Abstract
In plants, methylation is a common step in specialized metabolic pathways, leading to a vast diversity of natural products. The methylation of these small molecules is catalyzed by S-adenosyl-l-methionine (SAM)-dependent methyltransferases, which are categorized based on the methyl-accepting atom (O, N, C, S, or Se). These methyltransferases are responsible for the transformation of metabolites involved in plant defense response, pigments, and cell signaling. Plant natural product methyltransferases are part of the Class I methyltransferase-superfamily containing the canonical Rossmann fold. Recent advances in genomics have accelerated the functional characterization of plant natural product methyltransferases, allowing for the determination of substrate specificities and regioselectivity and further realizing the potential for enzyme engineering. This review compiles known biochemically characterized plant natural product methyltransferases that have contributed to our knowledge in the diversification of small molecules mediated by methylation steps.
1. Introduction
Methylations, a process universal within all organisms, are responsible for extensive cellular modulations. These reactions are typically catalyzed by methyltransferases that require cofactor S-adenosyl-l-methionine (SAM), resulting in the formation of the methylated product and S-adenosyl-l-homocysteine (SAH) [1]. More commonly known for their roles in epigenetics, methyltransferases are also involved in natural product biosynthesis in plants. Plant natural product methyltransferases (PNPMTs) contribute to the diversification of specialized metabolites with functions such as pigments [2,3], antioxidants [4], signal transducer, and plant defense response [5,6,7,8]. Phenolic compounds, specifically, support plant growth and provide additional advantages to tolerate environmental stresses against insects and microbial invasion [9,10].
PNPMTs and their subsequent methylated products also display pharmaceutical properties for humans. Compared to their non-methylated counterparts, methylated plant natural products exhibit different chemical properties, and thereby alter biological activity. For instance, methylated resveratrol was shown to decrease its genotoxicity level compared to non-methylated resveratrol, which has been reported to cause chromosome aberration [11]. Furthermore, methylated resveratrol displays improved bioavailability and overall bioactivity [12,13]. Methylation also influences the binding of small molecules to a human neurotransmitter receptor. For example, as an intermediate in the morphine pathway, the O-methylated benzylisoquinoline (BIA) thebaine exhibits stimulatory effects and weaker analgesic properties compared to morphine, which lacks O-methylated groups [14]. Similarly, the methylated monoterpene indole alkaloid (MIA) ibogaine is less polar than its non-methylated counterpart, noribogaine, and is far more readily sequestered to the lipophilic compartment of the brain [15,16].
In recent years, advanced -omics have enabled the discoveries of new plant natural product methyltransferases. A number of PNPMTs have been elucidated by X-ray crystallography, illuminating the SAM and substrate-binding domains. Canonically, based on structures, all natural-product methyltransferases across kingdoms belong to Classes I and III [1]. PNPMTs, bearing the Rossmann fold-binding site for SAM, are categorized as Class I, while Class III is typically found in bacteria and associated with membranes [1,17,18,19]. The remaining classes of methyltransferases have been structurally characterized by the domains that participate in macromolecular (DNA, RNA, and protein) methylation for epigenetic regulation and have been reviewed elsewhere [19,20,21]. In the past twenty years, the emergence of synthetic biology and biotechnological advancements in the heterologous production of plant enzymes in microbial systems have accelerated the gene identification and functional characterization of plant natural product enzymes, including methyltransferases. Here, we highlight how the biochemical characterizations of PNPMT genes integrated with the structural knowledge could lay the foundations for more extensive enzyme engineering efforts. In this review, we focus on Class I, SAM-dependent plant natural product methyltransferases (PNPMTs), summarizing their structure, function, and application for metabolic engineering to alter their enzyme specificity and diversification of plant specialized metabolites.
2. Classification of PNPMTs Based on Methyl Acceptor
Methyltransferases are categorized based on the substrate atoms O-, N-, C-, S-, and Se- that accept the methyl group. Though rare, a few reported PNMPTs were noted for their ability to transfer methyl groups to more than one acceptor, such as those methylating halides and thiocyanates [22]. Among all plant natural product methyltransferases, O-directed methyltransferases (OMTs) are the most abundant to date [1]. OMTs target the hydroxyl groups of small molecules, such as alkaloids, lignin precursors, simple phenols, phytoalexins, phytohormones, and flavonoids, to produce their methylated form. This also includes the hydroxyl moiety of the carboxyl group, which can be found in salicylic acid [5,23], jasmonic acid [7] and iridoid loganic acid [24]. OMTs are also further characterized into two subclasses based on their dependence on or independence of a divalent cation. The cation-dependent OMTs, also known as caffeoyl-CoA OMTs (CCoAOMTs), typically have a lower subunit molecular weight of 26–30 kDa. CCoAOMTs are mainly involved in lignin, phytohormone, and scent metabolism [16,25]. Comparatively, the cation-independent OMTs, also known as caffeic acid OMT (COMT), range between 37 and 43 kDa and are primarily active in phenylpropanoid and alkaloid biosynthesis [26].
The abundance of OMTs is also seen through the incredible diversity of their methoxylated products (Figure 1). For instance, plant alkaloid OMTs have been well-characterized, including the monoterpene indole alkaloid in Catharanthus roseus to synthesize vindoline, which is then dimerized with catharanthine to form anticancer vinblastine [27] and monoterpene isoquinoline alkaloid in Psychotria ipecacuanha [28]. Notably, out of all known alkaloid OMTs to date, the majority of them are involved in benzylisoquinoline alkaloid biosynthesis (Table 1). Further characterization of plant alkaloid OMTs, known as norbelladine 4′OMTs, has been reported in Amaryllidaceae alkaloid biosynthesis from Narcissus spp and Lycoris aurea [29,30]. Although heterocyclic nitrogenous methoxypyrazines are not characterized as alkaloids, the OMTs involved in their pathway have been reported [31,32,33].
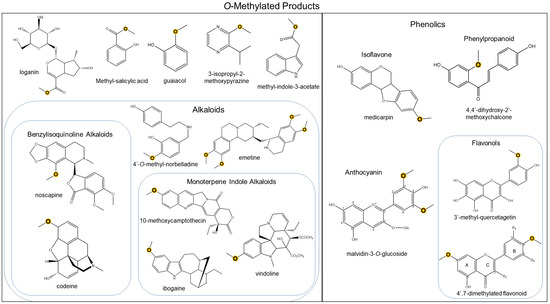
Figure 1.
Representation of chemical diversity generated by O-directed methyltransferases. Methylated atoms are highlighted in yellow.

Table 1.
Biochemically characterized plant natural product methyltransferases.
The majority of plant natural product OMTs have been characterized as part of phenylpropanoid-related biosynthetic pathways, which include flavonoids. Various types of flavonoids (flavonols, flavones, isoflavones, flavanols, and anthocyanins) play significant physiological roles in plants, typically in response to biotic and abiotic stresses [136]. For instance, the methoxylated flavonoid tricin in rice serves as a plant defense response against insect herbivores [137]. While flavonoid-OMTs generally do not require a cation for catalyzation, cation-dependent flavonoid-OMTs have been reported. Specific cation-dependent flavonoid-OMTs include the anthocyanin-OMT and an (iso) flavonol-6-OMT, which are multifunctional in methylating caffeoyl-CoA and associated flavonoids [70,90,95,138] (Table 1).
The position and number of methyl groups determine the structural diversity of flavonoids, though these are restricted by the placement of hydroxyl groups [136]. O-methylation can occur at any position theoretically, however, 7- and 4′-methoxylation are most prevalent among flavonoids; interestingly, a number of polymethoxylated flavonoids have also been reported [136]. Although the majority of flavonoid-OMTs are active towards aglycone substrates, anthocyanin 3′OMT and 5′OMT were later characterized and displayed strong preference for glycosylated substrates [138].
While OMTs are generally regiospecific, plant OMTs involved in phenylpropanoid and flavonoid biosynthesis are less selective and catalyze sequential methylations of almost identical substrates [1,65,107,139]. A small group of plant OMTs has demonstrated strong enzymatic specificity for a single structure, but overall, plant OMTs tend to accept a wide span of substrates, with varying substrate preferences, including flavonoids, lignin precursors, and alkaloids [48,140]. For example, an alkaloid OMT, 10-hydroxycamptothecin OMT in Camptotheca acuminata, was strongly preferred to methylate 10-hydroxycamptothecin but was also capable of methylating flavonoids, stilbenes, and caffeic acids [48].
Plant natural product OMTs have also been cloned and biochemically characterized from several BIA-producing species. In the most studied BIA-producing organism, Papaver somniferum, once (S)-norcoclaurine is formed by the condensation of dopamine and 4-hydroxyphenylacetaldehyde, the hydroxyl group in the isoquinoline moiety is methylated to produce (S)-coclaurine. Further characterization of similar cDNAs encoding this methyltransferase has been reported in six species [34,141,142,143,144]. Moreover, the crystal structure of norcoclaurine-6-O-methyltransferase from Thalictrum flavum (Tf6OMT) has been reported (PDB accession number: 5ICC) [143]. Additional OMTs in the BIA pathway contribute to the final step in the central pathway to produce (S)-reticuline by methylating 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT). The corresponding transcripts encoding 4′OMT-like enzymes have also been reported in three species [34,141,142,144,145,146]. A number of these BIA OMTs are remarkably substrate-specific, such as norreticuline 7-O-methyltransferase, which introduces a third methoxy group to produce (S)-norlaudanine in papaverine biosynthesis [38,141]. Multiple O-methylation is more common in BIA biosynthetic pathways, and the additional transcripts of similar enzymes have also been reported, including reticuline 7-O-methyltransferase and scoulerine 9-O-methyltransferase [39,141,142,144,147,148,149]. In other cases, a similar enzyme to biochemically characterized scoulerine 9-O-methyltransferase preferentially methylates the 2-hydroxyl of quaternary protoberberine and columbamine in only Coptis japonica [150].
Though not as abundant as the OMTs, N-directed methyltransferases (NMTs) still offer a respectable diversity of natural products. The N-methylated phytochemicals consist of primary amines, secondary amines found in indoles and imidazoles, tertiary amines, or more complex alkaloids [1] (Figure 2). Most N-directed methyltransferases have been cloned from alkaloid-producing plants. In particular, the caffeine biosynthetic pathway in xanthosine involves a series of N-methylation steps that serve as a model to study alkaloid biosynthesis [151,152]. The biosynthetic genes involved were cloned from Camellia sinensis [152,153], Coffea arabica [54,55,57,58,153] Paullinia cupana [59], Theobroma cacao [154], and Citrus sinensis [151]. Remarkably, caffeine and its xanthine precursors have evolved independently in a number of flowering plants using either of the two biochemical pathways through caffeine synthase or xanthine methyltransferase-like enzymes, as exhibited in chocolate, citrus, and guarana plants [151]. Based on coding sequences, xanthine NMTs were determined to belong to the SABATH (salicylic acid, benzoic acid, theobromine synthase) family of methyltransferases, which exclusively utilize the “proximity and desolvation effects” [1,5], rather than general acid-base catalysis or cation-dependent mechanisms. Since SABATH methyltransferases generally methylate oxygen atoms, it is likely that a recent change in the function of xanthine alkaloid-producing species occurred [16]. A recent study hypothesized that despite stemming from different lineages, xanthosine methyltransferases have convergent evolutionary histories [16,151].
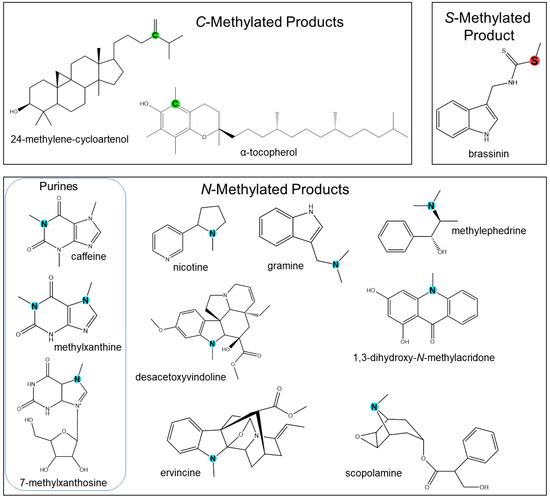
Figure 2.
Representation of chemical diversity generated by N-, C-, S-directed methyltransferases. Methylated atoms are highlighted in blue (N-), green (C-), and red (S-).
A number of NMTs involved in monoterpene indole alkaloid biosynthetic pathways have been characterized, including those involved in vindoline production in Catharanthus roseus [49], N-methylation of picrinine from the Apocynaceae family [51], and ajmaline synthesis [52]. NMTs have contributed significantly to the diversification of benzylisoquinoline alkaloids (BIA), and several NMTs have been characterized in nine Ranunculales species [16].
C-directed methyltransferases (CMTs) have been well characterized and primarily participate in specialized metabolism by methylating aliphatics, such as tocopherol [116,117,118] and sterol [120,121,122,123]. Notable examples highlighted in this review are illustrated in Figure 2.
S-directed methyltransferases (SMTs) are generally involved in the production of volatile halogen and sulfur compounds as well as thiocyanate [1]. A few examples of characterized SMTs in plants have been reported, including those from C. roseus [132]. A thiocyanate methyltransferase found in Arabidopsis thaliana that synthesizes methylthiocyanate [124] was later structurally analyzed [22]. Halide/bisulfide methyltransferase activity was also detected in the leaves of Brassica oleracea [155]. Some MTs exhibit high specificity for halides or bisulfides, known as halide ion methyltransferases (HMTs) and halide/thiol methyltransferases (HTMTs). HMTs and HTMTs are found in coastal trees, grasses, and several agricultural plants, with particularly high levels of activity in Raphanus sativus (daikon radish), Oryza sativa (paddy rice) Triticum aestivum (wheat), and Cyathea lepifera (fern) [125]. These HMT and HTMT genes were later cloned [156,157]. HMTs have also been biochemically characterized in broccoli with high substrate specificity toward homocysteine [158]. Lyi and colleagues also reported a selenocysteine methyltransferase that produces Se-methylselenocysteine [135]. Interestingly, these two genes play roles in sulfur and selenium metabolism in broccoli. Another SAM-dependent HTMT was cloned from Raphanus, and upon biochemical testing, exhibited high specificity for iodide, bisulfide, and thiocyanate [125]. Plant SMTs have also been characterized in the production of brassinin in cruciferous vegetables [131,159]. A comprehensive list of biochemically characterized PNPMTs with some structurally characterized features is detailed in Table 1.
3. The Identification of Plant Natural Product Methyltransferase (PNPMT) Genes
All known Class I PNPMTs consist of an alternating α-helix/ß-strand structure with associated monomeric molecular masses of 25 to 55 kDa [1]. Bioinformatic analyses to identify putative PNPMT genes are primarily based on a series of conserved motifs, with heavy reliance on the core Rossmann fold [1,25]. Within the Rossmann fold, a nine-residue amino acid incorporating a glycine-rich structure “GxGxG” signature sequence corresponding to a SAM-binding motif is found in SAM-dependent MTs (red boxes in Figure 3). Although these consensus sequences (V/I/L)(L/V)(D/E)(V/I)G(G/C)G(T/P)G [160] contain substitutions, the amino acid sequences are typically better aligned within the same subclasses (cation-dependent, cation-independent, SABATH, BIA).
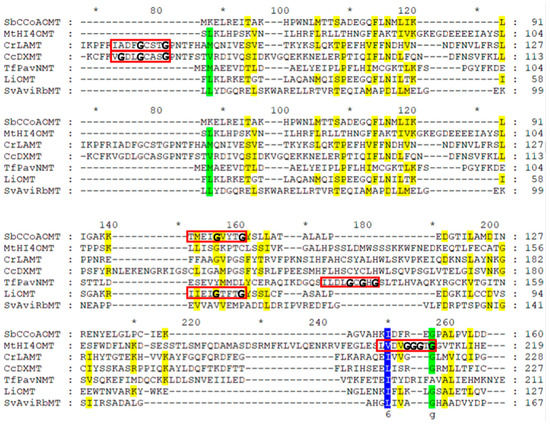
Figure 3.
Alignment of the protein sequences of plant NPMTs and bacterial MTs. The plant species and accession numbers of these MTs are listed in Table 1. Plant NPMTs include cation-dependent SbCCoAOMT, cation-independent MtHI4′OMT, SABATH CrLAMT, and CcDXMT, and BIA TfPavNMT, in comparison to the bacterial MT within Class I bacterial LiOMT (Leptospira interrogans, WP_000087781), and non-Class I bacterial SvAviRbMT (Streptomyces viridochromogenes, WP_003998250). The red boxes indicate the glycine-rich Rossmann fold consensus sequences with 0–3 mismatches. The amino acid residues highlighted in blue are highly conserved, those in green are conserved, and those in yellow are moderately conserved. “*” represents every 10th amino acid after each numerical indicator.
Amino acid sequences of PNPMTs from separate subclasses of cation-dependent, cation-independent, SABATH, and BIA MTs were chosen from the list of biochemically characterized plant natural product MTs (Table 1) to construct a phylogenetic tree (Figure 4). The genes within the same subclasses share high sequence similarity, while several of these highly homologous MTs have been reported to be involved in the biosynthesis of similar classes of metabolites, including clusters of BIA OMTs/NMTs and alkaloid biosynthetic genes (Ca10OMT, IpeOMT1, Vm16OMT, Cr16OMT) [16,48]. Sequence analysis and the phylogenetic results confirmed the functional classification of cation-dependent, cation-independent, and SABATH families. Class I cation-dependent bacteria OMT is included to compare with plant cation-dependent OMTs (Figure 4).
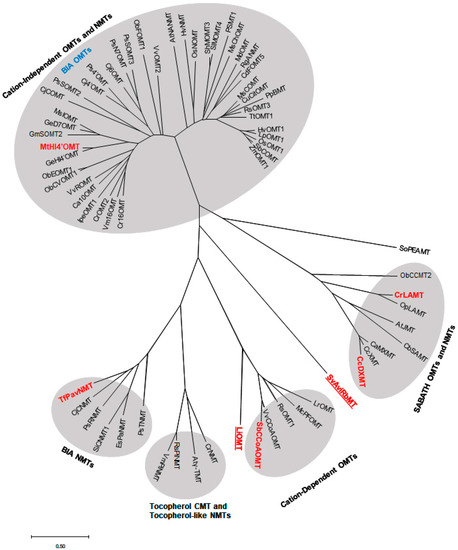
Figure 4.
Phylogenetic relationships among functionally characterized plant NPMTs and bacterial MTs. A phylogenetic tree was generated using the neighbor-joining method and the Poisson correlation method via MEGA X [161]. The scale bar shows the amino acids substituted per sequence alignment. Bolded sequences indicate the NPMTs further analyzed in this review. The underlined sequences are bacterial methyltransferases, showing the relationship between bacteria and plant NPMTs. The plant species and accession numbers can be found in Table 1. The bacterial MT species and accession numbers are: LiOMT, Leptospira interrogans O-methyltransferase (WP_000087781); SvAviRbMT, Streptomyces viridochromogenes antibiotic-resistant mediating O-methyltransferase (WP_003998250).
4. Structural Biochemistry of PNPMTs
Protein X-ray crystallography has allowed for the extensive structural elucidation of numerous Class I methyltransferases. The resulting X-ray crystal structures have offered great insight into the general structures of these methyltransferases and have determined that they remain highly consistent, despite the primary amino acid sequences having low similarity [162]. Nevertheless, the primary amino acid sequences are still necessary to conduct phylogenetic analysis as they yield clarity into the evolutionary relationships of all PNPMTs. Furthermore, increased genomic data and the availability of coding sequences facilitate the molecular characterization or determinants of function [16,151]. For example, characterized active sites that accept xanthine have distinguishing features that promote attachment with xanthine molecules [152,154]. In particular, the hydrophobic pocket and specific residues affecting substrate specificity, encourage such xanthine attachment, later confirmed by site-directed mutagenesis [16,151,154,163]. In plant OMTs, structural analysis often involves dimers. Most OMTs form homodimers [92], though a BIA OMT was reported to form a heterodimer [164]. Typically, a dimerization interface is present as several helices actively participate in substrate binding. This dimerization interface also constitutes a hydrophobic “back wall” [92], though its participation in substrate binding varies among plant OMTs [16]. The analysis of primary amino acid sequences revealed that the N-terminus domain contributes to the dimerization with partial involvement in substrate-binding within the hydrophobic pocket, while the C-terminus domain is involved in SAM-binding via the Rossmann fold.
Class I PNPMTs are characterized by a conserved domain known as the Rossmann fold. The Rossmann fold is defined by a seven-stranded ß-sheet, commonly ordered 3 2 1 4 5 7 6, with the seventh strand in an antiparallel position to the remaining strands and three α-helices on each side of the ß-sheet [17,165].
Recognized as the defining features of all SAM-dependent methyltransferases, six sequentially ordered motifs (I-VI) either contribute to the formation of the SAM-binding pocket or have interactions with SAM itself. [1,18,25,166] (Figure 5). Motif I consists of ß-strand I and the adjoining loop, and located at the N-terminus is a glycine-rich sequence (GxGxG) with one or two glycines interacting with SAM [18]. Generally, this glycine-rich sequence remains conserved throughout Class I, but there is a minority of cases where alanine residues replaced the glycines [18]. Motif II consists of ß-strand II and the following helix, and there is either an aspartate or glutamate residue located at the C-terminus [18,166]. The helix is structurally conserved and directly contributes to the SAM-binding pocket [18]. Motif III is involved in SAM binding. This motif possesses a hydrophilic amino acid at the N-terminus of ß-strand III, and a partially conserved glycine residue at the C-terminus [18]. Motif IV is composed of ß-strand IV and the adjoining loops. Located at the C-terminus is a partially conserved acidic residue [1]. Motif V consists of a helix following strand IV, although this motif does not typically interact with SAM [18]. However, in a few cases, this motif may act as a site for hydrophobic side chains, securing SAM’s adenine moiety [166]. Finally, motif VI, very uncommonly interacting with SAM, is characterized by strand V and its preceding loop, as well as a conserved glycine residue, followed by two hydrophobic residues at the beginning of the strand [18]. Interestingly, among all subclasses, the residues of the motifs were highly conserved, although SAM’s position with its binding pocket varied depending on the subclass [18,25,166] (Figure 5).
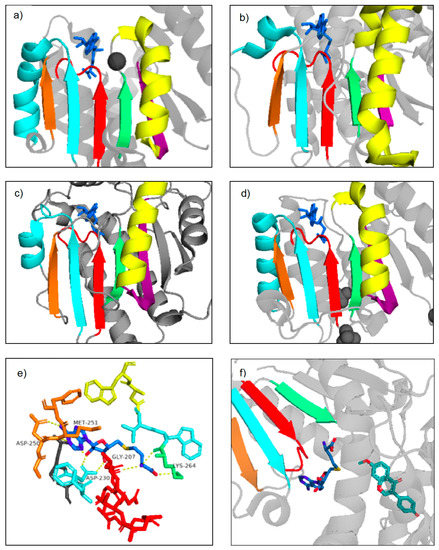
Figure 5.
Rossmann fold motifs highlighted in Class I plant NPMTs and Class I bacterial MTs. Three-dimensional visualizations were created using PDB crystal structures and PyMOL. (a) Cation-dependent, SbCCoAOMT, (b) Cation-independent MtHI4′OMT, (c) SABATH, CrLAMT, (d) bacterial Leptospira interrogans O-methyltransferase, LiOMT (PDB 2HNK), (e) closer look at the MtHI4′OMT SAM-binding pocket, with important residues and hydrogen bonding highlighted, (f) representation of the methyl transfer between SAM and MtH14′OMT. Species names and PDB numbers for the above-mentioned plant NPMTs can be found in Table 1. The colored beta sheets and alpha helices represent Motifs I-VI, which make up the characteristic MT Class I Rossmann fold. The red strand represents Motif I, the light blue strand and helix represents Motif II, the orange strand represents Motif III, the green strand represents Motif IV, the yellow helix represents Motif V, and the purple strand represents Motif VI. The darker blue structure is either SAM or its demethylated form, SAH. The lighter blue structure is the substrate.
The Rossmann fold is a multifunctional domain of the PNPMT and other Class I methyltransferases alike—not only is the Rossmann fold able to house the methyl donor SAM, but it also has influences on the substrate-binding pocket, and thus, the orientation of the methyl acceptor atom [5]. While the Rossmann-fold domain is widely conserved across all Class I methyltransferases, structural tweaks allow for some degree of promiscuity. In contrast to DNA methyltransferases, which are designed to identify the general features of macromolecular substrates due to their widely conserved active sites [19], PNPMTs generally exhibit higher levels of specificity [1]. A number of important amino acid residues that contribute to this specificity have been identified via structural-based analysis and confirmed by site-directed mutagenesis in several cases, including in caffeoyl-CoA methyltransferases [103], chalcone and isoflavone methyltransferase [92], and benzylisoquinoline [16,167].
In some alkaloid biosynthesis studies, heterodimers have been suggested to participate in the catalysis of new substrates. From Thalictrum tuberosum, the heterodimers of four OMTs accepted substrates, such as catechols, hydroxycinnamates, and alkaloids, exhibit far more promiscuity than their homodimer counterparts [47]. A missing step in noscapine biosynthesis in Papaver somniferum was filled with a then newly discovered heterodimer consisting of PsSOMT2 and PsSOMT/Ps6OMT [164]. Due to the degree of promiscuity within alkaloid heterodimers, understanding their functionalization has the potential to uncover new biochemical processes [136]. Overall, the advances in the structural biochemistry of PNMPTs have highlighted the degree of specificity that both conserves the SAM-binding pocket and provides structural determinants of functions and their involvements in plant natural product biosynthesis.
5. Engineering Plant Natural Product Methyltransferases (PNPMTs)
Methyltransferases are attractive enzymes to manipulate for the purpose of altering metabolic pathways. The regioselectivity of PNPMTs is relatively more favorable in comparison to organic synthesis in which protecting groups are often added to produce specific methylated products with a further disadvantage of low yield [1]. The structure–function analysis has been extensively applied for enzyme engineering, which involves mutagenesis approaches to substitute their amino acids to obtain the desired methylated products. For example, a poplar flavonoid-OMT mutant with amino acid Asp257Gly was reported to methylate the 3-hydroxyl group compared to its native selectivity in methylating the 7-hydroxyl group [168]. In another case, variants of Vitis vinifera resveratrol OMT with point mutations were generated and resulted in modified native substrate specificity to increase the yield of pinostilbene (monomethylated resveratrol), rather than to produce dimethylated resveratrol [169]. In addition to shifting in the regiospecificity of methylation and/or modified substrate specificity, the promiscuity of PNPMTs has been proven to be more versatile, as the biosynthetic pathway can be altered with a possibility of yielding an unknown molecule via de novo pathways, and thus, increasing the diversity of plant natural products [1,170]. In most cases, beneficial mutations are identified by in-silico methods combined with a high-throughput screening assay to accelerate the engineering process. Zhao and colleagues rationally designed Peucedanum praeruptorum bergaptol O-methyltransferase to increase the production of bergapten with improved pharmacological properties, especially for increasing pigmentation levels [171]. One Val320Ile mutant protein was particularly reported to be promising for further applications in treating depigmentation disorder [171]. In the absence of crystal structures, computer-guided analysis to analyze the active site and the catalytic mechanisms of plant natural product methyltransferases can provide alternative manufacturing strategies via metabolic engineering approaches in microbial systems.
A number of de novo biosynthesis pathways that produce methylated natural products have been successfully designed in microbial systems. For instance, dimethylated resveratrol (pterostilbene) was produced in an engineered Escherichia coli strain expressing Arabidopsis caffeic acid/resveratrol O-methyltransferase. The intracellular pool of l-tyrosine, a precursor molecule in the stilbene biosynthetic pathway, was improved in this engineered strain grown in l-methionine-rich media to increase the SAM supply [172]. This approach overcomes a central issue in engineering methyltransferases, which is cofactor regeneration, as SAH is a known inhibitor for methyltransferase activities and its accumulation can be toxic for the microbial hosts [1,170]. Although the incorporation of native SAM recycling pathways involving multiple enzymatic reactions can be challenging to integrate within the host systems, deregulation of the SAM pathway was shown to successfully increase the production of O-methylated products. MetJ or Met repressor in bacteria is often a target for silencing or deletion in order to regulate methionine production. Silencing of the MetJ gene via CRISPRi enhances the production of O-methylated anthocyanin in Escherichia coli [173]. Similarly, deleting MetJ was also effective in shuttling the pool of SAM in an engineered Escherichia coli strain that overexpressed methionine biosynthetic pathway genes, especially in improving vanillate production [174].
The engineering of methyltransferases has advanced beyond its ability in transferring a methyl group. The development of SAM analogs has allowed for the diversification of small molecules. In particular, a mutated halide methyltransferase from Arabidopsis thaliana efficiently transferred ethyl-, propyl-, and allyl-moieties to SAH in order to produce SAM analogs with the respective alkyl groups [175]. Recently, efforts in developing SAM analogs to diversify active biomolecules have been accelerated after the discovery of the naturally occurring carboxy-S-adenosylmethionine pathway involved in tRNA modification [176]. Interestingly, coclaurine N-methyltransferase from Coptis japonica was selected to test the possibility of plant methyltransferases catalyzing the reaction using an alternative cofactor of carboxy-SAM [177]. By comparing its SAM-binding residues with the crystal structure tRNA carboxymethyltransferase, which highly prefers carboxyl-SAM, coclaurine N-methyltranferase was successfully engineered to accept carboxy-SAM over SAM to produce carboxymethylated tetrahydroisoquinoline [177]. While the use of SAM analogs in the diversification of plant small molecules seems likely to be effective at smaller scales, more innovations are still required to increase efforts in their regeneration and efficient synthesis.
Although microbial systems remain excellent hosts for engineering plant methyltransferases, plant hosts are still relevant to consider, especially in obtaining natural products for industrial and specific agricultural applications. Engineered monolignol 4-O-methyltransferase, for example, was useful for reducing lignin content for the effective production of liquid biofuels, a desired utilization of cellulosic fiber, while it allows for the diversification of possible active compounds in transgenic plants [178]. Engineered plant host systems also provide additional platforms for extending the natural product biosynthetic pathways beyond methylated metabolites, as novel compounds with unique biological activities might be accumulated. Overall, the versatility of plant natural product methyltransferases can inspire future efforts in enzyme and metabolic engineering for improved capabilities in widening the diversity of bioactive compounds with pharmacological properties.
6. Conclusions and Future Directions
The structural elucidation of PNPMTs has heavily augmented our knowledge of how SAM-dependent methyltransferases contribute to the diversification of small molecules in the plant kingdom. The increasing number of structure determinations, along with the elucidation of their architecture, has significantly provided insights into their molecular evolution. A comprehensive representation of biochemically characterized PNPMTs demonstrates not only the structural diversity of Class I of SAM-dependent methyltransferases but also provides a point of reference to search for other PNPMTs that have yet to be discovered. With the continuous advances in next-generation sequencing and progress in -omics-based technologies, we anticipate more PNPMT genes will be discovered. While only less than 20% of the PNPMTs that we compiled have been structurally elucidated, we expect more reports on the protein crystal structures of PNPMTs. This knowledge of structural biochemistry will benefit the development of machine learning algorithms for more advanced prediction of protein folding, gene annotation, and functional validation. By delineating the substrate specificity and structural attributes of PNPMTs, we have provided an overview of the recently expanded strategies to facilitate the development of enzyme and metabolic engineering. We also anticipate that all these advancements will give rise to further diversification and increased production of methylated natural products with enhanced pharmacological properties as human therapeutics. Ultimately, the complete knowledge of molecular and structural determinants of PNPMT function will reveal important aspects of the catalytic mechanism and inform metabolic engineering efforts, thus generating novel biocatalysts for wide applications in biotechnology.
Author Contributions
Conceptualization, A.L., R.M., P.E. and V.S.; software, R.M., A.L., S.P. and S.-A.J.; writing—original draft preparation, A.L., R.M., S.P., S.-A.J., P.E. and V.S.; writing—review and editing, A.L., S.-A.J. and V.S.; visualization, R.M., S.P. and S.-A.J.; supervision, V.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH), United States of America, under grant number P20GM103424.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the Louisiana State University Shreveport (LSUS) Cyber Collaboratory staff for technical assistance and computational support. We also acknowledge Elahe Mahdavian for 3-D molecular visualization tools.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liscombe, D.K.; Louie, G.V.; Noel, J.P. Architectures, mechanisms and molecular evolution of natural plant product methyltransferases. Nat. Prod. Rep. 2012, 29, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, J.; Ji, K.-X.; Zeng, Q.-Y.; Bhuiya, M.-W.; Su, S.; Shu, Q.-Y.; Ren, H.-X.; Liu, Z.-A.; Wang, L.-S. Methylation mediated by an anthocyanin, O-methylation, is involved in purple flower coloration in Paeonia. J. Exp. Biol. 2015, 66, 6563–6577. [Google Scholar]
- Roldan, M.V.G.; Outchkourov, N.; van Houwelingen, A.; Lammers, M.; de la Fuente, I.R.; Ziklo, N.; Aharoni, A.; Hall, R.D.; Beekwilder, J. An O-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J. 2014, 80, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Sattler, S.; Maeda, H.; Sakuragi, Y.; Bryant, D.A.; DellaPenna, D. Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 2003, 15, 2343–2356. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Ross, J.R.; Koscheski, P.; Yang, Y.; Pichersky, E.; Noel, J.P. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 2003, 15, 1704–1716. [Google Scholar] [CrossRef]
- Farmer, E.E.; Ryan, C.A. Botany Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 1990, 87, 7713–7716. [Google Scholar] [CrossRef]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwanag, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef]
- Liu, C.-J.; Deavours, B.E.; Richard, S.B.; Ferrer, J.-L.; Blount, J.W.; Huhman, D.; Dixon, R.A.; Noel, J.P. Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 2006, 18, 3656–3669. [Google Scholar] [CrossRef]
- Ibrahim, R.K.; Bruneau, A.; Bantignies, B. Plant O-Methyltransferases: Molecular analysis, common signature, and classification. Plant Mol. Biol. 1998, 36, 1–10. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant secondary metabolites as defenses, regulators, and primary metabolites: The blurred functional trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Fukuhara, K.; Nakanishi, I.; Matsuoka, A.; Matsumura, T.; Honda, S.; Hayashi, M.; Ozawa, T.; Miyata, N.; Saito, S.; Ikota, N.; et al. Effect of methyl substitution on the antioxidative property and genotoxicity of resveratrol. Chem. Res. Toxicol. 2008, 21, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Mikstacka, R.; Pryzbylska, D.; Rimando, A.M.; Baer-Dubowska, W. Inhibition of human recombinant cytochromes P450 CYP1A1 and CYP1B1 by trans-resveratrol methyl ethers. Mol. Nutr. Food Res. 2007, 51, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-Y.; Lee, J.K.; Choi, O.; Kim, C.Y.; Jang, J.-H.; Hwang, B.Y.; Hong, Y.-S. Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol. 2014, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G.; Elliott, H.W. The effects of morphine, morphinone and thebaine on the EEG and behavior of rabbits and cats. Neuropharmacology 1971, 10, 367–377. [Google Scholar] [CrossRef]
- Zubaran, C. Ibogaine and noribogaine: Comparing parent compound to metabolite. CNS Drug Rev. 2000, 6, 219–240. [Google Scholar] [CrossRef]
- Morris, J.S.; Facchini, P.J. Molecular origins of functional diversity in benzylisoquinoline alkaloid methyltransferases. Front. Plant Sci. 2019, 10, 1058. [Google Scholar] [CrossRef]
- Chouhan, B.P.S.; Maimaiti, S.; Gade, M.; Laurino, P. Rossmann-fold methyltransferases: Taking a “β-turn” around their cofactor, S-adenosylmethionine. Biochemistry 2019, 58, 166–170. [Google Scholar] [CrossRef]
- Gana, R.; Rao, S.; Huang, H.; Wu, C.; Vasudevan, S. Structural and functional studies of S-adenosyl-L-methionine binding proteins: A ligand-centric approach. Struct. Biol. 2013, 13, 6. [Google Scholar] [CrossRef]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X. Many paths to methyltransferase: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef]
- He, X.-J.; Chen, T.; Zhu, J.-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Scheer, S.; Ackloo, S.; Medina, T.S.; Schapira, M.; Li, F.; Ward, J.A.; Lewis, A.M.; Northrop, J.P.; Richardson, P.L.; Kaniskan, H.U.; et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat. Commun. 2019, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Schmidberger, J.W.; James, A.B.; Edwards, R.; Naismith, J.H.; O’Hagan, D. Halomethan biosynthesis: Structure of a SAM-dependent halide methyltransferase from Arabidopsis thaliana. Angew. Chem. Int. Ed. 2010, 122, 3646–3648. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.R.; Nam, K.H.; D’Auria, J.C.; Pichersky, E. S-Adenosyl-l-Methionine: Salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 1999, 367, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Roepke, J.; Gordon, H.; De Luca, V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 2008, 20, 524–542. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.P.; Chiang, V.L. Conserved sequence motifs in plant S-adenosyl-l-methionine-dependent methyltransferases. Plant Mol. Biol. 1998, 37, 663–674. [Google Scholar] [CrossRef]
- Lam, K.C.; Ibrahim, R.K.; Behdad, B.; Dayanandan, S. Structure, function, and evolution of plant O-methyltransferases. CSP 2007, 50, 1001–1013. [Google Scholar]
- Levac, D.; Murata, J.; Kim, W.S.; De Luca, V. Application of carborundum abrasion for investigating the leaf epidermis: Molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 2008, 53, 225–236. [Google Scholar] [CrossRef]
- Nomura, T.; Kutchan, T.M. Three new O-methyltransferases are sufficient for all O-methylation reactions of ipecac alkaloid biosynthesis in root culture of Psychotria ipecacuanha. J. Biol. Chem. 2010, 285, 7722–7738. [Google Scholar] [CrossRef]
- Kilgore, M.B.; Augustin, M.M.; Starks, C.M.; O’Neil-Johnson, M.; May, G.D.; Crow, J.A.; Kutchan, T.M. Cloning and characterization of a norbelladine 4′-O-methyltransferase involved in the biosynthesis of the Alzheimer’s drug galanthamine in Narcissus sp. aff. pseudonarcissus. PLoS ONE 2014, 9, e103223. [Google Scholar] [CrossRef]
- Sun, J.; Wang, P.; Wang, R.; Li, Y.; Xu, S. Molecular cloning and characterization of a meta/para-O-Methyltransferase from Lycoris aurea. Int. J. Mol. Sci. 2018, 19, 1911. [Google Scholar] [CrossRef]
- Dunlevy, J.D.; Soole, K.L.; Perkins, M.V.; Dennis, E.G.; Keyzers, R.A.; Kalua, C.M.; Boss, P.K. Two O-methyltransferases involved in the biosynthesis of methoxypyrazines: Grape-derived aroma compounds important to wine flavor. Plant Mol. Biol. 2010, 74, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Dunlevy, J.D.; Dennis, E.G.; Soole, K.L.; Perkins, M.V.; Davies, C.; Boss, P.K. A methyltransferase essential for the methoxypyrazine-derived flavour of wine. Plant J. 2013, 75, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Guillaumie, S.; Ilg, A.; Réty, S.; Brette, M.; Trossat-Magnin, C.; Decroocq, S.; Léon, C.; Keime, C.; Ye, T.; Baltnweck-Guyot, R.; et al. Genetic analysis of the biosynthesis of 2-methoxy-3-isobutylpyrazine, a major grape-derived aroma compound impacting wine quality. Plant Physiol. 2013, 162, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Morishige, T.; Tsujita, T.; Yamada, Y.; Sato, F. Molecular characterization of the S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 2000, 275, 23398–23405. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.B.; Morishige, T.; Shitan, N.; Yazaki, K.; Sato, F. Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 2002, 277, 830–835. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Thompson, M.L.; Shepard, S.A.; Dunstan, M.S.; Herbert, A.J.; Smith, D.R.M.; Cronin, V.A.; Menon, B.R.K. Structure and biocatalytic scope of coclaurine N-methyltransferase. Angew. Chem. 2018, 57, 10600–10604. [Google Scholar] [CrossRef]
- Cabry, M.P.; Offen, W.A.; Saleh, P.; Li, Y.; Winzer, T.; Graham, I.A.; Davies, G.J. Structure of papaver somniferum O-methyltransferase 1 reveals initiation of noscapine biosynthesis with implications for plant natural product methylation. ACS Catal. 2019, 9, 3840–3848. [Google Scholar] [CrossRef]
- Pienkny, S.; Brandt, W.; Schmidt, J.; Kramell, R.; Ziegler, J. Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J. 2009, 60, 56–67. [Google Scholar] [CrossRef]
- Dang, T.-T.T.; Facchini, P.J. Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 2012, 159, 618–631. [Google Scholar] [CrossRef]
- Zhao, W.; Shen, C.; Zhu, J.; Ou, C.; Liu, M.; Dai, W.; Liu, X.; Liu, J. Identification and characterization of methyltransferases involved in benzylisoquinoline alkaloids biosynthesis from Stephania intermedia. Biotechnol. Lett. 2020, 42, 461–469. [Google Scholar] [CrossRef]
- Li, Q.; Bu, J.; Ma, Y.; Yang, J.; Hu, Z.; Lai, C.; Xu, Y.; Tang, J.; Cui, G.; Wang, Y.; et al. Characterization of O-methyltransferases involved in the biosynthesis of tetrandrine in Stephania tetrandra. J. Plant Physiol. 2020, 250, 153181. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Perdomo, I.M.; Facchini, P.J. Isolation and characterization of two O-methyltransferases involved in benzylisoquinoline alkaloid biosynthesis in sacred lotus (Nelumbo nucifera). J. Biol. Chem. 2020, 256, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.S.; Facchini, P.J. Isolation and characterization of reticuline N-methyltransferase involved in biosynthesis of the aporphine alkaloid magnoflorine in opium poppy. J. Biol. Chem. 2016, 291, 23416–23427. [Google Scholar] [CrossRef] [PubMed]
- Liscombe, D.K.; Facchini, P.J. Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 2007, 282, 14741–14751. [Google Scholar] [CrossRef]
- Torres, M.A.; Hoffarth, E.; Eugenio, L.; Savtchouk, J.; Chen, X.; Morris, J.S.; Facchini, P.J.; Ng, K.K. Structural and functional studies of pavine N-methyltransferase from Thalictrum flavum reveal novel insights into substrate recognition and catalytic mechanism. J. Biol. Chem. 2016, 291, 23403–23415. [Google Scholar] [CrossRef]
- Lang, D.E.; Morris, J.S.; Rowley, M.; Torres, M.A.; Maksimovich, V.A.; Facchini, P.J.; Ng, K.K.S. Structure-function studies of tetrahydroprotoberberine N-methyltransferase reveal the molecular basis of stereoselective substrate recognition. J. Biol. Chem. 2019, 294, 14482–14498. [Google Scholar] [CrossRef]
- Frick, S.; Kutchan, T.M. Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J. 1999, 17, 329–339. [Google Scholar] [CrossRef]
- Salim, V.; Jones, A.D.; DellaPenna, D. Camptotheca acuminata 10-hydroxycamptothecin O-methyltransferase: An alkaloid biosynthetic enzyme co-opted from flavonoid metabolism. Plant J. 2018, 95, 112–125. [Google Scholar] [CrossRef]
- Liscombe, D.K.; Usera, A.R.; O’Connor, S.E. Homolog of tocopherol C-methyltransferases catalyzes N-methylation in anticancer alkaloid biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 18793–18798. [Google Scholar] [CrossRef]
- Farrow, S.C.; Kamileen, M.O.; Meades, J.; Ameyaw, B.; Xiao, Y.; O’Connor, S.E. Cytochrome P450 and O-methyltransferase catalyze the final steps in the biosynthesis of the anti-addictive alkaloid ibogaine from Tabernanthe iboga. J. Biol. Chem. 2018, 293, 13821–13833. [Google Scholar] [CrossRef]
- Levac, D.; Cázares, P.; Yu, F.; De Luca, V. A picrinine N-methyltransferase belongs to a new family of γ-tocopherol-like methyltransferases found in medicinal plants that make biologically active monoterpenoid indole alkaloids. Plant Physiol. 2016, 170, 1935–1944. [Google Scholar] [CrossRef] [PubMed]
- Cázares-Flores, P.; Levac, D.; De Luca, V. Rauvolfia serpentina N-methyltransferases involved in ajmaline and N-methylajmaline biosynthesis belong to a gene family derived from γ-tocopherol C-methyltransferase. Plant J. 2016, 87, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Stander, E.A.; Sepúlveda, L.J.; Dugé de Bernonville, T.; Carqueijeiro, I.; Koudounas, K.; Lemos Cruz, P.; Besseau, S.; Lanoue, A.; Papon, N.; Giglioli-Guivarc’h, N.; et al. Identifying genes involved in alkaloid biosynthesis in Vinca minor through transcriptomics and gene co-expression analysis. Biomolecules 2020, 10, 1595. [Google Scholar] [CrossRef] [PubMed]
- Uefuji, H.; Ogita, S.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol. 2003, 132, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Herai, Y.; Koizumi, N.; Kusano, T.; Sano, H. 7-Methylxanthine methyltransferase of coffee plants. J. Biol. Chem. 2001, 276, 8213–8218. [Google Scholar] [CrossRef]
- McCarthy, A.A.; Biget, L.; Lin, C.; Petiard, V.; Tanksley, S.D.; McCarthy, J.G. Cloning, expression, crystallization and preliminary X-ray analysis of the XMT and DXMT N-methyltransferases from Coffea canephora (robusta). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 304–307. [Google Scholar] [CrossRef]
- Mizuno, K.; Kato, M.; Irino, F.; Yomeyama, N.; Fujimura, T.; Ashhara, H. The first committed step reaction of caffeine biosynthesis: 7-methylxanthosine synthase is closely homologous to caffeine synthase in coffee (Coffea arabica L.). FEBS 2003, 547, 56–60. [Google Scholar] [CrossRef]
- Mizuno, K.; Okuda, A.; Kato, M.; Yoneyama, N.; Tanaka, H.; Ashihara, H.; Fujimura, T. Isolation of a new dual-function caffeine synthase gene encoding an enzyme for the conversion of 7-methylxanthine to caffeine from coffee (Coffea arabica L.). FEBS 2003, 534, 75–81. [Google Scholar] [CrossRef]
- Schimpl, F.C.; Kiyota, E.; Mayer, J.L.S.; Gonçalves, J.F.d.C.; da Silva, J.F.; Mazzafera, P. Molecular and biochemical characterization of caffeine synthase and purine alkaloid concentration in guarana fruit. Phytochemistry 2014, 105, 25–36. [Google Scholar] [CrossRef]
- Li, W.; Qiao, C.; Pang, J.; Zhang, G.; Luo, Y. The versatile O-methyltransferase LrOMT catalyzes multiple O-methylation reactions in Amaryllidaceae alkaloids biosynthesis. Int. J. Biol. Macromol. 2019, 141, 680–692. [Google Scholar] [CrossRef]
- Hibi, N.; Higashiguchi, S.; Hashimoto, T.; Yamada, Y. Gene expression in tobacco low-nicotine mutants. Plant Cell 1994, 6, 723–735. [Google Scholar] [PubMed]
- Stenzel, O.; Teuber, M.; Drager, B. Putrescine N-methyltransferase in Solanum tuberosum L., a calystegine-forming plant. Planta 2006, 233, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Tueber, M.; Azeni, M.E.; Namjoyan, F.; Meier, A.; Wodak, A.; Brandt, W.; Drager, B. Putrescine N-methyltransferase—A structure-function analysis. Plant Mol. Biol. 2007, 63, 787–801. [Google Scholar] [CrossRef]
- Junker, A.; Fischer, J.; Sichhart, Y.; Brandt, W.; Drager, B. Evolution of the key alkaloid enzyme putrescine N-methyltransferase from spermidine synthase. Front. Plant Sci. 2013, 29, 260. [Google Scholar] [CrossRef]
- Itoh, N.; Iwata, C.; Toda, H. Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa. BMC Plant Biol. 2016, 16, 180. [Google Scholar] [CrossRef] [PubMed]
- Cacace, S.; Schröder, G.; Wehinger, E.; Strack, D.; Schmidt, J.; Schröder, J. A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry. 2003, 62, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Schröder, G.; Wehinger, E.; Lukacin, R.; Wellmann, F.; Seefelder, W.; Schwab, W.; Schröder, J. Flavonoid methylation: A novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 2004, 65, 1085–1094. [Google Scholar] [CrossRef]
- Seoka, M.; Ma, G.; Zhang, L.; Yahata, M.; Yamawaki, K.; Kan, T.; Kato, M. Expression and functional analysis of the nobiletin biosynthesis-related gene CitOMT in citrus fruit. Sci. Rep. 2020, 10, 15288. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Fukushi, Y.; Wollenweber, E.; Ibrahim, R.K. Characterization of two O-methyltransferase-like genes in barley and maize. Pharm. Biol. 2008, 46, 26–34. [Google Scholar] [CrossRef]
- Berim, A.; Hyatt, D.C.; Gang, D.R. A set of regioselective O-methyltransferases gives rise to the complex pattern of methoxylated flavones in sweet basil. Plant Physiol. 2012, 160, 1052–1069. [Google Scholar] [CrossRef]
- Shimizu, T.; Lin, F.; Hasegawa, M.; Okada, K.; Nojiri, H.; Yamane, H. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 2012, 287, 19315–19325. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Li, C.; Jones, D.; Pichersky, E. Characterization of a flavonol 3-O-methyltransferase in the trichomes of the wild tomato species Solanum habrochaites. Planta 2012, 236, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Matsuba, Y.; Ning, J.; Schilmiller, A.L.; Hammar, D.; Jones, A.D.; Pichersky, E.; Last, R.L. Analysis of natural and induced variation in tomato glandular trichome flavonoids identifies a gene not present in the reference genome. Plant Cell 2014, 26, 3272–3285. [Google Scholar] [CrossRef] [PubMed]
- Willits, M.G.; Giovanni, M.; Prata, R.T.N.; Kramer, C.M.; De Luca, V.; Steffens, J.C.; Graser, G. Bio-fermentation of modified flavonoids: An example of in vivo diversification of secondary metabolites. Phytochemistry 2004, 65, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, R.; Gao, S.; Cheng, A. The functional characterization of a site-specific apigenin 4′-O-methyltransferase synthesized by the liverwort species Plagiochasma appendiculatum. Molecules 2017, 22, 759. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Li, C.; Shi, F.; Jones, A.D.; Pichersky, E. Polymethylated myricetin in trichomes of the wild tomato species Solanum habrochaites and characterization of trichome-specific 3′/5′ and 7/4′-myricetin O-methyltransferases. Plant Physiol. 2011, 155, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Wils, C.R.; Brandt, W.; Manke, K.; Vogt, T. A single amino acid determines position specificity of an Arabidopsis thaliana CCoAOMT-like O-methyltransferase. FEBS 2013, 587, 683–689. [Google Scholar] [CrossRef]
- Muzac, I.; Wang, J.; Anzellotti, D.; Zhang, H.; Ibrahim, R.K. Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch. Biochem. Biophys. 2000, 375, 385–388. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Goodman, H.M. An Arabidopsis gene encoding a putative 14-3-3-interacting protein, caffeic acid/5-hydroxyferulic acid O-methyltransferase. Biochim. Biophys. Acta. 1997, 1353, 199–202. [Google Scholar] [CrossRef]
- Gauthier, A.; Gulick, P.J.; Ibrahim, R.K. cDNA cloning and characterization of a 3′/5′-O-methyltransferase for partially methylated flavonols from Chrysosplenium americanum. Short Comm. 1996, 32, 1163–1169. [Google Scholar] [CrossRef]
- Gauthier, A.; Gulick, P.J.; Ibrahim, R.K. Characterization of Two cDNA Clones Which Encode O-Methyltransferases for the Methylation of both Flavonoid and Phenylpropanoid Compounds. Arch. Biochem. Biophys. 1998, 351, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Tobimatsu, Y.; Matsumoto, N.; Suzuki, S.; Lan, W.; Takeda, Y.; Yamamura, M.; Sakamoto, M.; Ralph, J.; Lo, C.; et al. OsCAldOMT1 is a bifunctional O-methyltransferase involved in the biosynthesis of tricin-liginins in rice cell walls. Sci. Rep. 2019, 9, 11597. [Google Scholar] [CrossRef] [PubMed]
- Chandran, K.S.; Humphries, J.; Goodger, J.Q.D.; Woodrow, I.E. Molecular characterisation of flavanone O-methylation in Eucalyptus. Int. J. Mol. Sci. 2022, 23, 3190. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, B.-G.; Lee, Y.; Ryu, J.Y.; Lim, Y.; Hur, H.-G.; Ahn, J.-H. Regiospecific methylation of naringenin to ponciretin by soybean O-methyltransferase expressed in Escherichia coli. J. Biotechnol. 2005, 119, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Fournier-Level, A.; Hugueney, P.; Verriès, C.; This, P.; Ageorges, A. Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol. 2011, 11, 179. [Google Scholar] [CrossRef]
- Busam, G.; Junghanns, K.T.; Kneusel, R.E.; Kassemeyer, H.H.; Matern, U. Characterization and expression of caffeoyl-coenzyme A 3-O-methyltransferase proposed for the induced resistance response of Vitis vinifera L. Plant Physiol. 1997, 115, 1039–1048. [Google Scholar] [CrossRef]
- Hugueney, P.; Provenzano, S.; Verries, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef]
- Giordano, D.; Provenzano, S.; Ferrandino, A.; Vitali, M.; Pagliarani, C.; Roman, F.; Cardinale, F.; Castellarin, S.D.; Schubert, A. Characterization of a multifunctional caffeoyl-CoA O-methyltransferase activated in grape berries upon drought stress. Plant Physiol. Biochem. 2016, 101, 23–32. [Google Scholar] [CrossRef]
- Akashi, T.; Sawada, Y.; Shimada, N.; Sakurai, N.; Aoki, T.; Ayabe, S. cDNA Cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol. 2003, 44, 103–112. [Google Scholar] [CrossRef]
- Uchida, K.; Sawada, Y.; Ochiai, K.; Sato, M.; Inaba, J.; Hirai, M.Y. Identification of a unique type of isoflavone O-methyltransferase, GmIOMT1, based on multi-omics analysis of soybean under biotic stress. Plant Cell Physiol. 2020, 61, 1974–1985. [Google Scholar] [CrossRef]
- He, X.-Z.; Reddy, J.T.; Dixon, R.A. Stress responses in alfalfa (Medicago sativa L). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol. Biol. 1998, 36, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; He, X.-Z.; Dixon, R.A.; Noel, J.P. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 2001, 8, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Gou, J.; Wang, X.; Fan, R.; Zhang, Y. An alternative pathway for formononetin biosynthesis in Pueraria lobata. Front. Plant Sci. 2016, 7, 861. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Gou, J.; Zhang, Y. Molecular cloning and functional characterization of a novel isoflavone 3′-O-methyltransferase from Pueraria lobata. Front. Plant Sci. 2016, 7, 793. [Google Scholar] [CrossRef] [PubMed]
- Ibdah, M.; Zhang, X.-H.; Schmidt, J.; Vogt, T. A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J. Biol. Chem. 2003, 278, 43961–43972. [Google Scholar] [CrossRef]
- Kim, B.G.; Lee, H.J.; Park, Y.; Lim, Y.; Ahn, J.-H. Characterization of an O-methyltransferase from soybean. Plant Physiol. Biochem. 2006, 44, 236–241. [Google Scholar] [CrossRef]
- Zubieta, C.; Kota, P.; Ferrer, J.-L.; Dixon, R.A.; Noel, J.P. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 2002, 14, 1265–1277. [Google Scholar] [CrossRef]
- Gang, D.R.; Lavid, N.; Zubieta, C.; Chen, F.; Beuerle, T.; Lewinsohn, E.; Noel, J.P.; Pichersky, E. Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 2002, 14, 505–519. [Google Scholar] [CrossRef]
- Kim, B.G.; Lee, Y.; Hur, H.G.; Lim, Y.; Ahn, J.H. Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 2006, 67, 387–394. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Fukushi, Y.; Wang, X.-F.; Ibrahim, R.K. Characterization of a novel flavone O-methyltransferase gene in rice. Nat. Prod. Commun. 2006, 1, 981–984. [Google Scholar] [CrossRef]
- Widiez, T.; Hartman, T.G.; Dudai, N.; Yan, Q.; Lawton, M.; Havkin-Frenkel, D.; Belanger, F.C. Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol Biol. 2011, 76, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Wiens, B.; Luca, V.D. Molecular and biochemical characterization of a benzenoid/phenylpropanoid meta/para-O-methyltransferase from Rauwolfia serpentina roots. Phytochemistry 2016, 132, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.-L.; Zubieta, C.; Dixon, R.A.; Noel, J.P. Crystal structures of alfalfa caffeoyl coenzyme a 3-O-methyltransferase. Plant Physiol. 2005, 137, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.M.; Sattler, S.A.; Regner, M. The structure and catalytic mechanism of sorghum bicolor caffeoyl-CoA O-methyltransferase. Plant Physiol. 2016, 172, 78–92. [Google Scholar] [CrossRef]
- Green, A.R.; Lewis, K.M.; Barr, J.T.; Jones, J.P.; Lu, F.; Ralph, J.; Vermerris, W.; Sattler, S.E.; Kang, C. Determination of the structure and catalytic mechanism of sorghum bicolor caffeic acid O-methyltransferase and the structural impact of three brown midrib12 mutations. Plant Physiol. 2014, 165, 1440–1456. [Google Scholar] [CrossRef]
- Kapteyn, J.; Qualley, A.V.; Xie, Z.; Fridman, E.; Dudareva, N.; Gang, D.R. Evolution of cinnamate/p-coumarate carboxyl methyltransferases and their role in the biosynthesis of methylcinnamate. Plant Cell 2007, 19, 3212–3229. [Google Scholar] [CrossRef]
- Louie, G.V.; Bowman, M.E.; Tu, Y.; Mouradov, A.; Spangenberg, G.; Noel, J.P. Structure-function analyses of a caffeic acid O-methyltransferase from perennial ryegrass reveal the molecular basis for substrate preference. Plant Cell 2010, 22, 4114–4127. [Google Scholar] [CrossRef]
- Wang, J.; Pichersky, E. Characterization of S-adenosyl-L-methionine: (iso)eugenol O-methyltranferase involved in floral scent production in Clarkia breweri. Arch. Biochem. Biophys. 1998, 349, 153–160. [Google Scholar] [CrossRef]
- Nagel, J.; Culley, L.K.; Lu, Y.; Liu, E.; Matthews, P.D.; Stevens, J.F.; Page, J.E. EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 2008, 20, 186–200. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Harrison, M.J.; Dixon, R.A. Molecular characterization and expression of alfalfa isoliquiritigenin 2′O-methyltransferase, an enzyme specifically involved in the biosynthesis of an inducer of Rhizobium meliloti nodulation genes. Plant J. 1993, 4, 971–981. [Google Scholar] [CrossRef]
- Koeduka, T.; Hatada, M.; Suzuki, H.; Suzuki, S.; Matsui, K. Molecular cloning and functional characterization of an O-methyltransferase catalyzing 4′-O-methylation of resveratrol in Acorus calamus. J. Biosci. Bioeng. 2019, 127, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Schmidlin, L.; Poutaraud, A.; Claudel, P.; Mestre, P.; Prado, E.; Santos-Rosa, M.; Wiedemann-Merdinoglu, S.; Karst, F.; Merdinoglu, D.; Hugueney, P. A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008, 148, 1630–1639. [Google Scholar] [CrossRef]
- Chen, F.; D’Auria, J.C.; Tholl, D.; Ross, J.R.; Gershenzon, J.; Noel, J.P.; Pichersky, E. An Arabidopsis gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003, 36, 577–588. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Q.; Liu, Y.; Hao, X.; Wang, C.; Liang, Y.; Chen, J.; Xiao, Y.; Kai, G. Divergent camptothecin biosynthetic pathway in Ophiorrhiza pumila. BMC Biol. 2021, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, N.; Zeng, Z.; Xu, S.; Huang, C.; Wang, W.; Liu, T.; Luo, J.; Kong, L. Cloning, functional characterization, and catalytic mechanism of a bergaptol O-methyltransferase from Peucedanum praeruptorum Dunn. Front. Plant Sci. 2016, 7, 722. [Google Scholar] [CrossRef] [PubMed]
- Shintani, D.; DellaPenna, D. Elevating the vitamin E content of plants through metabolic engineering. Science 1998, 282, 2098–2100. [Google Scholar] [CrossRef]
- Koch, M.; Lemke, R.; Heise, K.-P.; Mock, H.-P. Characterization of γ-tocopherol methyltransferase from Capsicum annuum L and Arabidopsis thaliana. Eur. J. Biochem. 2003, 270, 84–92. [Google Scholar] [CrossRef]
- Tavva, V.S.; Kim, Y.-H.; Kagan, I.A.; Dinkins, R.D.; Kim, K.-H.; Collins, G.B. Increased alpha-tocopherol content in soybean seed overexpressing the Perilla frutescens gamma-tocopherol methyltransferase gene. Plant Cell Rep. 2007, 26, 61–70. [Google Scholar] [CrossRef]
- Sarkate, A.; Saini, S.S.; Gaid, M.; Teotia, D.; Mir, J.I.; Agrawal, P.K.; Beerhues, L.; Sircar, D. Molecular cloning and functional analysis of a biphenyl phytoalexin-specific O-methyltransferase from apple cell suspension cultures. Planta 2019, 249, 677–691. [Google Scholar] [CrossRef]
- Shi, J.; Gonzales, R.A.; Bhattacharyya, M.K. Identification and characterization of an S-adenosyl-L-methionine: Δ24-sterol-C-methyltransferase cDNA from soybean. J. Biol. Chem. 1996, 271, 9384–9389. [Google Scholar] [CrossRef]
- Guan, H.; Zhao, Y.; Su, P.; Tong, Y.; Liu, Y.; Hu, T.; Zhang, Y.; Zhang, X.; Li, J.; Wu, X.; et al. Molecular cloning and functional identification of sterol C24-methyltransferase gene from Tripterygium wilfordii. Acta Pharm. Sin. B 2017, 7, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Bouvier-Nave, P.; Husselstein, T.; Desprez, T.; Benveniste, P. Identification of cDNAs encoding sterol methyltransferases involved in the second methylation step of plant sterol biosynthesis. Eur. J. Biochem. 1997, 246, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Bouvier-Nave, P.; Husselstein, T.; Benveniste, P. Two families of sterol methyltransferases are involved in the first and the second methylation steps of plant sterol biosynthesis. Eur. J. Biochem. 1998, 256, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Nagatoshi, Y.; Nakamura, T. Arabidopsis HARMLESS TO OZONE LAYER Protein methylates a glucosinolate breakdown product and functions in resistance to Pseudomonas syringae pv. maculicola. J. Biol. Chem. 2009, 284, 19301–19309. [Google Scholar] [CrossRef]
- Itoh, N.; Toda, H.; Matsuda, M.; Negishi, T.; Taniguchi, T.; Ohsawa, N. Involvement of S-adenosylmethionine-dependent halide/thiol methyltransferase (HTMT) in methyl halide emissions from agricultural plants: Isolation and characterization of an HTMT-coding gene from Raphanus sativus (daikon radish). BMC Plant Biol. 2009, 9, 116. [Google Scholar] [CrossRef]
- Rohde, B.; Hans, J.; Martens, S.; Baumert, A.; Hunziker, P.; Matern, U. Anthranilate N-methyltransferase, a branch-point enzyme of acridone biosynthesis. Plant J. 2008, 53, 541–553. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.-J.; Lee, H.Y.; Back, K. Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J. Pineal Res. 2016, 60, 65–73. [Google Scholar] [CrossRef]
- Morris, J.S.; Groves, R.A.; Hagel, J.M.; Facchini, P.J. An N-methyltransferase from Ephedra sinica catalyzing the formation of ephedrine and pseudoephedrine enables microbial phenylalkylamine production. J. Biol. Chem. 2018, 293, 13364–13376. [Google Scholar] [CrossRef]
- Larsson, K.A.E.; Zetterlund, I.; Delp, G.; Jonsson, L.M.V. N-methyltransferase involved in gramine biosynthesis in barley:cloning and characterization. Phytochem. 2006, 67, 2002–2008. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Ziemak, M.J.; Henry, S.A.; Weretilnyk, E.A.; Hanson, A.D. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 2000, 275, 14095–14101. [Google Scholar] [CrossRef]
- Klein, A.P.; Satterly, E.S. Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proc. Natl. Acad. Sci. USA 2017, 114, 1910–1915. [Google Scholar] [CrossRef]
- Coiner, H.; Schröder, G.; Wehinger, E.; Liu, C.-J.; Noel, J.P.; Schwab, W.; Schröder, J. Methylation of sulfhydryl groups: A new function for a family of small molecule plant O-methyltransferases. Plant J. 2006, 46, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Koeduka, T.; Kajiyama, M.; Suzuki, H.; Furuta, T.; Tsuge, T.; Matsui, K. Benzoid biosynthesis in the flowers of Eriobotrya japonica: Molecular cloning and functional characterization of p-methoxybenzoic carboxyl methyltransferase. Planta 2016, 244, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ferrer, J.-L.; Ross, J.; Guan, J.; Yang, Y.; Pichersky, E.; Noel, J.P.; Chen, F. Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol. 2008, 146, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lyi, S.; Heller, L.I.; Rutzke, M.; Welch, R.M.; Kochian, L.V.; Li, L. Molecular and biochemical characterization of the selenocysteine Se-methyltransferase gene and Se-methylselenocysteine synthesis in broccoli. Plant Physiol. 2005, 138, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fernie, A.R.; Tohge, T. Diversification of chemical structures of methoxylated flavonoids and genes encoding flavonoid-O-methyltransferases. Plants 2022, 11, 564. [Google Scholar] [CrossRef]
- Gong, G.; Yuan, L.-Y.; Li, Y.-F.; Xiao, H.-X.; Li, Y.-F.; Zhang, Y.; Wu, W.-J.; Zhang, Z.-F. Salivary protein 7 of the brown planthopper functions as an effector for mediating tricin metabolism in rice plants. Sci. Rep. 2022, 12, 3205. [Google Scholar] [CrossRef] [PubMed]
- Lücker, J.; Martens, S.; Lund, S.T. Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3′,5′-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry 2010, 71, 1474–1484. [Google Scholar] [CrossRef]
- Kopycki, J.G.; Rauh, D.; Chumanevich, A.A.; Neumann, P.; Vogt, T.; Stubbs, M.T. Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J. Mol. Biol. 2008, 378, 154–164. [Google Scholar] [CrossRef]
- Roje, S. S-adenosyl-l-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef]
- Desgagné-Penix, I.; Facchini, P.J. Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 2012, 72, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Hagel, J.M.; Facchini, P.J. Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol. 2015, 169, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Robin, A.Y.; Giustini, C.; Graindorge, M.; Matringe, M.; Dumas, R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 2016, 87, 641–653. [Google Scholar] [CrossRef] [PubMed]
- He, S.-M.; Liang, Y.-L.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.-M.; Zhang, J.-J.; Wang, X.; Dong, Y.; Yang, J.-L.; et al. Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in Coptis species. Front. Plant Sci. 2018, 9, 731. [Google Scholar] [CrossRef]
- Facchini, P.J.; Park, S.-U. Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 2003, 64, 177–186. [Google Scholar] [CrossRef]
- Ziegler, J.; Diaz-Chávez, M.L.; Kramell, R.; Ammer, C.; Kutchan, T.M. Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta 2005, 222, 458–471. [Google Scholar] [CrossRef]
- Ounaroon, A.; Decker, G.; Schmidt, J.; Lottspeich, F.; Kutchan, T.M. (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum—cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 2003, 36, 808–819. [Google Scholar] [CrossRef]
- Fujii, N.; Inui, T.; Iwasa, K.; Morishige, T.; Sato, F. Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 2007, 16, 363–375. [Google Scholar] [CrossRef]
- Purwanto, R.; Hori, K.; Yamada, Y.; Sato, F. Unraveling additional O-methylation steps in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica). Plant Cell Physiol. 2017, 58, 1528–1540. [Google Scholar] [CrossRef]
- Morishige, T.; Dubouzet, E.; Choi, K.-B.; Yazaki, K.; Sato, F. Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 2002, 269, 5659–5667. [Google Scholar] [CrossRef]
- Huang, R.; O’Donnell, A.J.; Barboline, J.J.; Barkman, T.J. Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. USA 2016, 113, 10613–10618. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.A.; McCarthy, J.G. The structure of two N-Methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 2007, 144, 879–889. [Google Scholar] [CrossRef]
- Kato, M.; Mizuno, K.; Crozier, A.; Fujimura, T.; Ashihara, H. Caffeine synthase gene from tea leaves. Nature 2000, 406, 956–957. [Google Scholar] [CrossRef]
- Yoneyama, N.; Morimoto, H.; Ye, C.; Ashihara, H.; Mizuno, K.; Kato, M. Substrate specificity of N-methyltransferase involved in purine alkaloids synthesis is dependent upon one amino acid residue of the enzyme. Mol. Gen. Genom. 2006, 275, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Attieh, J.M.; Hanson, A.D.; Saini, H.S. Purification and characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J. Biol. Chem. 1995, 270, 9250–9257. [Google Scholar] [CrossRef] [PubMed]
- Attieh, J.; Sparace, S.A.; Saini, H.S. Purification and properties of multiple isoforms of a novel thiol methyltransferase involved in the production of volatile sulfur compounds from Brassica oleracea. Arch. Biochem. Biophys. 2000, 380, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Attieh, J.; Djiana, R.; Koonjul, P.; Étienne, C.; Sparace, S.A.; Saini, H.S. Cloning and functional expression of two plant thiol methyltransferases: A new class of enzymes involved in the biosynthesis of sulfur volatiles. Plant Mol. Biol. 2002, 50, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Lyi, S.M.; Zhou, X.; Kochian, L.V.; Li, L. Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 2007, 68, 1112–1119. [Google Scholar] [CrossRef]
- Dunbar, K.L.; Scharf, D.H.; Litomska, A.; Hertweck, C. Enzymatic carbon-sulfur bond formation in natural product biosynthesis. Chem. Rev. 2017, 117, 5521–5577. [Google Scholar] [CrossRef]
- Kagan, R.M.; Clarke, S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. 1994, 310, 417–427. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Struck, A.-W.; Thompson, M.L.; Wong, L.S.; Micklefield, J. S-Adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem 2012, 13, 2642–2655. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-Q.; Yao, M.-Z.; Ma, C.-L.; Ma, J.-Q.; Chen, L. Natural allelic variations of TCS1 play a crucial role in caffeine biosynthesis of tea plant and its related species. Plant Physiol. Biochem. 2016, 100, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Chen, X.; Lang, D.E.; Ng, K.K.S.; Facchini, P.J. Heterodimeric O-methyltransferases involved in the biosynthesis of noscapine in opium poppy. Plant J. 2018, 95, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L.; McMillan, F.M. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 19. [Google Scholar] [CrossRef]
- Morris, J.S.; Yu, L.; Facchini, P.J. A single residue determines substrate preference in benzylisoquinoline alkaloid N-methyltransferases. Phytochemistry 2020, 170, 112193. [Google Scholar] [CrossRef]
- Joe, E.J.; Kim, B.-G.; An, B.-C.; Chong, Y.; Ahn, J.-H. Engineering of flavonoid O-methyltransferase for a novel regioselectivity. Mol. Cells 2010, 30, 137–141. [Google Scholar] [CrossRef]
- Herrera, D.P.; Chánique, A.M.; Martínez-Márquez, A.; Bru-Martínez, R.; Kourist, R.; Parra, L.P.; Schüller, A. Rational design of resveratrol O-methyltransferase for the production of pinostilbene. Int. J. Mol. Sci. 2021, 22, 4345. [Google Scholar] [CrossRef]
- Zhang, C.; Sultan, S.A.; Rehka, T.; Chen, X. Biotechnological applications of S-adenosyl-methionine-dependent methyltransferases for natural products biosynthesis and diversification. Bioresour.Bioprocess 2021, 8, 72. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, N.; Wu, H.; Zhou, Y.; Huang, C.; Luo, J.; Zeng, Z.; Kong, L. Structure-based tailoring of the first coumarins-specific bergaptol O-methyltransferase to synthesize bergapten for depigmentation disorder treatment. J. Adv. Res. 2020, 21, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.T.; Kang, S.-Y.; Hong, Y.-S. De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis. Microb. Cell Fact. 2017, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Cress, B.E.; Leitz, Q.D.; Kim, D.C.; Amore, T.D.; Suzuki, J.Y.; Linhardt, R.J.; Koffas, M.A.G. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Kunjapur, A.M.; Hyun, J.C.; Prather, K.L.J. Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Fact. 2016, 15, 1–17. [Google Scholar] [CrossRef]
- Tang, Q.; Grathwol, C.W.; Asianüzel, A.S.; Wu, S.; Link, A.; Pavlidis, I.V.; Badenhorst, C.P.S.; Bornscheuer, U.T. Directed evolution of a halide methyltransferase enables biocatalytic synthesis of diverse SAM analogs. Angew. Chem. Int. Ed. 2021, 60, 1524–1527. [Google Scholar] [CrossRef]
- Kim, J.; Xiao, H.; Bonanno, J.B.; Kalyanaraman, C.; Brown, S.; Tang, X.; Al-Obaidi, N.F.; Patskovsky, Y.; Babbitt, P.C.; Jacobson, M.P.; et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 2013, 498, 123–126. [Google Scholar] [CrossRef]
- Herbert, A.J.; Shepherd, S.A.; Cronin, V.A.; Bennett, M.R.; Sung, R.; Micklefield, J. Engineering orthogonal methyltransferases to create alternative bioalkylation pathways. Angew. Chem. Int. Ed. 2020, 59, 14950–14956. [Google Scholar] [CrossRef]
- Zhang, K.; Bhuiya, M.-W.; Pazo, J.R.; Miao, Y.; Kim, H.; Ralph, J.; Liu, C.-J. An Engineered Monolignol 4-O-Methyltransferase Depresses Lignin Biosynthesis and Confers Novel Metabolic Capability in Arabidopsis. Plant Cell 2012, 24, 3135–3152. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).