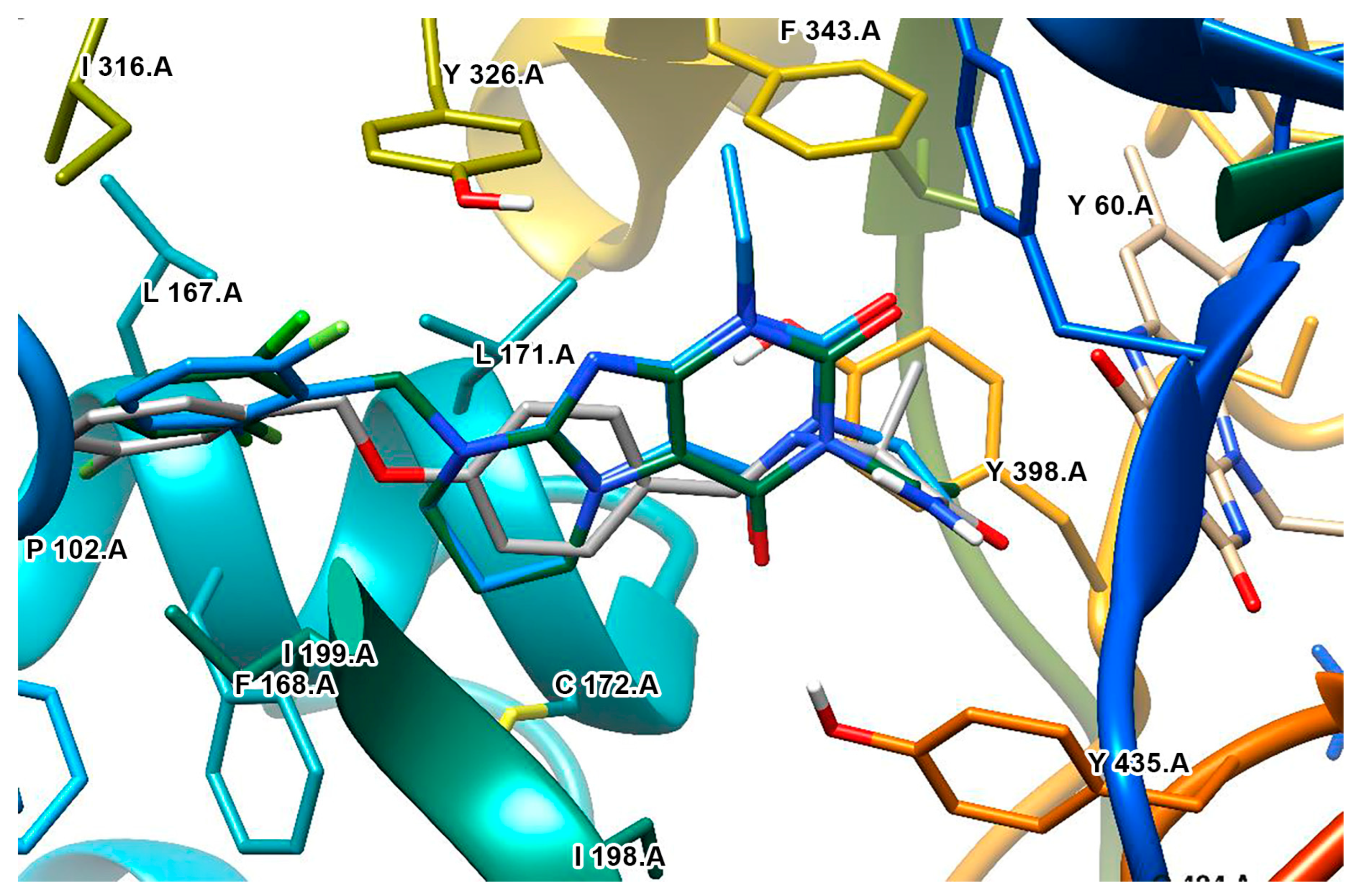
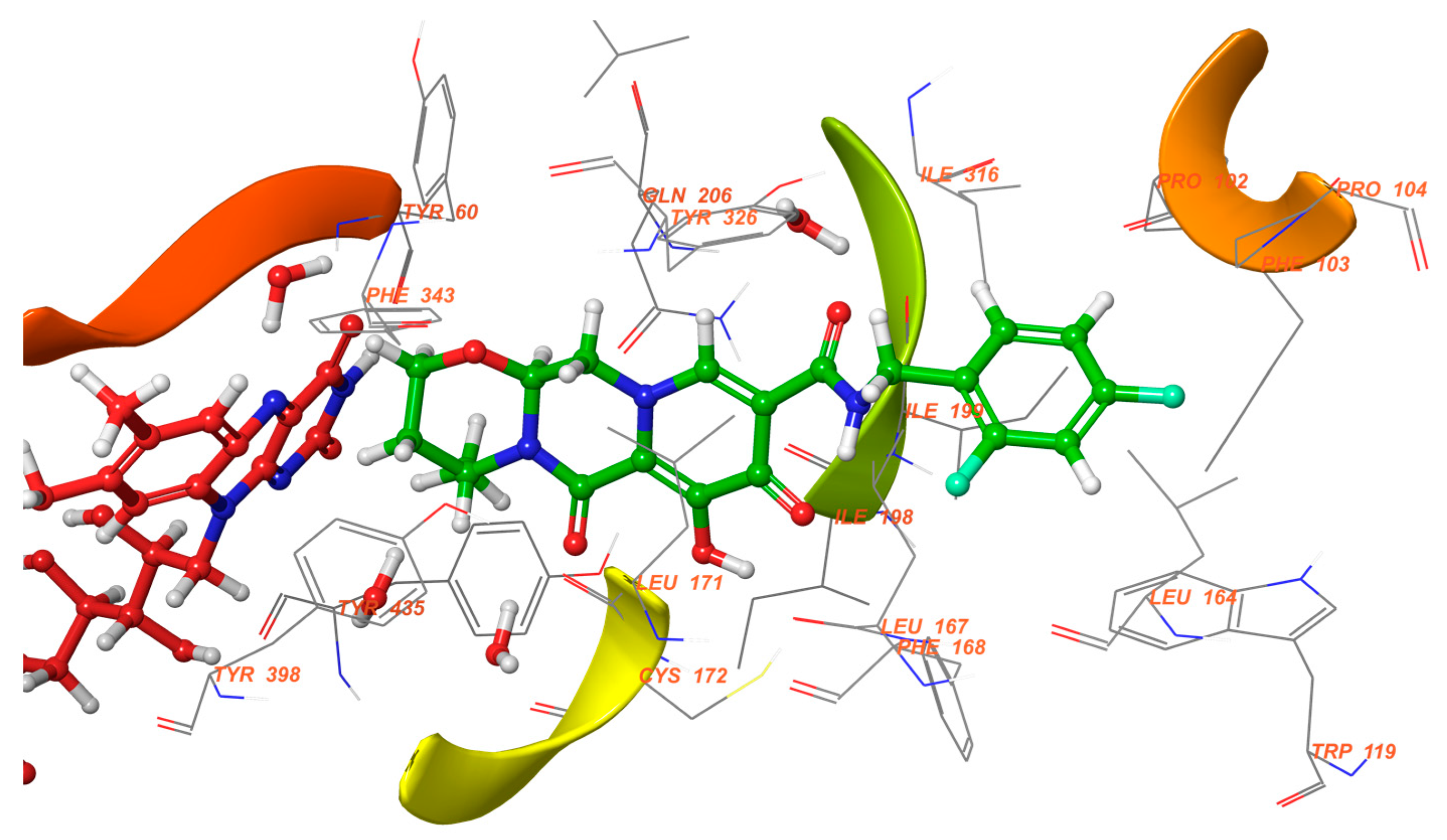
2.6. Xanthines
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 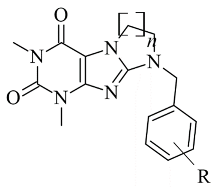 | IC50 0.083—111 (μM) | Glide | 2V5Z | [112] |
| 2. | 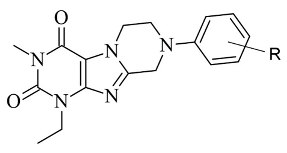 | IC50 106—136 (nM) | Glide | 2V5Z | [113] |
| 3. | 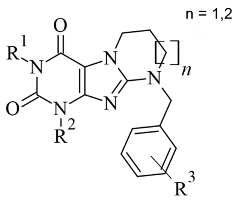 | IC50 83—1241 (nM) | Glide | 2V5Z | [115] |
2.7. Miscellaneous Compounds
3. Conclusions
Funding
Conflicts of Interest
References
- Durães, F.; Pinto, M.; Sousa, E. Old Drugs as New Treatments for Neurodegenerative Diseases. Pharmaceuticals 2018, 11, 44. [Google Scholar] [CrossRef]
- Gordon, R.; Woodruff, T.M. Neuroinflammation as a therapeutic target in neurodegenerative diseases. In Disease-Modifying Targets in Neurodegenerative Disorders; Elsevier: Amsterdam, The Netherlands, 2017; pp. 49–80. [Google Scholar]
- Raza, C.; Anjum, R.; Shakeel, N.A. Parkinson’s disease: Mechanisms, translational models and management strategies. Life Sci. 2019, 226, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Oertel, W.; Schulz, J.B. Current and experimental treatments of Parkinson disease: A guide for neuroscientists. J. Neurochem. 2016, 139, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Favieri, F.; Boncompagni, I.; Agostini, F.; Cantone, M.; Casagrande, M. Executive Functions in Alzheimer Disease: A Systematic Review. Front. Aging Neurosci. 2019, 10, 437. [Google Scholar] [CrossRef] [PubMed]
- Schedin-Weiss, S.; Inoue, M.; Hromadkova, L.; Teranishi, Y.; Yamamoto, N.G.; Wiehager, B.; Bogdanovic, N.; Winblad, B.; Sandebring-Matton, A.; Frykman, S.; et al. Monoamine oxidase B is elevated in Alzheimer disease neurons, is associated with γ-secretase and regulates neuronal amyloid β-peptide levels. Alzheimer’s Res. Ther. 2017, 9, 57. [Google Scholar] [CrossRef]
- Carradori, S.; Petzer, J.P. Novel monoamine oxidase inhibitors: A patent review (2012–2014). Expert Opin. Ther. Pat. 2014, 25, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Uehara, S.; Uno, Y.; Yamazaki, H. Molecular cloning, sequence analysis, and tissue distribution of marmoset monoamine oxidases A and B. Drug Metab. Pharmacokinet. 2020, 35, 479–482. [Google Scholar] [CrossRef]
- Schain, M.; Kreisl, W.C. Neuroinflammation in Neurodegenerative Disorders—A Review. Curr. Neurol. Neurosci. Rep. 2017, 17, 25. [Google Scholar] [CrossRef]
- Riederer, P.; Müller, T. Use of monoamine oxidase inhibitors in chronic neurodegeneration. Expert Opin. Drug Metab. Toxicol. 2017, 13, 233–240. [Google Scholar] [CrossRef]
- Tipton, K.F.; Boyce, S.; O’Sullivan, J.; Davey, G.P.; Healy, J. Monoamine Oxidases: Certainties and Uncertainties. Curr. Med. Chem. 2004, 11, 1965–1982. [Google Scholar] [CrossRef]
- Bonivento, D.; Milczek, E.M.; McDonald, G.R.; Binda, C.; Holt, A.; Edmondson, D.E.; Mattevi, A. Potentiation of ligand binding through cooperative effects in monoamine oxidase B. J. Biol. Chem. 2010, 285, 36849–36856. [Google Scholar] [CrossRef]
- Carradori, S.; Secci, D.; Petzer, J.P. MAO inhibitors and their wider applications: A patent review. Expert Opin. Ther. Pat. 2018, 28, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Pratama, M.R.F.; Poerwono, H.; Siswodihardjo, S. Molecular docking of novel 5-O-benzoylpinostrobin derivatives as wild type and L858R/T790M/V948R mutant EGFR inhibitor. J. Basic Clin. Physiol. Pharmacol. 2019, 30. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Huang, Y.; Wang, C.; Chen, F.; Yang, L.; Ling, L.; Che, Z.; Chen, X. Recent developments in molecular docking technology applied in food science: A review. Int. J. Food Sci. Technol. 2019, 55, 33–45. [Google Scholar] [CrossRef]
- Kumar, R.; Långström, B.; Darreh-Shori, T. Novel ligands of Choline Acetyltransferase designed by in silico molecular docking, hologram QSAR and lead optimization. Sci. Rep. 2016, 6, 31247. [Google Scholar] [CrossRef] [PubMed]
- Doak, B.C.; Norton, R.S.; Scanlon, M.J. The ways and means of fragment-based drug design. Pharmacol. Ther. 2016, 167, 28–37. [Google Scholar] [CrossRef]
- Crisan, L.; Istrate, D.; Bora, A.; Pacureanu, L. Virtual screening and drug repurposing experiments to identify potential novel selective MAO-B inhibitors for Parkinson’s disease treatment. Mol. Divers. 2020, 25, 1775–1794. [Google Scholar] [CrossRef]
- Mezei, M. A new method for mapping macromolecular topography. J. Mol. Graph. Model. 2003, 21, 463–472. [Google Scholar] [CrossRef]
- Koshland, D.E. The Key–Lock Theory and the Induced Fit Theory. Angew. Chem. Int. Ed. Engl. 1995, 33, 2375–2378. [Google Scholar] [CrossRef]
- DesJarlais, R.L.; Sheridan, R.P.; Seibel, G.L.; Dixon, J.S.; Kuntz, I.D.; Venkataraghavan, R. Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J. Med. Chem. 1988, 31, 722–729. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Bankaitis, V.A. Molecular Docking: From Lock and Key to Combination Lock. J. Mol. Med. Clin. Appl. 2017, 2. [Google Scholar] [CrossRef]
- Williams, D.; Kita, D.; Chang, E.; Pahel, D.; Wang, Q. Robust ligand-protein docking using an advanced genetic algorithm. Theor. Comput. Chem. 2021. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995, 245, 43–53. [Google Scholar] [CrossRef]
- Huang, S.-Y. Comprehensive assessment of flexible-ligand docking algorithms: Current effectiveness and challenges. Brief. Bioinform. 2017, 19, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Matthews, N.; Kitao, A.; Laycock, S.; Hayward, S. Haptic-Assisted Interactive Molecular Docking Incorporating Receptor Flexibility. J. Chem. Inf. Model. 2019, 59, 2900–2912. [Google Scholar] [CrossRef]
- Falcon, W.; Ellingson, S.; Smith, J.; Baudry, J. Ensemble Docking in Drug Discovery: How Many Protein Configurations from Molecular Dynamics Simulations are Needed To Reproduce Known Ligand Binding? J. Phys. Chem. B 2019, 123, 5189–5195. [Google Scholar] [CrossRef] [PubMed]
- Najmanovich, R.; Kuttner, J.; Sobolev, V.; Edelman, M. Side-chain flexibility in proteins upon ligand binding. Proteins: Struct. Funct. Genet. 2000, 39, 261–268. [Google Scholar] [CrossRef]
- Mateev, E.; Valkova, I.V.A.; Georgieva, M.; Zlatkov, A. Database Enrichments of Mao-B through Ensemble Docking. Int. J. Pharm. Pharm. Sci. 2021, 13, 32–35. [Google Scholar] [CrossRef]
- Awuni, Y.; Mu, Y. Reduction of false positives in structure-based virtual screening when receptor plasticity is considered. Molecules 2015, 20, 5152–5164. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. Computer Simulation of Molecular Dynamics: Methodology, Applications, and Perspectives in Chemistry. Angew. Chem. Int. Ed. Engl. 1990, 29, 992–1023. [Google Scholar] [CrossRef]
- Shoichet, B.K. Virtual screening of chemical libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Perez-Sanchez, H.; Lightstone, F.C. A Comprehensive Docking and MM/GBSA Rescoring Study of Ligand Recognition upon Binding Antithrombin. Curr. Top. Med. Chem. 2017, 17, 1631–1639. [Google Scholar] [CrossRef]
- Thilagavathi, R.; Mancera, R.L. Ligand−Protein Cross-Docking with Water Molecules. J. Chem. Inf. Model. 2010, 50, 415–421. [Google Scholar] [CrossRef]
- Murphy, R.B.; Repasky, M.P.; Greenwood, J.R.; Tubert-Brohman, I.; Jerome, S.; Annabhimoju, R.; Boyles, N.A.; Schmitz, C.D.; Abel, R.; Farid, R.; et al. WScore: A Flexible and Accurate Treatment of Explicit Water Molecules in Ligand–Receptor Docking. J. Med. Chem. 2016, 59, 4364–4384. [Google Scholar] [CrossRef]
- Kumar, A.; Zhang, K.Y.J. Investigation on the Effect of Key Water Molecules on Docking Performance in CSARdock Exercise. J. Chem. Inf. Model. 2013, 53, 1880–1892. [Google Scholar] [CrossRef] [PubMed]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fu, A.; Zhang, L. An Overview of Scoring Functions Used for Protein–Ligand Interactions in Molecular Docking. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef]
- Kinnings, S.L.; Liu, N.; Tonge, P.J.; Jackson, R.M.; Xie, L.; Bourne, P.E. A machine learning-based method to improve docking scoring functions and its application to drug repurposing. J. Chem. Inf. Model. 2011, 51, 408–419. [Google Scholar] [CrossRef]
- Wang, R.; Lu, Y.; Wang, S. Comparative Evaluation of 11 Scoring Functions for Molecular Docking. J. Med. Chem. 2003, 46, 2287–2303. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten docking programs on a diverse set of protein–ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Automated docking of substrates to proteins by simulated annealing. Proteins Struct. Funct. Genet. 1990, 8, 195–202. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Kramer, B.; Rarey, M.; Lengauer, T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins Struct. Funct. Genet. 1999, 37, 228–241. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.Z.; Cheng, F.; Zhang, Y.J.; Ge, J.Y.; Yao, S.Q.; Zhu, Q. Rapid synthesis of flavone-based monoamine oxidase (MAO) inhibitors targeting two active sites using click chemistry. Chem. Biol. Drug Des. 2016, 89, 141–151. [Google Scholar] [CrossRef]
- Murugan, N.A.; Zaleśny, R. Multiscale Modeling of Two-Photon Probes for Parkinson’s Diagnostics Based on Monoamine Oxidase B Biomarker. J. Chem. Inf. Model. 2020, 60, 3854–3863. [Google Scholar] [CrossRef]
- Binda, C.; Li, M.; Hubalek, F.; Restelli, N.; Edmondson, D.E.; Mattevi, A. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA 2003, 100, 9750–9755. [Google Scholar] [CrossRef] [PubMed]
- Hudspith, L.; Shmam, F.; Dalton, C.F.; Princivalle, A.; Turega, S.M. Neurotransmitter selection by monoamine oxidase isoforms, dissected in terms of functional groups by mixed double mutant cycles. Org. Biomol. Chem. 2019, 17, 8871–8877. [Google Scholar] [CrossRef] [PubMed]
- Geha, R.M.; Chen, K.; Wouters, J.; Ooms, F.; Shih, J.C. Analysis of conserved active site residues in monoamine oxidase A and B and their three-dimensional molecular modeling. J. Biol. Chem. 2002, 277, 17209–17216. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Abodabos, H.S.; Taban, I.M.; Rfieda, A.R.; Mahmood, D.; Anwar, M.J.; Khan, S.; Sizochenko, N.; Poli, G.; Tuccinardi, T.; et al. Rutin as promising drug for the treatment of Parkinson’s disease: An assessment of MAO-B inhibitory potential by docking, molecular dynamics and DFT studies. Mol. Simul. 2019, 45, 1563–1571. [Google Scholar] [CrossRef]
- Shetnev, A.; Osipyan, A.; Baykov, S.; Sapegin, A.; Chirkova, Z.; Korsakov, M.; Petzer, A.; Engelbrecht, I.; Petzer, J.P. Novel monoamine oxidase inhibitors based on the privileged 2-imidazoline molecular framework. Bioorganic Med. Chem. Lett. 2019, 29, 40–46. [Google Scholar] [CrossRef]
- Hamulakova, S.; Kozurkova, M.; Kuca, K. Coumarin Derivatives in Pharmacotherapy of Alzheimer’s Disease. Curr. Org. Chem. 2017, 21, 602–612. [Google Scholar] [CrossRef]
- Hubálek, F.; Binda, C.; Khalil, A.; Li, M.; Mattevi, A.; Castagnoli, N.; Edmondson, D.E. Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors. J. Biol. Chem. 2005, 280, 15761–15766. [Google Scholar] [CrossRef]
- Łażewska, D.; Olejarz-Maciej, A.; Reiner, D.; Kaleta, M.; Latacz, G.; Zygmunt, M.; Doroz-Płonka, A.; Karcz, T.; Frank, A.; Stark, H.; et al. Dual Target Ligands with 4-tert-Butylphenoxy Scaffold as Histamine H(3) Receptor Antagonists and Monoamine Oxidase B Inhibitors. Int. J. Mol. Sci. 2020, 21, 3411. [Google Scholar] [CrossRef]
- Li, M.; Binda, C.; Mattevi, A.; Edmondson, D.E. Functional role of the “aromatic cage” in human monoamine oxidase B: Structures and catalytic properties of Tyr435 mutant proteins. Biochemistry 2006, 45, 4775–4784. [Google Scholar] [CrossRef]
- Akyüz, M.A.; Erdem, S.S.; Edmondson, D.E. The aromatic cage in the active site of monoamine oxidase B: Effect on the structural and electronic properties of bound benzylamine and p-nitrobenzylamine. J. Neural Transm. 2007, 114, 693–698. [Google Scholar] [CrossRef]
- Son, S.-Y.; Ma, J.; Kondou, Y.; Yoshimura, M.; Yamashita, E.; Tsukihara, T. Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA 2008, 105, 5739–5744. [Google Scholar] [CrossRef] [PubMed]
- Moraes, F.P.; de Azevedo, W.F. Targeting imidazoline site on monoamine oxidase B through molecular docking simulations. J. Mol. Model. 2012, 18, 3877–3886. [Google Scholar] [CrossRef]
- Ramsay, R. Molecular aspects of monoamine oxidase B. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2016, 69, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, A.; Yabanoglu, S.; Sinha, B.N.; Ucar, G.; Basu, A.; Jayaprakash, V. Towards development of selective and reversible pyrazoline based MAO-inhibitors: Synthesis, biological evaluation and docking studies. Bioorganic Med. Chem. Lett. 2010, 20, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Royo, J.L.; Castellano-Castillo, D.; Ruiz-Galdon, M.; Molina-Vega, M.; Cardona, F.; Tinahones, F.J.; Fernández-García, J.C.; Reyes-Engel, A. Monoamino oxidase alleles correlate with the presence of essential hypertension among hypogonadic patients. Mol. Genet. Genomic. Med. 2020, 8, e1040. [Google Scholar] [CrossRef] [PubMed]
- De Castro Julve, M.; Miralles Albors, P.; Ortonobes Roig, S.; Vives, R.; Falgueras, L.; Gómez-Valent, M. Hypertensive crisis following the administration of tedizolid: Possible serotonin syndrome. Eur. J. Hosp. Pharm. 2020, 27, 52–54. [Google Scholar] [CrossRef]
- Colibus, L.; Li, M.; Binda, C.; Lustig, A.; Edmondson, D.; Mattevi, A. Three-dimensional structure of human monoamine oxidase A (MAO A): Relation to the structures of rat MAO A and human MAO B. Proc. Natl. Acad. Sci. USA 2005, 102, 12684–12689. [Google Scholar] [CrossRef]
- Tsugeno, Y.; Ito, A. A Key Amino Acid Responsible for Substrate Selectivity of Monoamine Oxidase A and B. J. Biol. Chem. 1997, 272, 14033–14036. [Google Scholar] [CrossRef]
- Park, J.-H.; Ju, Y.H.; Choi, J.W.; Song, H.J.; Jang, B.K.; Woo, J.; Chun, H.; Kim, H.J.; Shin, S.J.; Yarishkin, O.; et al. Newly developed reversible MAO-B inhibitor circumvents the shortcomings of irreversible inhibitors in Alzheimer’s disease. Sci. Adv. 2019, 5, eaav0316. [Google Scholar] [CrossRef]
- Sano, M.; Ernesto, C.; Thomas, R.G.; Klauber, M.R.; Schafer, K.; Grundman, M.; Woodbury, P.; Growdon, J.; Cotman, C.W.; Pfeiffer, E.; et al. A Controlled Trial of Selegiline, Alpha-Tocopherol, or Both as Treatment for Alzheimer’s Disease. N. Engl. J. Med. 1997, 336, 1216–1222. [Google Scholar] [CrossRef]
- Kar Mahapatra, D.; Asati, V.; Bharti, S.K. An updated patent review of therapeutic applications of chalcone derivatives (2014-present). Expert Opin. Ther. Pat. 2019, 29, 385–406. [Google Scholar] [CrossRef]
- Guglielmi, P.; Mathew, B.; Secci, D.; Carradori, S. Chalcones: Unearthing their therapeutic possibility as monoamine oxidase B inhibitors. Eur. J. Med. Chem. 2020, 205, 112650. [Google Scholar] [CrossRef]
- Mathew, B.; Haridas, A.; Uçar, G.; Baysal, I.; Adeniyi, A.A.; Soliman, M.E.S.; Joy, M.; Mathew, G.E.; Lakshmanan, B.; Jayaprakash, V. Exploration of chlorinated thienyl chalcones: A new class of monoamine oxidase-B inhibitors. Int. J. Biol. Macromol. 2016, 91, 680–695. [Google Scholar] [CrossRef]
- Mathew, B.; Haridas, A.; Uçar, G.; Baysal, I.; Joy, M.; Mathew, G.E.; Lakshmanan, B.; Jayaprakash, V. Synthesis, Biochemistry, and Computational Studies of Brominated Thienyl Chalcones: A New Class of Reversible MAO-B Inhibitors. ChemMedChem 2016, 11, 1161–1171. [Google Scholar] [CrossRef]
- Lakshminarayanan, B.; Baek, S.C.; Lee, J.P.; Kannappan, N.; Mangiatordi, G.F.; Nicolotti, O.; Subburaju, T.; Kim, H.; Mathew, B. Ethoxylated Head of Chalcones as a New Class of Multi-Targeted MAO Inhibitors. ChemistrySelect 2019, 4, 6614–6619. [Google Scholar] [CrossRef]
- Oh, J.M.; Kang, M.-G.; Hong, A.; Park, J.-E.; Kim, S.H.; Lee, J.P.; Baek, S.C.; Park, D.; Nam, S.-J.; Cho, M.-L.; et al. Potent and selective inhibition of human monoamine oxidase-B by 4-dimethylaminochalcone and selected chalcone derivatives. Int. J. Biol. Macromol. 2019, 137, 426–432. [Google Scholar] [CrossRef]
- Parambi, D.G.T.; Oh, J.M.; Baek, S.C.; Lee, J.P.; Tondo, A.R.; Nicolotti, O.; Kim, H.; Mathew, B. Design, synthesis and biological evaluation of oxygenated chalcones as potent and selective MAO-B inhibitors. Bioorganic Chem. 2019, 93, 103335. [Google Scholar] [CrossRef] [PubMed]
- Stefanachi, A.; Leonetti, F.; Pisani, L.; Catto, M.; Carotti, A. Coumarin: A Natural, Privileged and Versatile Scaffold for Bioactive Compounds. Molecules 2018, 23, 250. [Google Scholar] [CrossRef] [PubMed]
- Pisani, L.; Muncipinto, G.; Miscioscia, T.F.; Nicolotti, O.; Leonetti, F.; Catto, M.; Caccia, C.; Salvati, P.; Soto-Otero, R.; Mendez-Alvarez, E.; et al. Discovery of a Novel Class of Potent Coumarin Monoamine Oxidase B Inhibitors: Development and Biopharmacological Profiling of 7-[(3-Chlorobenzyl)oxy]-4-[(methylamino)methyl]-2H-chromen-2-one Methanesulfonate (NW-1772) as a Highly Potent, Selective, Reversible, and Orally Active Monoamine Oxidase B Inhibitor. J. Med. Chem. 2009, 52, 6685–6706. [Google Scholar] [CrossRef]
- Joubert, J.; Foka, G.B.; Repsold, B.P.; Oliver, D.W.; Kapp, E.; Malan, S.F. Synthesis and evaluation of 7-substituted coumarin derivatives as multimodal monoamine oxidase-B and cholinesterase inhibitors for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2017, 125, 853–864. [Google Scholar] [CrossRef]
- He, Q.; Liu, J.; Lan, J.-S.; Ding, J.; Sun, Y.; Fang, Y.; Jiang, N.; Yang, Z.; Sun, L.; Jin, Y.; et al. Coumarin-dithiocarbamate hybrids as novel multitarget AChE and MAO-B inhibitors against Alzheimer’s disease: Design, synthesis and biological evaluation. Bioorganic Chem. 2018, 81, 512–528. [Google Scholar] [CrossRef]
- Jeong, G.S.; Kang, M.-G.; Lee, J.Y.; Lee, S.R.; Park, D.; Cho, M.; Kim, H. Inhibition of Butyrylcholinesterase and Human Monoamine Oxidase-B by the Coumarin Glycyrol and Liquiritigenin Isolated from Glycyrrhiza uralensis. Molecules 2020, 25, 3896. [Google Scholar] [CrossRef]
- Repsold, B.P.; Malan, S.F.; Joubert, J.; Oliver, D.W. Multi-targeted directed ligands for Alzheimer’s disease: Design of novel lead coumarin conjugates. SAR QSAR Environ. Res. 2018, 29, 231–255. [Google Scholar] [CrossRef] [PubMed]
- Mateev, E.V.; Valkova, I.; Georgieva, M.; Zlatkov, A. Suitable Docking Protocol for the Design of Novel Coumarin Derivatives with Selective MAO-B Effects. J. Mol. Docking 2021, 1, 40–47. [Google Scholar] [CrossRef]
- Tao, D.; Wang, Y.; Bao, X.-Q.; Yang, B.-B.; Gao, F.; Wang, L.; Zhang, D.; Li, L. Discovery of coumarin Mannich base derivatives as multifunctional agents against monoamine oxidase B and neuroinflammation for the treatment of Parkinson’s disease. Eur. J. Med. Chem. 2019, 173, 203–212. [Google Scholar] [CrossRef]
- Rauhamäki, S.; Postila, P.A.; Niinivehmas, S.; Kortet, S.; Schildt, E.; Pasanen, M.; Manivannan, E.; Ahinko, M.; Koskimies, P.; Nyberg, N.; et al. Structure-Activity Relationship Analysis of 3-Phenylcoumarin-Based Monoamine Oxidase B Inhibitors. Front. Chem. 2018, 6, 41. [Google Scholar] [CrossRef]
- Gaspar, A.; Milhazes, N.; Santana, L.; Uriarte, E.; Borges, F.; Matos, M. Oxidative Stress and Neurodegenerative Diseases: Looking for a Therapeutic Solution Inspired on Benzopyran Chemistry. Curr. Top. Med. Chem. 2015, 15, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Mathew, G.E.; Petzer, J.P.; Petzer, A. Structural Exploration of Synthetic Chromones as Selective MAO-B Inhibitors: A Mini Review. Comb. Chem. High Throughput Screen. 2017, 20, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.; Reis, J.; Fonseca, A.; Milhazes, N.; Viña, D.; Uriarte, E.; Borges, F. Chromone 3-phenylcarboxamides as potent and selective MAO-B inhibitors. Bioorganic Med. Chem. Lett. 2011, 21, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.; Cagide, F.; Chavarria, D.; Silva, T.; Fernandes, C.; Gaspar, A.; Uriarte, E.; Remião, F.; Alcaro, S.; Ortuso, F.; et al. Discovery of New Chemical Entities for Old Targets: Insights on the Lead Optimization of Chromone-Based Monoamine Oxidase B (MAO-B) Inhibitors. J. Med. Chem. 2016, 59, 5879–5893. [Google Scholar] [CrossRef]
- Reis, J.; Cagide, F.; Valencia, M.E.; Teixeira, J.; Bagetta, D.; Pérez, C.; Uriarte, E.; Oliveira, P.J.; Ortuso, F.; Alcaro, S.; et al. Multi-target-directed ligands for Alzheimer’s disease: Discovery of chromone-based monoamine oxidase/cholinesterase inhibitors. Eur. J. Med. Chem. 2018, 158, 781–800. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.J.; Abhijit, K.; Mallikarjun, G.; Hemasri, Y. Design and synthesis of novel benzyloxy-tethered-chromone-carboxamide derivatives as potent and selective human monoamine oxidase-b inhibitors. Chem. Pap. 2020, 75, 703–716. [Google Scholar] [CrossRef]
- Wang, X.-B.; Yin, F.-C.; Huang, M.; Jiang, N.; Lan, J.-S.; Kong, L.-Y. Chromone and donepezil hybrids as new multipotent cholinesterase and monoamine oxidase inhibitors for the potential treatment of Alzheimer’s disease. RSC Med. Chem. 2020, 11, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.; Reis, J.; Silva, T.; Matos, M.; Bagetta, D.; Ortuso, F.; Alcaro, S.; Borges, F. Coumarin versus Chromone Monoamine Oxidase B Inhibitors: Quo Vadis? J. Med. Chem. 2017, 60, 7206–7212. [Google Scholar] [CrossRef]
- Pachón-Angona, I.; Refouvelet, B.; Andrýs, R.; Martin, H.; Luzet, V.; Iriepa, I.; Moraleda, I.; Diez-Iriepa, D.; Oset-Gasque, M.-J.; Marco-Contelles, J.; et al. Donepezil + chromone + melatonin hybrids as promising agents for Alzheimer’s disease therapy. J. Enzym. Inhib. Med. Chem. 2019, 34, 479–489. [Google Scholar] [CrossRef]
- Patil, S.A.; Patil, R.; Patil, S.A. Recent developments in biological activities of indanones. Eur. J. Med. Chem. 2017, 138, 182–198. [Google Scholar] [CrossRef]
- Huang, L.; Lu, C.; Sun, Y.; Mao, F.; Luo, Z.; Su, T.; Jiang, H.; Shan, W.; Li, X. Multitarget-Directed Benzylideneindanone Derivatives: Anti-β-Amyloid (Aβ) Aggregation, Antioxidant, Metal Chelation, and Monoamine Oxidase B (MAO-B) Inhibition Properties against Alzheimer’s Disease. J. Med. Chem. 2012, 55, 8483–8492. [Google Scholar] [CrossRef]
- Nel, M.S.; Petzer, A.; Petzer, J.P.; Legoabe, L.J. 2-Benzylidene-1-indanone derivatives as inhibitors of monoamine oxidase. Bioorganic Med. Chem. Lett. 2016, 26, 4599–4605. [Google Scholar] [CrossRef]
- Mostert, S.; Petzer, A.; Petzer, J.P. Indanones As High-Potency Reversible Inhibitors of Monoamine Oxidase. ChemMedChem 2015, 10, 862–873. [Google Scholar] [CrossRef]
- Nel, M.S.; Petzer, A.; Petzer, J.P.; Legoabe, L.J. 2-Heteroarylidene-1-indanone derivatives as inhibitors of monoamine oxidase. Bioorganic Chem. 2016, 69, 20–28. [Google Scholar] [CrossRef]
- Manna, F.; Chimenti, F.; Bolasco, A.; Secci, D.; Bizzarri, B.; Befani, O.; Turini, P.; Mondovi, B.; Alcaro, S.; Tafi, A. Inhibition of Amine Oxidases Activity by 1-Acetyl-3,5-diphenyl-4,5-dihydro-(1H)-pyrazole Derivatives. ChemInform 2003, 34, 3629–3633. [Google Scholar] [CrossRef]
- Carradori, S.; Silvestri, R. New Frontiers in Selective Human MAO-B Inhibitors. J. Med. Chem. 2015, 58, 6717–6732. [Google Scholar] [CrossRef] [PubMed]
- Tok, F.; Koçyiğit-Kaymakçıoğlu, B.; Sağlık, B.N.; Levent, S.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis and biological evaluation of new pyrazolone Schiff bases as monoamine oxidase and cholinesterase inhibitors. Bioorganic Chem. 2019, 84, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, P.; Carradori, S.; Poli, G.; Secci, D.; Cirilli, R.; Rotondi, G.; Chimenti, P.; Petzer, A.; Petzer, J.P. Design, Synthesis, Docking Studies and Monoamine Oxidase Inhibition of a Small Library of 1-acetyl- and 1-thiocarbamoyl-3,5-diphenyl-4,5-dihydro-(1H)-pyrazoles. Molecules 2019, 24, 484. [Google Scholar] [CrossRef] [PubMed]
- Grewal, A.S.; Sharma, S.K.; Pandita, D.; Lather, V. Synthesis, Docking and Evaluation of Novel Pyrazole Carboxamide Derivatives as Multifunctional Anti-Alzheimer’s Agents. J. Med. Chem. Toxicol. 2017, 2, 47–54. [Google Scholar] [CrossRef]
- Upadhyay, S.; Tripathi, A.C.; Paliwal, S.; Saraf, S.K. Facile One-Pot Synthesis Methodology for Nitrogen-Containing Heterocyclic Derivatives of 3,5-Disubstituted 4,5-Dihydro-1H-Pyrazole, Their Biological Evaluation and Molecular Docking Studies. Pharm. Chem. J. 2017, 51, 564–575. [Google Scholar] [CrossRef]
- Vlok, N.; Malan, S.F.; Castagnoli, N.; Bergh, J.J.; Petzer, J.P. Inhibition of monoamine oxidase B by analogues of the adenosine A2A receptor antagonist (E)-8-(3-chlorostyryl)caffeine (CSC). Bioorganic Med. Chem. 2006, 14, 3512–3521. [Google Scholar] [CrossRef]
- Jasiewicz, B.; Sierakowska, A. Caffeine and its analogs, antioxidants and applications. In Aging; Elsevier: Amsterdam, The Netherlands, 2020; pp. 155–164. [Google Scholar] [CrossRef]
- Van den Berg, D.; Zoellner, K.R.; Ogunrombi, M.O.; Malan, S.F.; Terre’Blanche, G.; Castagnoli Jr, N.; Bergh, J.J.; Petzer, J.P. Inhibition of monoamine oxidase B by selected benzimidazole and caffeine analogues. Bioorganic Med. Chem. 2007, 15, 3692–3702. [Google Scholar] [CrossRef]
- Rivara, S.; Piersanti, G.; Bartoccini, F.; Diamantini, G.; Pala, D.; Riccioni, T.; Stasi, M.A.; Cabri, W.; Borsini, F.; Mor, M.; et al. Synthesis of (E)-8-(3-Chlorostyryl)caffeine Analogues Leading to 9-Deazaxanthine Derivatives as Dual A2A Antagonists/MAO-B Inhibitors. J. Med. Chem. 2013, 56, 1247–1261. [Google Scholar] [CrossRef]
- Załuski, M.; Schabikowski, J.; Schlenk, M.; Olejarz-Maciej, A.; Kubas, B.; Karcz, T.; Kuder, K.; Latacz, G.; Zygmunt, M.; Synak, D.; et al. Novel multi-target directed ligands based on annelated xanthine scaffold with aromatic substituents acting on adenosine receptor and monoamine oxidase B. Synthesis, in vitro and in silico studies. Bioorganic Med. Chem. 2019, 27, 1195–1210. [Google Scholar] [CrossRef]
- Koch, P.; Brunschweiger, A.; Namasivayam, V.; Ullrich, S.; Maruca, A.; Lazzaretto, B.; Küppers, P.; Hinz, S.; Hockemeyer, J.; Wiese, M.; et al. Probing Substituents in the 1- and 3-Position: Tetrahydropyrazino-Annelated Water-Soluble Xanthine Derivatives as Multi-Target Drugs With Potent Adenosine Receptor Antagonistic Activity. Front. Chem. 2018, 6, 206. [Google Scholar] [CrossRef]
- Binda, C.; Wang, J.; Pisani, L.; Caccia, C.; Carotti, A.; Salvati, P.; Edmondson, D.E.; Mattevi, A. Structures of Human Monoamine Oxidase B Complexes with Selective Noncovalent Inhibitors: Safinamide and Coumarin Analogs. J. Med. Chem. 2007, 50, 5848–5852. [Google Scholar] [CrossRef]
- Kuder, K.J.; Załuski, M.; Schabikowski, J.; Latacz, G.; Olejarz-Maciej, A.; Jaśko, P.; Doroz-Płonka, A.; Brockmann, A.; Müller, C.E.; Kieć-Kononowicz, K. Novel, Dual Target-Directed Annelated Xanthine Derivatives Acting on Adenosine Receptors and Monoamine Oxidase B. ChemMedChem 2020, 15, 772–786. [Google Scholar] [CrossRef]
- Li, F.; Wang, Z.-M.; Wu, J.-J.; Wang, J.; Xie, S.-S.; Lan, J.-S.; Xu, W.; Kong, L.-Y.; Wang, X.-B. Synthesis and pharmacological evaluation of donepezil-based agents as new cholinesterase/monoamine oxidase inhibitors for the potential application against Alzheimer’s disease. J. Enzym. Inhib. Med. Chem. 2016, 31 (Suppl. 3), 41–53. [Google Scholar] [CrossRef] [PubMed]
- Tavari, M.; Malan, S.F.; Joubert, J. Design, synthesis, biological evaluation and docking studies of sulfonyl isatin derivatives as monoamine oxidase and caspase-3 inhibitors. MedChemComm 2016, 7, 1628–1639. [Google Scholar] [CrossRef]
- Tripathi, R.K.P.; Ayyannan, S.R. Design, Synthesis, and Evaluation of 2-Amino-6-nitrobenzothiazole-Derived Hydrazones as MAO Inhibitors: Role of the Methylene Spacer Group. ChemMedChem 2016, 11, 1551–1567. [Google Scholar] [CrossRef] [PubMed]
- Mateev, E.; Kondeva-Burdina, M.; Georgieva, M.; Zlatkov, A. Repurposing of FDA-approved drugs as dual-acting MAO-B and AChE inhibitors against Alzheimer’s disease: An in silico and in vitro study. J. Mol. Graph. Model. 2023, 122, 108471. [Google Scholar] [CrossRef] [PubMed]








| Program | Company/Designer | License Terms | Search Algorithm | Score Function |
|---|---|---|---|---|
| AutoDock [44] | D. S. Good sell and A. J. Olson The Scripps Research Institute | Free for Academic use | Genetic algorithm Lamarckian genetic algorithm | Auto Dock (Force-field) |
| AutoDock Vina [45] | Dr. Oleg Trott Molecular Graphics Lab (CCSB), The Scripps Research Institute | Free for Academic use | Gradient optimization algorithm | United-atom scoring function |
| DOCK [46] | I. Kuntz University of California, San Francisco | Free for Academic use | Shape fitting; AMBER/GBSA and AMBER/PBSA for Dock 6.0 | Chem Score, GB/SA solvation Scoring |
| FlexX [47] | T. Lengauer and M. Rarey Bio Solve IT | Commercial Free evaluation | Incremental Build | FlexXScore, PLP, Screen Score, Drug Score |
| Glide [48] | Schrödinger Inc. | Commercial | Monte Carlo Sampling | Glide Score, Glide Comp |
| GOLD [49] | Cambridge Crystallographic Data Centre | Commercial | Genetic Algorithm | ChemPLP, GoldScore, ASP, Chemscore |
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 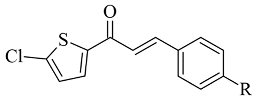 | Ki 5.52—0.49 (μM) | AutoDock 4.2 | 2BYB | [74] |
| 2. |  | Ki 0.11—9.30 (μM) | AutoDock 4.2 | 2BYB | [75] |
| 3. | 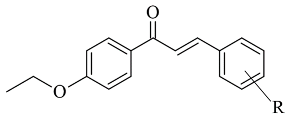 | IC50 0.68—28.50 (μM) | Glide | 2V5Z | [76] |
| 4 |  | IC50 0.0029—13.2 (μM) | AutoDock Vina | 4A79 | [77] |
| 5. | 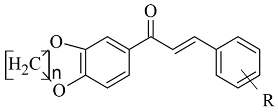 | IC50 0.00021—0.068 (μM) | Glide | 2V5Z | [78] |
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 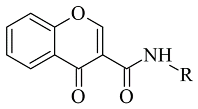 | IC50 1.03—7396 (nM) | Glide | 2V5Z | [91] |
| 2. | 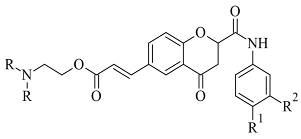 | IC50 0.63—3.81 (μM) | Glide | 2V5Z | [92] |
| 3. | 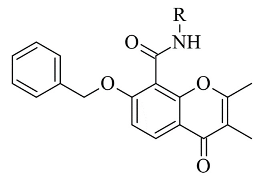 | IC50 0.078—2.22 (μM) | GOLD | 1GOS | [93] |
| 4. | 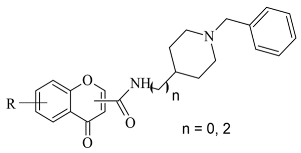 | IC50 0.035—52.45 (μM) | Molecular Operating Environment (MOE) | 2V60 | [94] |
| 5. | 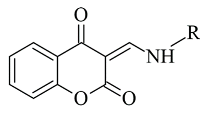 | IC50 5.07—621 (nM) | Discovery Studio (DS) | 2Z5X | [95] |
| 6. | 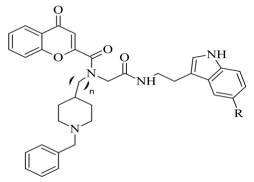 | IC50 15.17—80.59 (μM) | AutoDock Vina | 2V5Z | [96] |
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 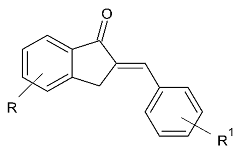 | IC50 0.0052—2.74 (μM) | Discovery Studio (DS) | 2V5Z | [99] |
| 2. | 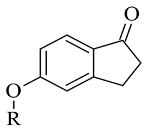 | IC50 0.015—0.298 (μM) | Discovery Studio (DS) | 2V5Z | [100] |
| 3. | 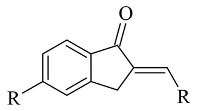 | IC50 0.0044—1.53 (μM) | Glide | 2V5Z | [101] |
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 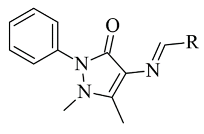 | IC50 0.049—0.114 (μM) | Glide | 2V5Z | [104] |
| 2. | 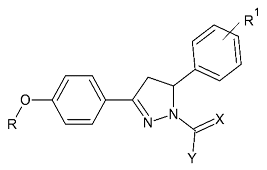 | IC50 0.38—>100 (μM) | AutoDock 4.2 | 4A79 | [105] |
| 3. | 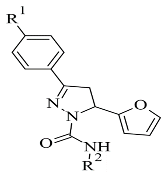 | - | AutoDock Vina | 2V61 | [106] |
| 4. | 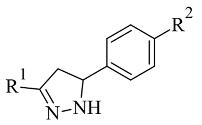 | - | Glide | 2V5Z | [107] |
| S. No. | Structure | MAO-B Inhibitor Activity | Docking Program | PDB Code | Ref. |
|---|---|---|---|---|---|
| 1. | 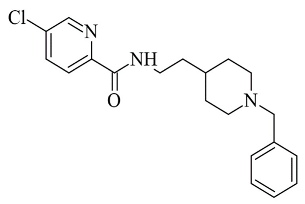 | IC50 2.53—94.1 (μM) | Molecular Operating Environment (MOE) | 2V60 | [116] |
| 2. | 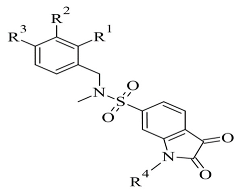 | Ki 2.57—55.18 (μM) | Molecular Operating Environment (MOE) | 2V5Z | [117] |
| 3. | 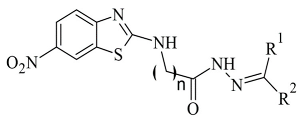 | IC50 0.00018—137 (μM) | AutoDock 4.2 | 2V5Z | [118] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateev, E.; Georgieva, M.; Mateeva, A.; Zlatkov, A.; Ahmad, S.; Raza, K.; Azevedo, V.; Barh, D. Structure-Based Design of Novel MAO-B Inhibitors: A Review. Molecules 2023, 28, 4814. https://doi.org/10.3390/molecules28124814
Mateev E, Georgieva M, Mateeva A, Zlatkov A, Ahmad S, Raza K, Azevedo V, Barh D. Structure-Based Design of Novel MAO-B Inhibitors: A Review. Molecules. 2023; 28(12):4814. https://doi.org/10.3390/molecules28124814
Chicago/Turabian StyleMateev, Emilio, Maya Georgieva, Alexandrina Mateeva, Alexander Zlatkov, Shaban Ahmad, Khalid Raza, Vasco Azevedo, and Debmalya Barh. 2023. "Structure-Based Design of Novel MAO-B Inhibitors: A Review" Molecules 28, no. 12: 4814. https://doi.org/10.3390/molecules28124814
APA StyleMateev, E., Georgieva, M., Mateeva, A., Zlatkov, A., Ahmad, S., Raza, K., Azevedo, V., & Barh, D. (2023). Structure-Based Design of Novel MAO-B Inhibitors: A Review. Molecules, 28(12), 4814. https://doi.org/10.3390/molecules28124814










