Abscisic Acid—Defensive Player in Flax Response to Fusarium culmorum Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material and Conditions of Flax Seedling Growth
2.2. H2O2 Content Determination in Flax Seedlings after Infection
2.3. Reverse Transcription Real-Time PCR
2.4. Isolation and Analysis of Terpenoids
2.5. Isolation and Analysis of Sterols
2.6. Abscisic Acid Isolation and Quantification
2.7. Callose Isolation and Quantification
2.8. Flax Seedling Treatment with Abscisic Acid
2.9. Treatment of F. culmorum with ABA
2.10. Treatment of F. culmorum with Carotenoids and Tocopherol
2.11. Statistical Analysis
3. Results
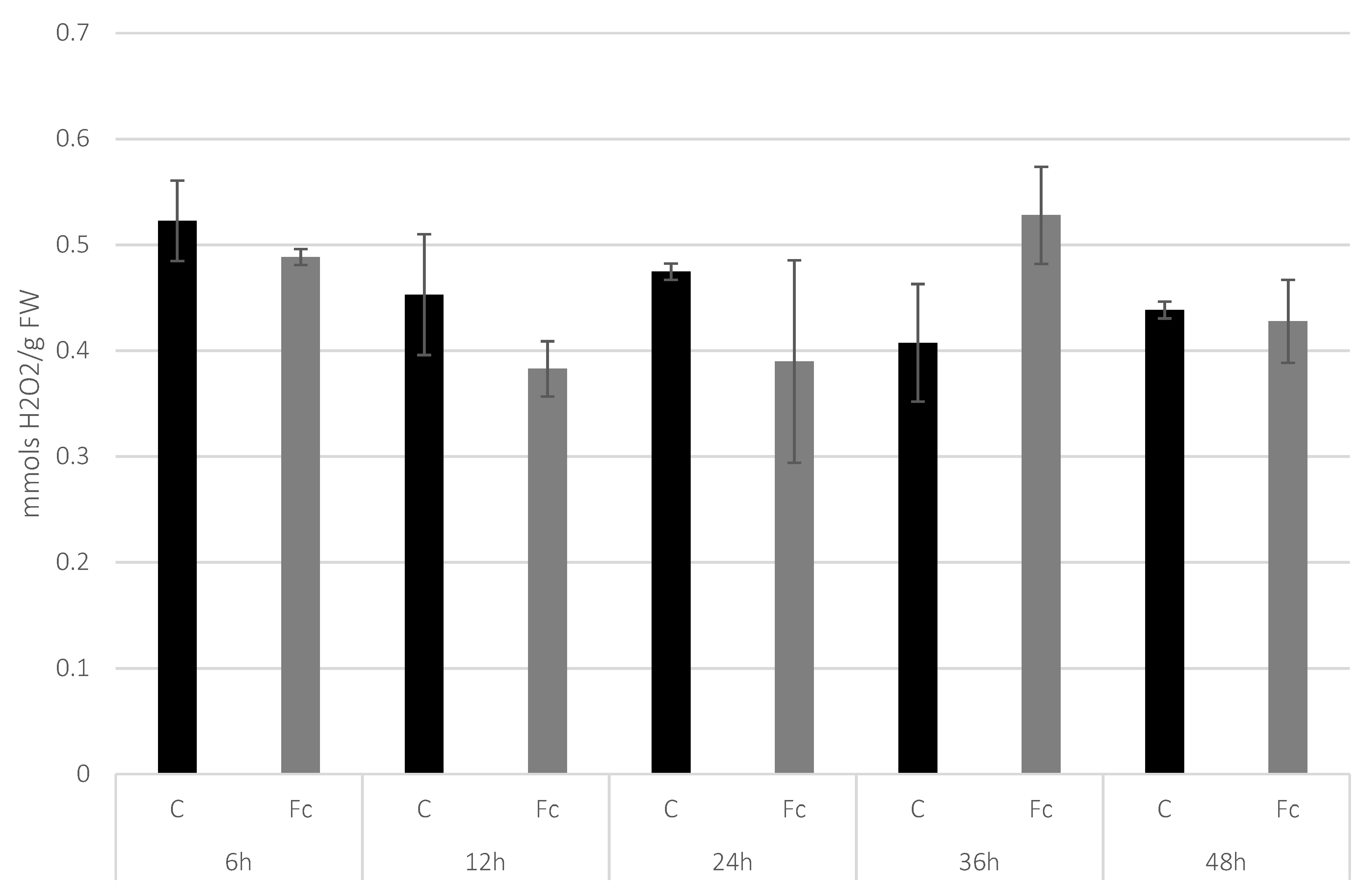
3.1. Determination of H2O2 Level in Flax Seedlings after F. culmorum Infection
3.2. Analysis of Transcript Levels of Genes Involved in ROS Processing
3.3. Analysis of Transcript Levels of Genes Involved in Isoprenoid Biosynthesis
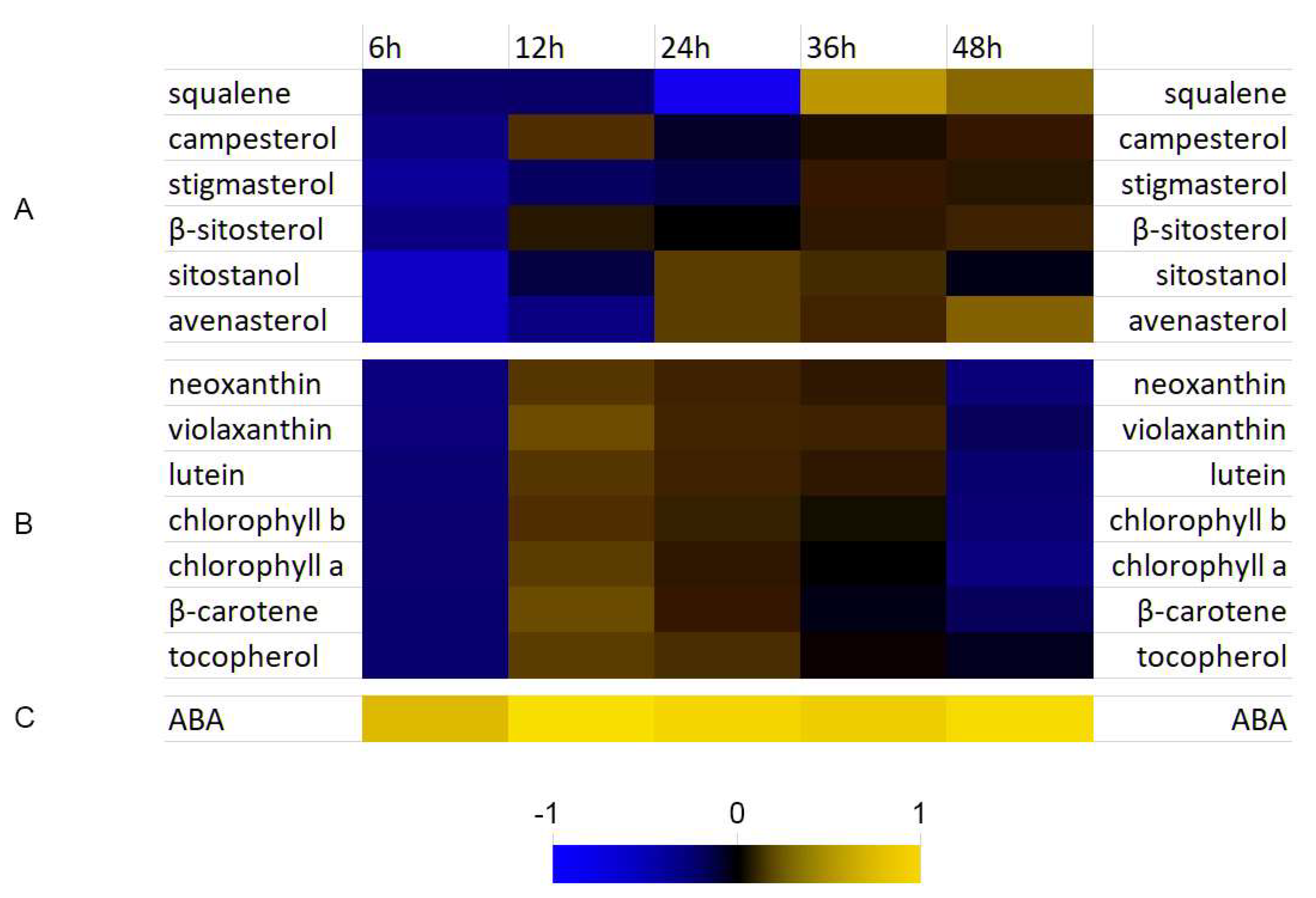
3.4. Analysis of Selected Isoprenoid Levels
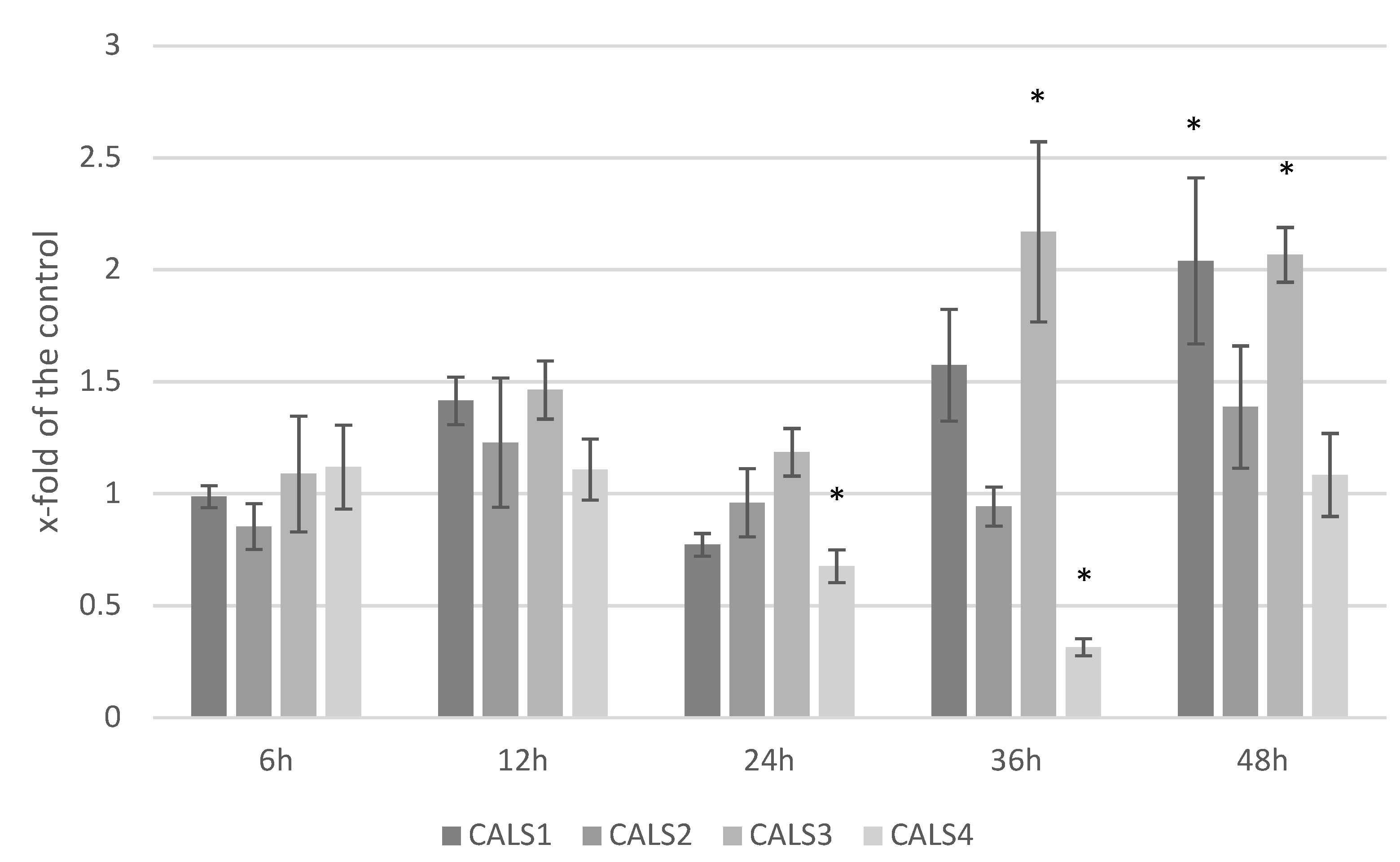
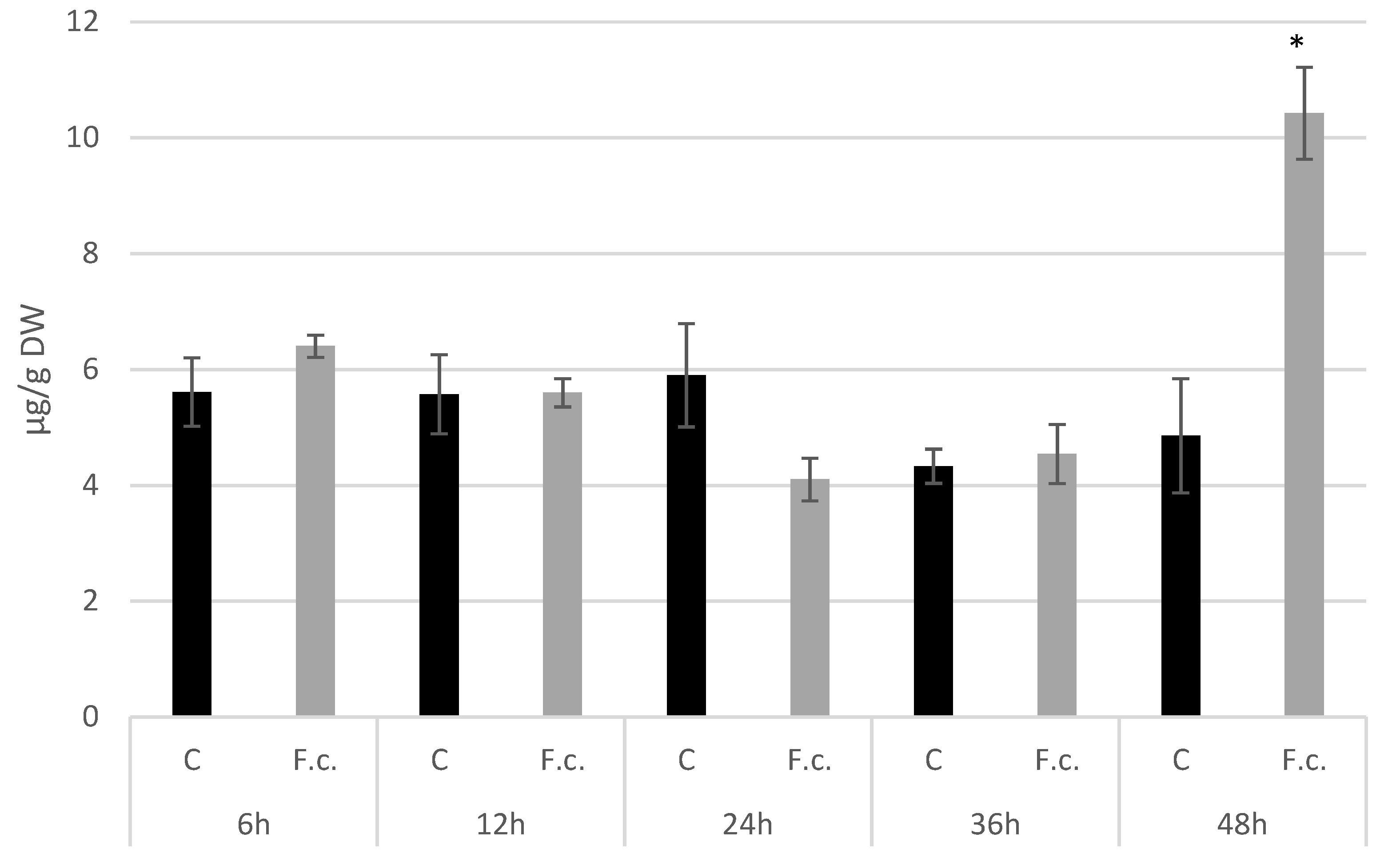
3.5. Analysis of the Expression Level of Callose Synthase Gene and Callose Content in Response to F. culmorum Infection
3.6. Influence of Carotenoids on F. culmorum Growth
3.7. Influence of ABA on F. culmorum Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| PR | pathogenesis related |
| TF | transcription factor |
| ET | ethylene |
| JA | jasmonic acid |
| SA | salicylic acid |
| ABA | abscisic acid |
| SAR | systemic acquired resistance |
| ISR | induced systemic resistance |
| Rboh | NADPH/respiratory burst oxidase homolog protein |
| NOX | NADPH oxidase |
| PAL | phenylalanine ammonia lyase |
| PTFE | polytetrafluoroethylene |
| BSTFA | N,O-Bis(trimethylsilyl)trifluoroacetamide |
| TMCS | trimethylsilyl chloride |
| ROS | reactive oxygen species |
| RNS | reactive nitrogen species |
| hpi | hours post infection |
| APx | ascorbate peroxidase |
| CAT | catalase |
| SOD | superoxide dismutase |
| MP | mevalonate pathway |
| NMP | non-mevalonate pathway |
| GA | gibberellic acid |
| IPP | isopentenyl pyrophosphate |
| DMAPP | dimethylallyl pyrophosphate |
| DXS | deoxy-d-xylulose 5-phosphate synthase |
| DXR | deoxy-d-xylulose 5-phosphate reductase |
| IDI | isopentenyl pyrophosphate isomerase |
| GPPS | geranyl pyrophosphate synthase |
| HMGS | 3-hydroxy-3-methylglutaryl-CoA synthase |
| HMGR | 3-hydroxy-3-methylglutaryl-CoA reductase |
| MVK | mevalonate kinase |
| FPPS | farnesyl pyrophosphate synthase |
| SMT2 | sterol methyltransferase 2 |
| GGR | geranyl-geranyl diphosphate reductase |
| VTE2 | homogentisate phytyl transferase |
| MPBQ | 2-methyl-6-phytyl-1,4-benzoquinol |
| VTE3 | MPBQ methyltransferase |
| VTE1 | tocopherol cyclase |
| VTE4 | γ-tocopherol methyltransferase |
| PSY | phytoene synthase |
| PDS | phytoene desaturase |
| ZCD | ζ-carotene desaturase |
| Z-ISO | ζ-carotene isomerase |
| CRTISO | carotenoid isomerase |
| LCYB | lycopene β-cyclase |
| LCYE | lycopene ε-cyclase |
| βHY | β-carotene hydroxylase |
| LUT1 | ε-ring hydroxylase |
| LUT5 | β-ring hydroxylase |
| ZEP | zeaxantin epoxidase |
| VDE | violaxanthin de-epoxidase |
| NCED3 | 3-cis-epoxycarotenoid dioxygenases |
| NCED9 | 9-cis-epoxycarotenoid dioxygenases |
| ABA2 | xanthoxin dehydrogenase |
| AAO3 | abscisic aldehyde oxidase |
| CCD | carotenoid cleavage dioxygenase |
| CALS | callose synthase |
References
- FAO. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 1 February 2022).
- Novakovskiy, R.O.; Dvorianinova, E.M.; Rozhmina, T.A.; Kudryavtseva, L.P.; Gryzunov, A.A.; Pushkova, E.N.; Povkhova, L.V.; Snezhkina, A.V.; Krasnov, G.S.; Kudryavtseva, A.V.; et al. Data on genetic polymorphism of flax (Linum usitatissimum L.) pathogenic fungi of Fusarium, Colletotrichum, Aureobasidium, Septoria, and Melampsora genera. Data Brief 2020, 31, 105710. [Google Scholar] [CrossRef] [PubMed]
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.M.; Lysh, D.; Dudek, B.; Szopa, J.; et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastuszak, J.; Szczerba, A.; Dziurka, M.; Hornyák, M.; Kopeć, P.; Szklarczyk, M.; Płażek, A. Physiological and Biochemical Response to Fusarium culmorum Infection in Three Durum Wheat Genotypes at Seedling and Full Anthesis Stage. Int. J. Mol. Sci. 2021, 22, 7433. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Summerell, B.A.; Bullock, S. The Fusarium Laboratory Manual; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Wagacha, J.M. Fusarium culmorum: Infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Prot. 2007, 26, 877–885. [Google Scholar] [CrossRef]
- Jaroszuk-Ściseł, J.; Kurek, E.; Winiarczyk, K.; Baturo, A.; Łukanowski, A. Colonization of root tissues and protection against Fusarium wilt of rye (Secale cereale) by nonpathogenic rhizosphere strains of Fusarium culmorum. Biol. Control 2008, 45, 297–307. [Google Scholar] [CrossRef]
- Urban, M.; Daniels, S.; Mott, E.; Hammond-Kosack, K. Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant J. 2002, 32, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Magan, N.; Hope, R.; Aldred, D. Ecophysiology of Fusarium culmorum and mycotoxin production. Adv. Exp. Med. Biol. 2006, 571, 123–136. [Google Scholar] [CrossRef]
- Motallebi, P.; Alkadri, D.; Pisi, A.; Nipoti, P.; Tonti, S.; Niknam, V.; Hashemi, M.; Prodi, A. Pathogenicity and mycotoxin chemotypes of Iranian Fusarium culmorum isolates on durum wheat, and comparisons with Italian and Syrian isolates. Phytopathol. Mediterr. 2015, 54, 437–445. [Google Scholar] [CrossRef]
- Winter, M.; Samuels, P.L.; Dong, Y.; Dill-Macky, R. Trichothecene production is detrimental to early root colonization by Fusarium culmorum and F. graminearum in fusarium crown and root rot of wheat. Plant Pathol. 2019, 68, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Spanic, V.; Zdunic, Z.; Drezner, G.; Vuletic, M. Differences in physiological traits at the initial stage of Fusarium head blight infection in wheat. Biol. Plant 2020, 64, 185–192. [Google Scholar] [CrossRef]
- Uluhan, E.; Keleş, E.; Tufan, F. Analysis of WRKY Transcription Factors in Barley Cultivars Infected with Fusarium culmorum. Int. J. Life Sci. Biotechnol. 2019, 2, 165–174. [Google Scholar] [CrossRef]
- Kostyn, K.; Czemplik, M.; Kulma, A.; Bortniczuk, M.; Skała, J.; Szopa, J. Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci. 2012, 190, 103–115. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhou, F.; Ji, B.-P.; Pei, R.-S.; Xu, N. The antibacterial mechanism of carvacrol and thymol against Escherichia coli. Lett. Appl. Microbiol. 2008, 47, 174–179. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; Coppola, R.; Feo, V.D. Essential Oils and Antifungal Activity. Pharmaceuticals 2017, 10, 86. [Google Scholar] [CrossRef] [Green Version]
- Perczak, A.; Gwiazdowska, D.; Marchwińska, K.; Juś, K.; Gwiazdowski, R.; Waśkiewicz, A. Antifungal activity of selected essential oils against Fusarium culmorum and F. graminearum and their secondary metabolites in wheat seeds. Arch. Microbiol. 2019, 201, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Métraux, J.P. Systemic Acquired Resistance. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 627–629. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2013, 65, 1229–1240. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamle, M.; Borah, R.; Bora, H.; Jaiswal, A.K.; Singh, R.K.; Kumar, P. Systemic Acquired Resistance (SAR) and Induced Systemic Resistance (ISR): Role and Mechanism of Action against Phytopathogens. In Fungal Biotechnology and Bioengineering; Hesham, A.E.-L., Upadhyay, R.S., Sharma, G.D., Manoharachary, C., Gupta, V.K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 457–470. [Google Scholar] [CrossRef]
- Motallebi, P.; Tonti, S.; Niknam, V.; Ebrahimzadeh, H.; Pisi, A.; Nipoti, P.; Hashemi, M.; Prodi, A. Induction of basal resistance by methyl jasmonate against Fusarium culmorum in bread wheat. Cereal Res. Commun. 2017, 45, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Makandar, R.; Nalam, V.J.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Salicylic Acid Regulates Basal Resistance to Fusarium Head Blight in Wheat. Mol. Plant-Microbe Interact. 2011, 25, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulferts, S.; Delventhal, R.; Splivallo, R.; Karlovsky, P.; Schaffrath, U. Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 2015, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vleesschauwer, D.; Yang, Y.; Vera Cruz, C.; Höfte, M. Abscisic Acid-Induced Resistance against the Brown Spot Pathogen Cochliobolus miyabeanus in Rice Involves MAP Kinase-Mediated Repression of Ethylene Signaling. Plant Physiol. 2010, 152, 2036–2052. [Google Scholar] [CrossRef] [Green Version]
- Sivakumaran, A.; Akinyemi, A.; Mandon, J.; Cristescu, S.M.; Hall, M.A.; Harren, F.J.M.; Mur, L.A.J. ABA Suppresses Botrytis cinerea Elicited NO Production in Tomato to Influence H2O2 Generation and Increase Host Susceptibility. Front. Plant Sci. 2016, 7, 709. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, H.; Yang, Y.; Zhang, Z.; Zhang, H.; Hu, X.; Chen, J.; Wang, X.-C.; Huang, R. Abscisic acid regulates TSRF1-mediated resistance to Ralstonia solanacearum by modifying the expression of GCC box-containing genes in tobacco. J. Exp. Bot. 2008, 59, 645–652. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Hill, L.; Crooks, C.; Doerner, P.; Lamb, C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009, 150, 1750–1761. [Google Scholar] [CrossRef] [Green Version]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef]
- Thaler, J.S.; Bostock, R.M. Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 2004, 85, 48–58. [Google Scholar] [CrossRef]
- Boba, A.; Kostyn, K.; Preisner, M.; Wojtasik, W.; Szopa, J.; Kulma, A. Expression of heterologous lycopene β-cyclase gene in flax can cause silencing of its endogenous counterpart by changes in gene-body methylation and in ABA homeostasis mechanism. Plant Physiol. Biochem. 2018, 127, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Stuper-Szablewska, K.; Kurasiak-Popowska, D.; Nawracała, J.; Perkowski, J. Response of non-enzymatic antioxidative mechanisms to stress caused by infection with Fusarium fungi and chemical protection in different wheat genotypes. Chem. Ecol. 2017, 33, 949–962. [Google Scholar] [CrossRef]
- Hanifei, M.; Dehghani, H.; Choukan, R. The role of antioxidant enzymes and phenolic compounds in disease resistance to Fusarium oxysporum f. sp. Melonis Race 1.2. Int. J. Agron. Plant Prod. 2013, 4, 1985–1996. [Google Scholar]
- Kurniawan, A.; Chuang, H.-W. Rhizobacterial Bacillus mycoides functions in stimulating the antioxidant defence system and multiple phytohormone signalling pathways to regulate plant growth and stress tolerance. J. Appl. Microbiol. 2022, 132, 1260–1274. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, D.; Nath , M.; Sharma , M.; Bhatt , M.D.; Bisht, D.S.; Butani, N.V. Role of growth regulators and phytohormones in overcoming environmental stress. In Protective Chemical Agents in the Amelioration of Plant Abiotic Stress; Roychoudhury, A., Tripathi, D.K., Eds.; Wiley: Hoboken, NJ, USA, 2020; pp. 254–279. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Khangwal, I.; Shukla, P. Combinatorial Interactions of Biotic and Abiotic Stresses in Plants and Their Molecular Mechanisms: Systems Biology Approach. Mol. Biotechnol. 2018, 60, 636–650. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Xu, J.; Höfte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Chen, Z.; Ji, L.; Wang, J.; Jin, J.; Yang, X.; Rao, P.; Gao, K.; Liao, W.; Ye, M.; An, X. Dynamic changes in the transcriptome of Populus hopeiensis in response to abscisic acid. Sci. Rep. 2017, 7, 42708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, Q.; Li, W.; Zhang, S.; Xi, W. Identification of key genes and regulators associated with carotenoid metabolism in apricot (Prunus armeniaca) fruit using weighted gene coexpression network analysis. BMC Genom. 2019, 20, 876. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Zhang, Y.; Zhou, J.; Wei, F.; Feng, Z.; Zhao, L.; Shi, Y.; Feng, H.; Zhu, H. The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae. Int. J. Mol. Sci. 2021, 22, 13041. [Google Scholar] [CrossRef] [PubMed]
- Shin, L.J.; Huang, H.E.; Chang, H.; Lin, Y.H.; Feng, T.Y.; Ger, M.J. Ectopic ferredoxin I protein promotes root hair growth through induction of reactive oxygen species in Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 434–440. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Tan, L.; Liu, Q.; Song, Y.; Zhou, G.; Luan, L.; Weng, Q.; He, C. Differential Function of Endogenous and Exogenous Abscisic Acid during Bacterial Pattern-Induced Production of Reactive Oxygen Species in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2544. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.W.; Hwang, B.K. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta 2012, 235, 1369–1382. [Google Scholar] [CrossRef]
- Niu, L.; Liao, W. Hydrogen Peroxide Signaling in Plant Development and Abiotic Responses: Crosstalk with Nitric Oxide and Calcium. Front. Plant Sci. 2016, 7, 230. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Seo, P.J. Ca2+talyzing Initial Responses to Environmental Stresses. Trends Plant Sci. 2021, 26, 849–870. [Google Scholar] [CrossRef]
- Iakimova, E.T.; Woltering, E.J. Nitric oxide prevents wound-induced browning and delays senescence through inhibition of hydrogen peroxide accumulation in fresh-cut lettuce. Innov. Food Sci. Emerg. Technol. 2015, 30, 157–169. [Google Scholar] [CrossRef]
- Barber, M.S.; Bertram, R.E.; Ride, J.P. Chitin oligosaccharides elicit lignification in wounded wheat leaves. Physiol. Mol. Plant Pathol. 1989, 34, 3–12. [Google Scholar] [CrossRef]
- Jaber, R.; Planchon, A.; Mathieu-Rivet, E.; Kiefer-Meyer, M.-C.; Zahid, A.; Plasson, C.; Pamlard, O.; Beaupierre, S.; Trouvé, J.-P.; Guillou, C.; et al. Identification of two compounds able to improve flax resistance towards Fusarium oxysporum infection. Plant Sci. 2020, 301, 110690. [Google Scholar] [CrossRef] [PubMed]
- García-Andrade, J.; Ramírez, V.; Flors, V.; Vera, P. Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J. 2011, 67, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Oide, S.; Bejai, S.; Staal, J.; Guan, N.; Kaliff, M.; Dixelius, C. A novel role of PR2 in abscisic acid (ABA) mediated, pathogen-induced callose deposition in Arabidopsis thaliana. New Phytol. 2013, 200, 1187–1199. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Stein, M.; Hou, B.-H.; Vogel, J.P.; Edwards, H.; Somerville, S.C. Loss of a Callose Synthase Results in Salicylic Acid-Dependent Disease Resistance. Science 2003, 301, 969–972. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boba, A.; Kostyn, K.; Kochneva, Y.; Wojtasik, W.; Mierziak, J.; Prescha, A.; Augustyniak, B.; Grajzer, M.; Szopa, J.; Kulma, A. Abscisic Acid—Defensive Player in Flax Response to Fusarium culmorum Infection. Molecules 2022, 27, 2833. https://doi.org/10.3390/molecules27092833
Boba A, Kostyn K, Kochneva Y, Wojtasik W, Mierziak J, Prescha A, Augustyniak B, Grajzer M, Szopa J, Kulma A. Abscisic Acid—Defensive Player in Flax Response to Fusarium culmorum Infection. Molecules. 2022; 27(9):2833. https://doi.org/10.3390/molecules27092833
Chicago/Turabian StyleBoba, Aleksandra, Kamil Kostyn, Yelyzaveta Kochneva, Wioleta Wojtasik, Justyna Mierziak, Anna Prescha, Beata Augustyniak, Magdalena Grajzer, Jan Szopa, and Anna Kulma. 2022. "Abscisic Acid—Defensive Player in Flax Response to Fusarium culmorum Infection" Molecules 27, no. 9: 2833. https://doi.org/10.3390/molecules27092833
APA StyleBoba, A., Kostyn, K., Kochneva, Y., Wojtasik, W., Mierziak, J., Prescha, A., Augustyniak, B., Grajzer, M., Szopa, J., & Kulma, A. (2022). Abscisic Acid—Defensive Player in Flax Response to Fusarium culmorum Infection. Molecules, 27(9), 2833. https://doi.org/10.3390/molecules27092833





