Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Composition Analysis of CSEO
2.2. Antibacterial Activity
2.3. Compounds Toxicity Evaluation by In Silico Tools
2.3.1. Computational COMPOUND Toxicity Prediction by VEGA HUB Software
2.3.2. Rodent Oral Toxicity and Cytotoxicity of (CSEO) Compounds Predicted by PROTOX II Tool
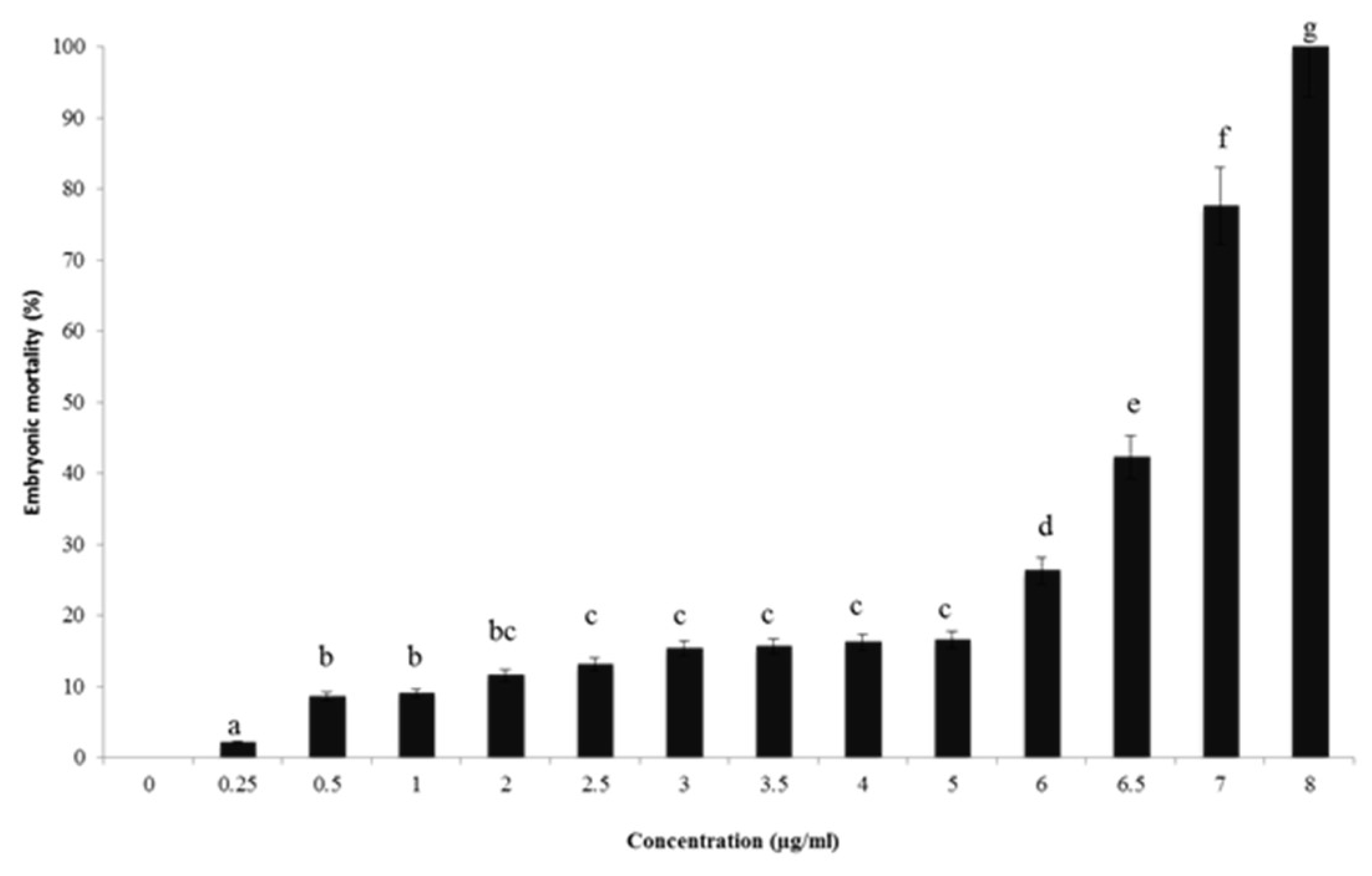
2.4. In Vivo Toxicity Assessment Using Zebrafish Model
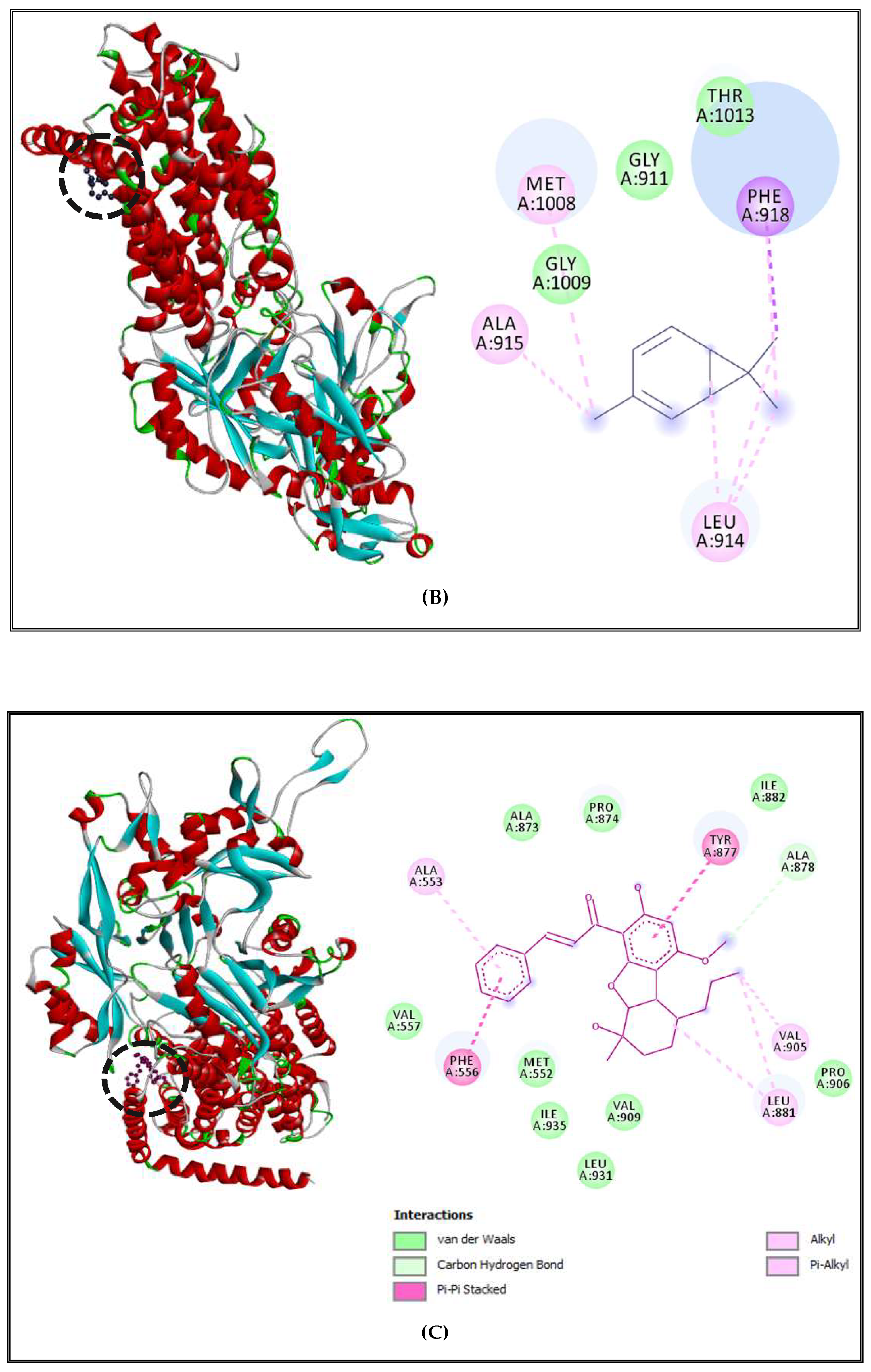
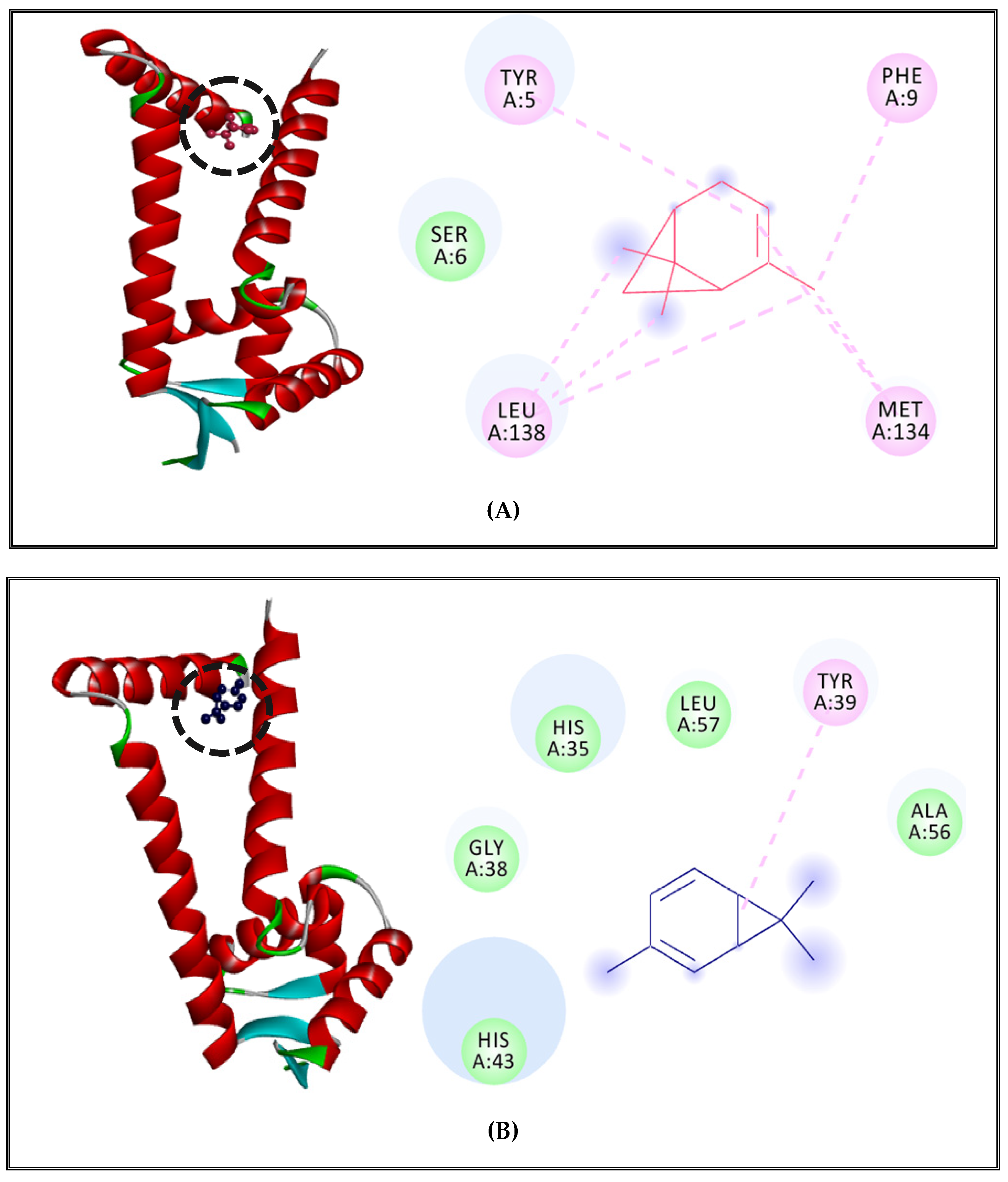
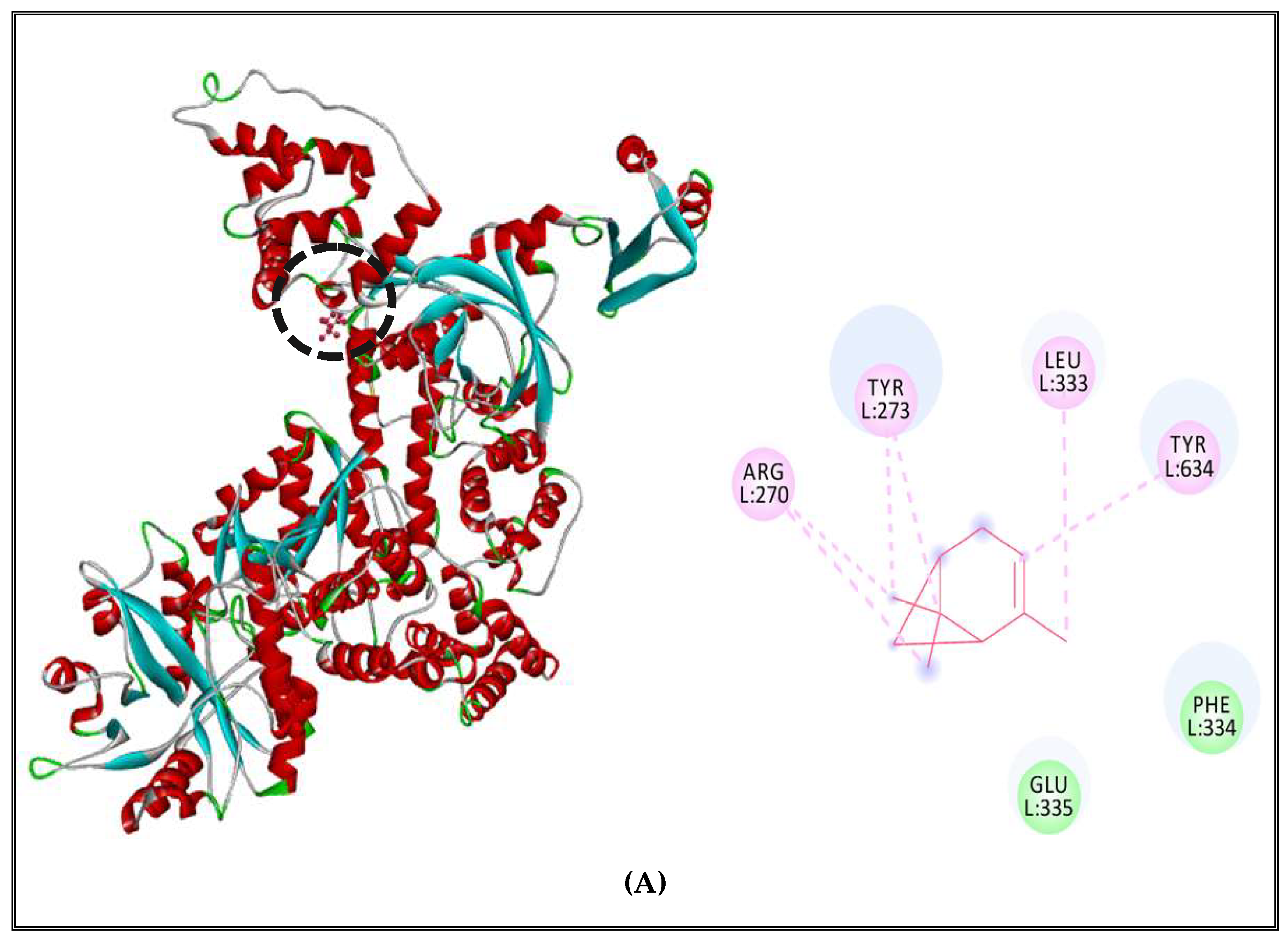
2.5. Antibacterial Mechanisms of (CSEO)
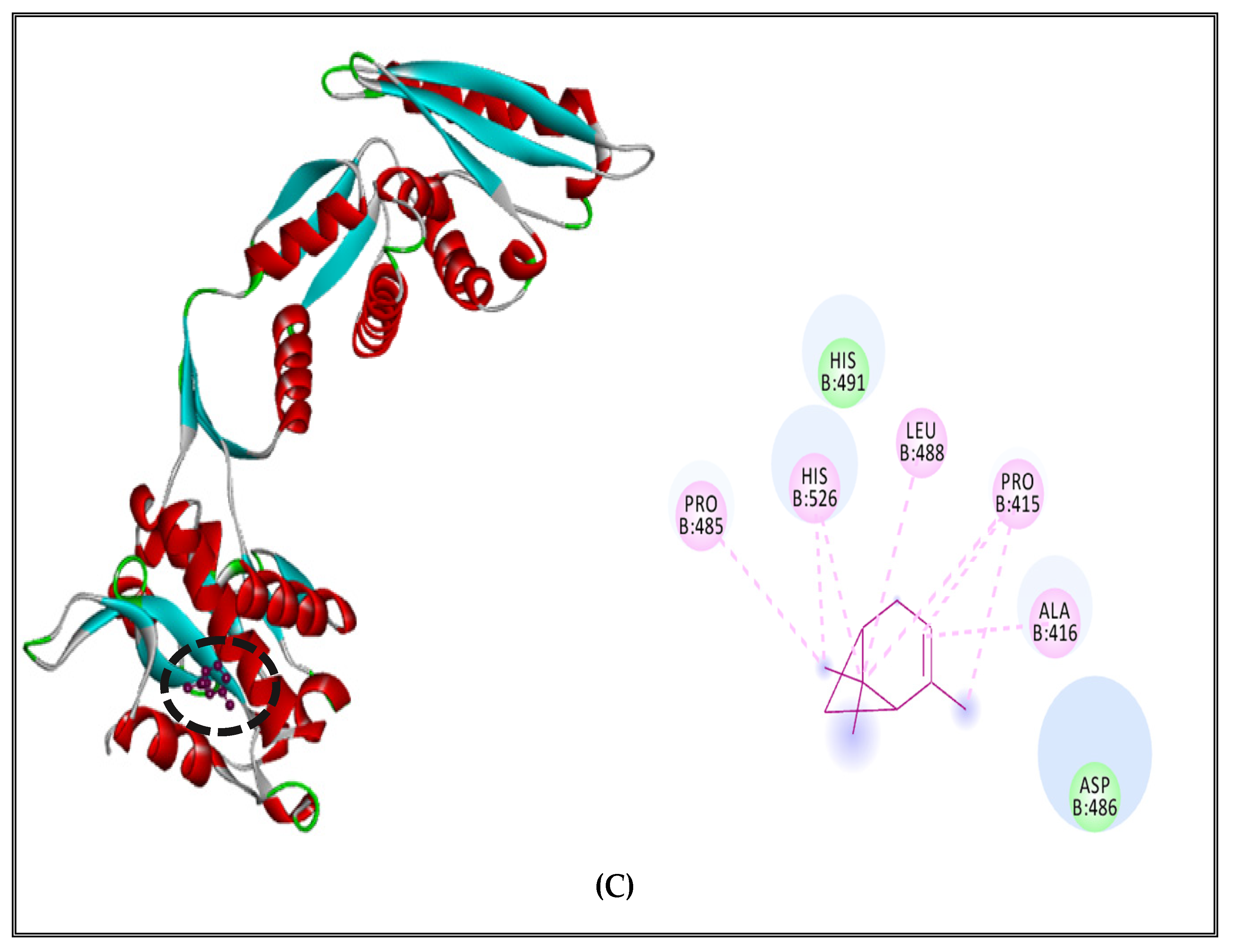
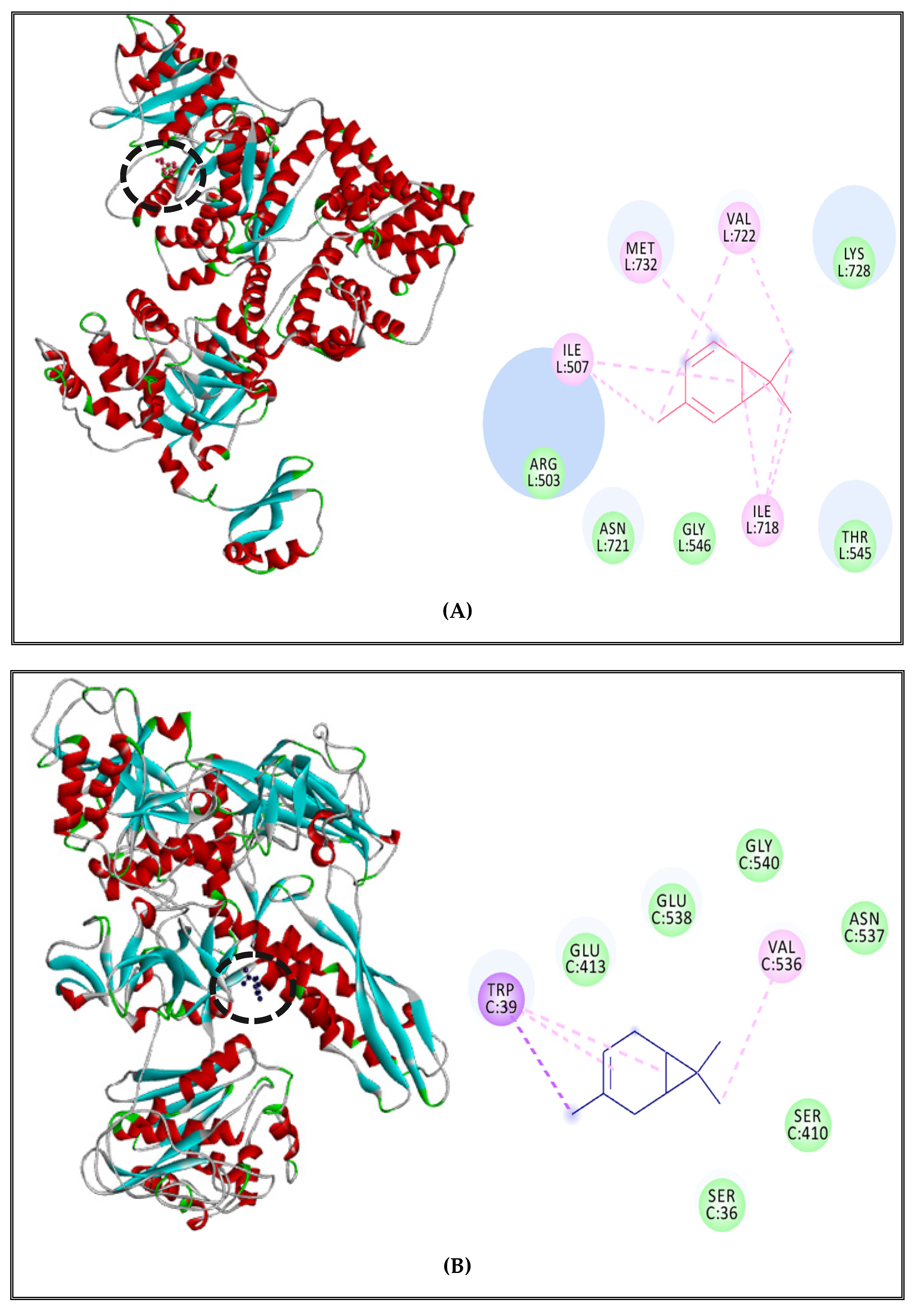
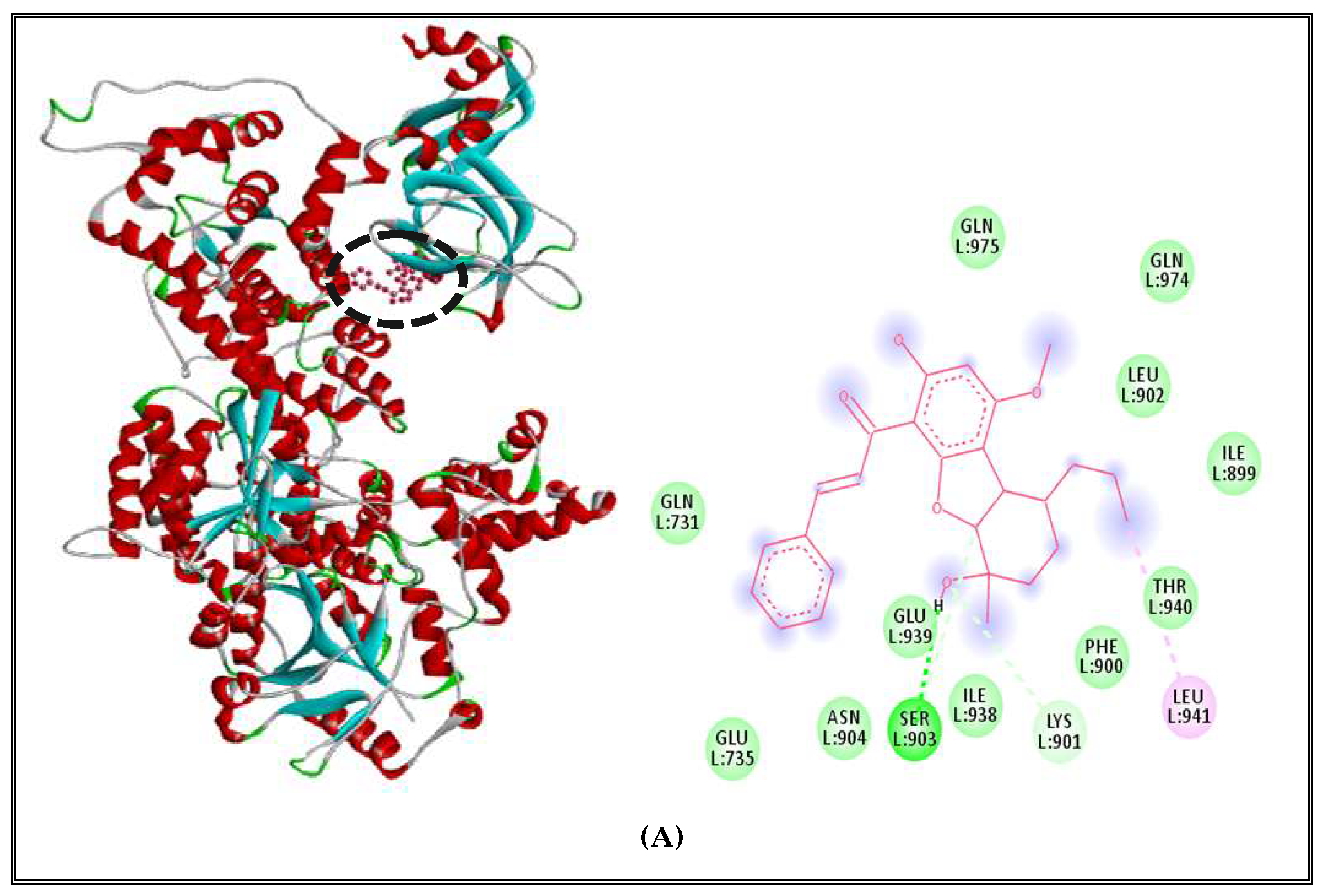
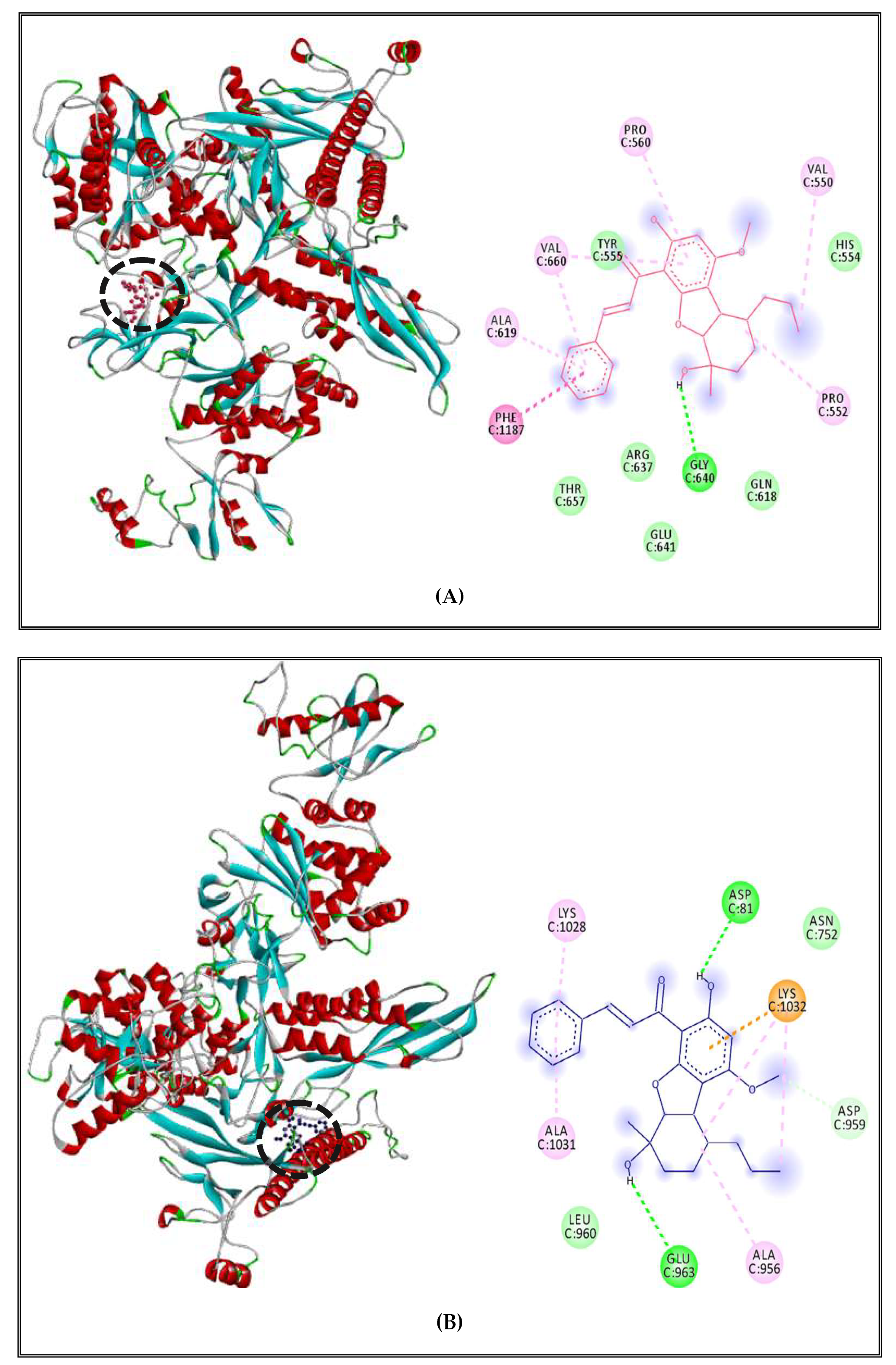
2.5.1. Alteration of Bacterial Cell Permeability: Inhibition of Efflux Pumps by (CSEO)
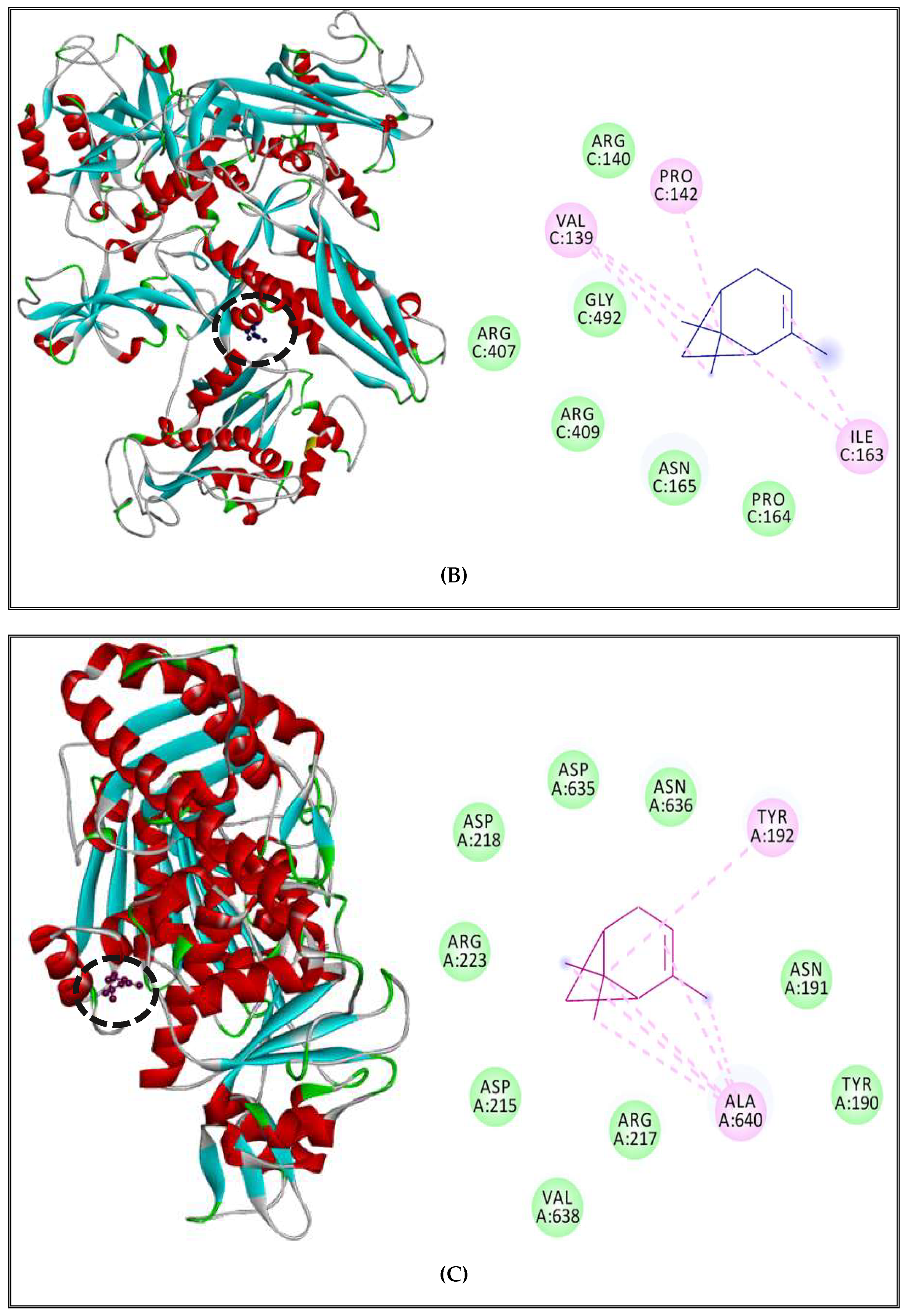
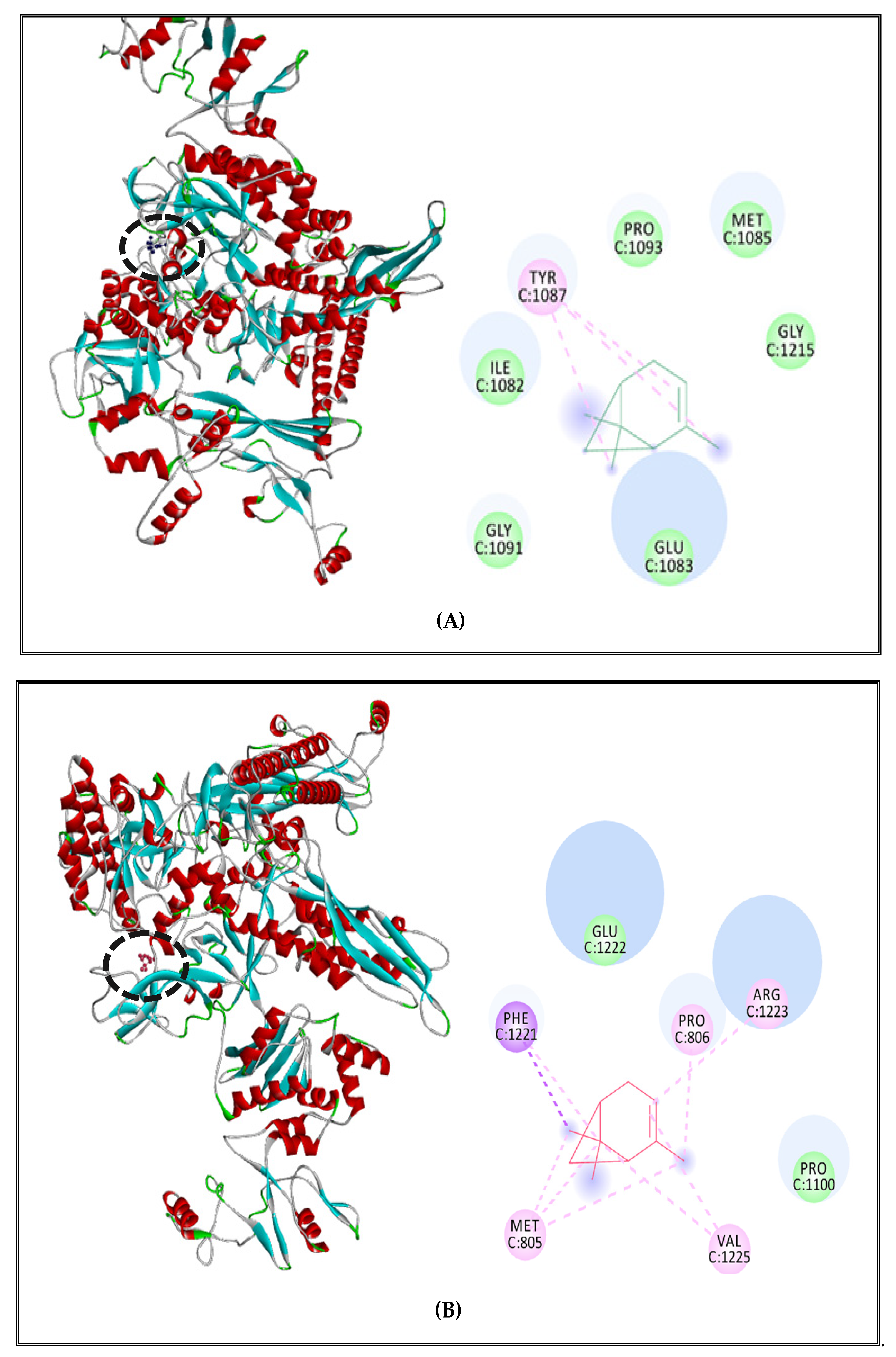
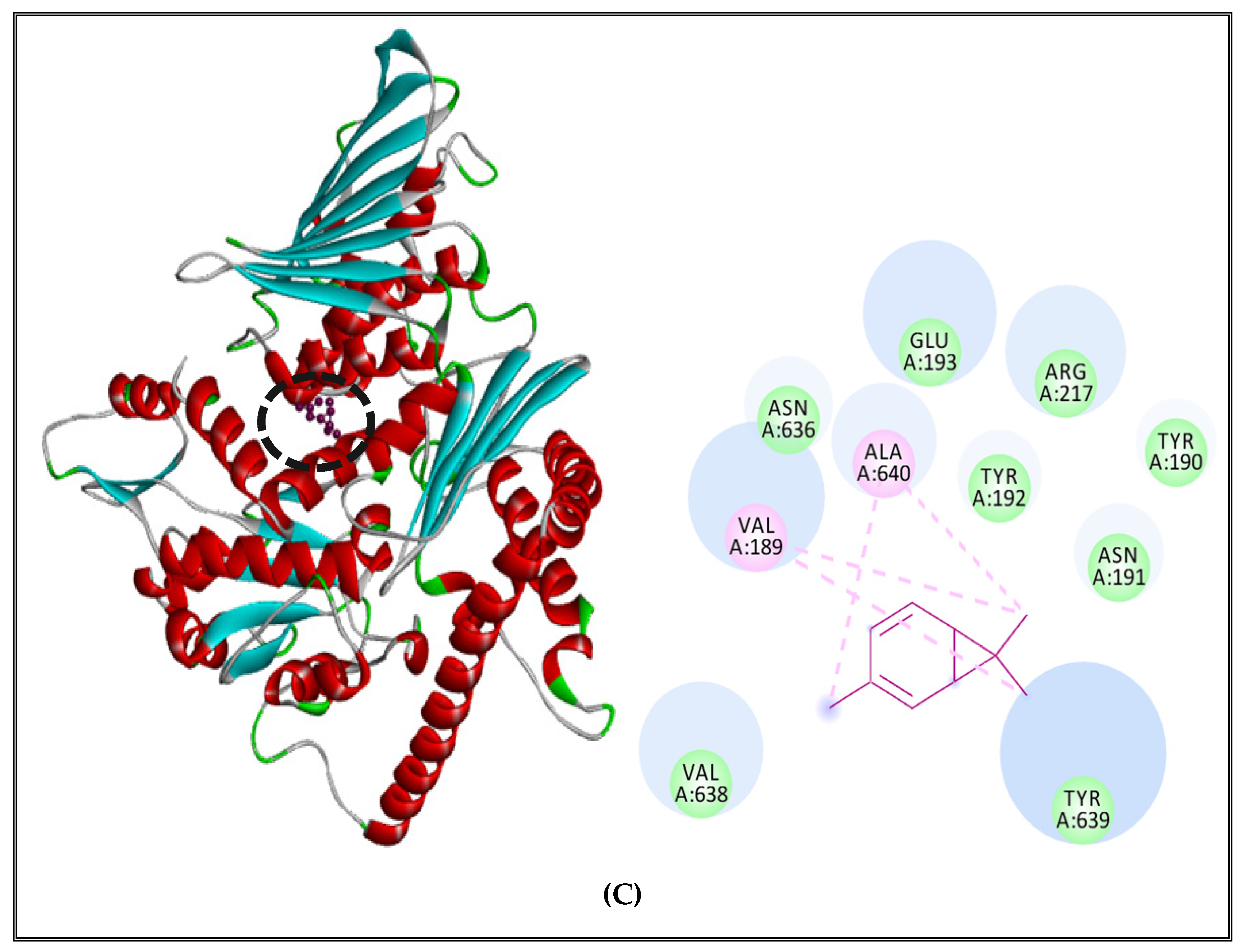
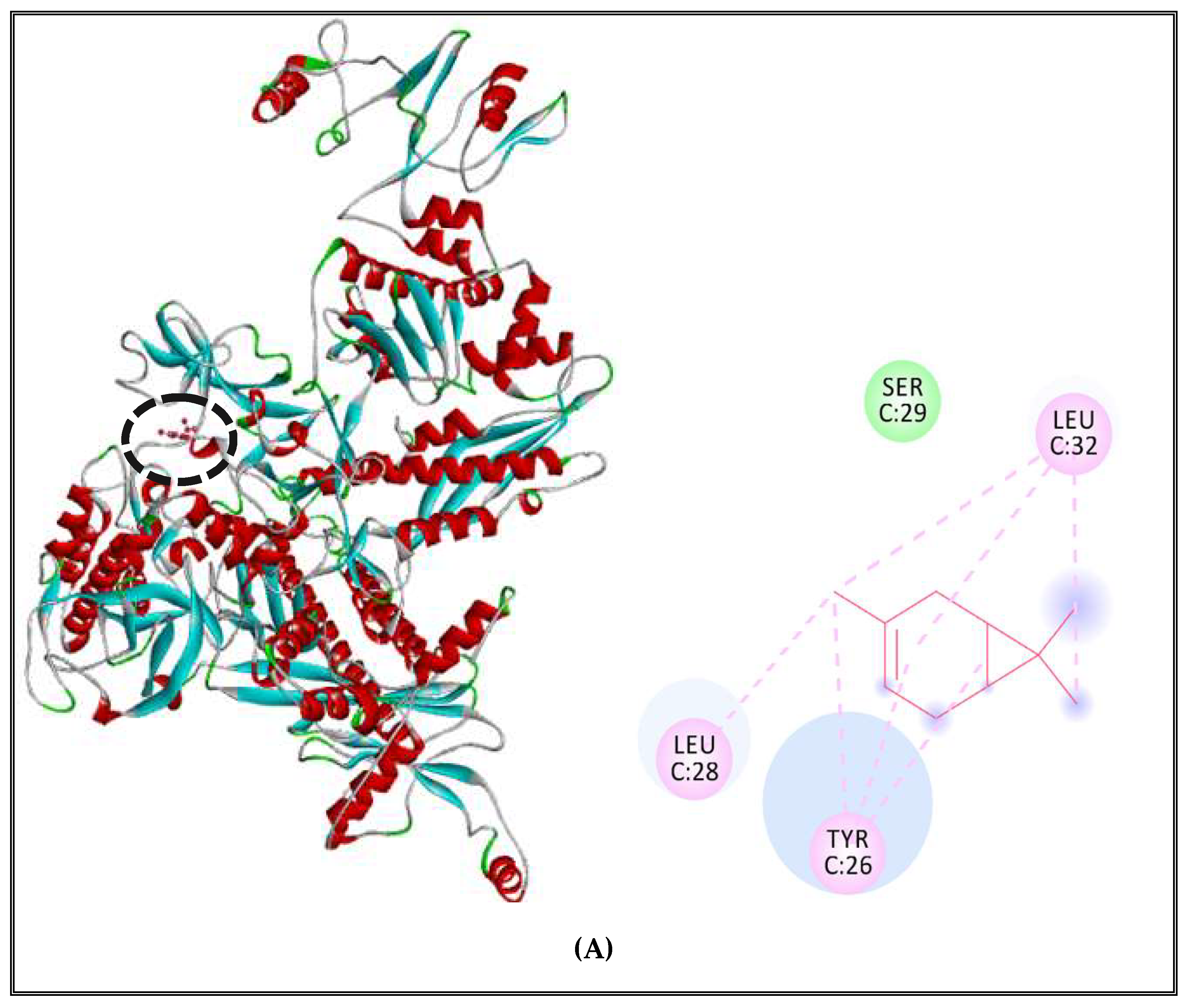
2.5.2. Interactions between CSEO Molecules and Bacterial Topoisomerase II, DNA and RNA Polymerases
3. Materials and Methods
3.1. Plant Material, Essential Oil Extraction, and Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
3.1.1. Plant Material
3.1.2. Extraction and Analysis of (CSEO)
3.2. Antibacterial Activity
3.2.1. Microorganisms and Growth Conditions
3.2.2. Agar Diffusion Method
3.2.3. Minimal Inhibitory Concentration (MIC)
3.3. Chemo-Computational Toxicity Evaluation Using In Silico Tools
3.3.1. Toxicity Prediction of Compounds by VEGA HUB Software Using QSAR Method
3.3.2. Rodent Oral Toxicity and Cytotoxicity of Selected Compounds Predicted by PROTOX II
3.4. In Vivo Toxicity Assessment Using Zebrafish Model
3.4.1. Zebrafish Maintenance and Embryos’ Collection
3.4.2. Zebrafish Embryonic Toxicity Test and Determination of LC50
3.5. Interaction Study between the (CSEO) Molecules and Bacterial Protein Targets by Molecular Docking
3.5.1. Homology Modeling of the Proteins
3.5.2. Validation of Protein Models
3.5.3. Binding Site Prediction
3.5.4. Selection of the Compounds
3.5.5. Molecular Docking by Autodock Vina
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Appendix A
| Bacterial Strain | Bacterial Target | Receptor (UniprotKB) | Template | Identity (%) | Ramachandran Plot | QMEAN | |
|---|---|---|---|---|---|---|---|
| Favoured Regions (%) | Additional Allowed Regions (%) | ||||||
| Staphylococcus aureus (strain Mu50/ATCC 700699) | DNA polymerase | P63979 | 4IQJ.1.L | 34.77 | 87.9 | 10.1 | −3.26 |
| RNA polymerase | Q932F8 | 6WVK.1.C | 81.09 | 85.5 | 12.7 | −2.36 | |
| Topoisomerase II | P66936 | 6GAV.1.A | 54.42 | 88.4 | 10.7 | −1.69 | |
| Salmonella Typhimurium (strain LT2/SGSC1412/ATCC 700720) | DNA polymerase | P14567 | 5FKU.1.A | 96.72 | 88.0 | 10.0 | −2.35 |
| RNA polymerase | P06173 | 4LLG.2.C | 98.66 | 88.0 | 11.0 | −1.11 | |
| Topoisomerase II | P0A213 | 4TMA.2.B | 95.41 | 90.2 | 9.2 | −1.82 | |
References
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Comparison of antibiotic resistance mechanisms in antibiotic-producing and pathogenic bacteria. Molecules 2019, 24, 3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngaruka, G.B.; Neema, B.B.; Mitima, T.K.; Kishabongo, A.S.; Kashongwe, O.B. Animal source food eating habits of outpatients with antimicrobial resistance in Bukavu, DR Congo. Antimicrob. Resist. Infect. Control 2021, 10, 124. [Google Scholar] [CrossRef]
- Kebede, T.; Gadisa, E.; Tufa, A. Antimicrobial activities evaluation and phytochemical screening of some selected medicinal plants: A possible alternative in the treatment of multidrug-resistant microbes. PLoS ONE 2021, 16, e0249253. [Google Scholar] [CrossRef]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef]
- Kosakowska, O.; Węglarz, Z.; Pióro-Jabrucka, E.; Przybył, J.L.; Kraśniewska, K.; Gniewosz, M.; Bączek, K. Antioxidant and antibacterial activity of essential oils and hydroethanolic extracts of Greek oregano (O. vulgare L. subsp. hirtum (Link) Ietswaart) and common oregano (O. vulgare L. subsp. vulgare). Molecules 2021, 26, 988. [Google Scholar] [CrossRef]
- Zaharieva, M.M.; Zheleva-Dimitrova, D.; Rusinova-Videva, S.; Ilieva, Y.; Brachkova, A.; Balabanova, V.; Gevrenova, R.; Kim, T.C.; Kaleva, M.; Georgieva, A.; et al. Antimicrobial and Antioxidant Potential of Scenedesmus obliquus Microalgae in the Context of Integral Biorefinery Concept. Molecules 2022, 27, 519. [Google Scholar] [CrossRef]
- Capatina, L.; Napoli, E.M.; Ruberto, G.; Hritcu, L. Origanum vulgare ssp. hirtum (Lamiaceae) Essential Oil Prevents Behavioral and Oxidative Stress Changes in the Scopolamine Zebrafish Model. Molecules 2021, 26, 7085. [Google Scholar] [CrossRef]
- Smaoui, S.; Hlima, H.B.; Tavares, L.; Ennouri, K.; Braiek, O.B.; Mellouli, L.; Abdelkafi, S.; Khaneghah, A.M. Application of essential oils in meat packaging: A systemic review of recent literature. Food Control 2022, 132, 108566. [Google Scholar] [CrossRef]
- Ni, Z.J.; Wang, X.; Shen, Y.; Thakur, K.; Han, J.; Zhang, J.G.; Hu, F.; Wei, Z.J. Recent updates on the chemistry, bioactivities, mode of action, and industrial applications of plant essential oils. Trends Food Sci. Technol. 2021, 110, 78–89. [Google Scholar] [CrossRef]
- Jugreet, B.S.; Suroowan, S.; Rengasamy, R.K.; Mahomoodally, M.F. Chemistry, bioactivities, mode of action and industrial applications of essential oils. Trends Food Sci. Technol. 2020, 101, 89–105. [Google Scholar] [CrossRef]
- Arsalani, M.; Griessinger, J.; Pourtahmasi, K.; Bräuning, A. Multi-centennial reconstruction of drought events in South-Western Iran using tree rings of Mediterranean cypress (Cupressus sempervirens L.). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2021, 567, 110296. [Google Scholar] [CrossRef]
- Fadel, H.; Benayache, F.; Chalchat, J.C.; Figueredo, G.; Chalard, P.; Hazmoune, H.; Benayache, S. Essential oil constituents of Juniperus oxycedrus L. and Cupressus sempervirens L. (Cupressaceae) growing in Aures region of Algeria. Nat. Prod. Res. 2021, 35, 2616–2620. [Google Scholar] [CrossRef] [PubMed]
- Argui, H.; Youchret-Zalleza, O.B.; Suner, S.C.; Periz, Ç.D.; Türker, G.; Ulusoy, S.; Ben-Attia, M.; Büyükkaya, F.; Oral, A.; Coskun, Y.; et al. Isolation, Chemical Composition, Physicochemical Properties, and Antibacterial Activity of Cupressus sempervirens L. Essential Oil. J. Essent. Oil-Bear. Plants 2021, 24, 439–452. [Google Scholar] [CrossRef]
- Rguez, S.; Djébali, N.; Slimene, I.B.; Abid, G.; Hammemi, M.; Chenenaoui, S.; Bachkouel, S.; Daami-Remadi, M.; Ksouri, R.; Hamrouni-Sellami, I. Cupressus sempervirens essential oils and their major compounds successfully control postharvest grey mould disease of tomato. Ind. Crops Prod. 2018, 123, 135–141. [Google Scholar] [CrossRef]
- Selim, S.A.; Adam, M.E.; Hassan, S.M.; Albalawi, A.R. Chemical composition, antimicrobial and antibiofilm activity of the essential oil and methanol extract of the Mediterranean cypress (Cupressus sempervirens L.). BMC Complement. Altern. Med. 2014, 14, 179. [Google Scholar] [CrossRef]
- Alimi, D.; Hajri, A.; Jallouli, S.; Sebai, H. Phytochemistry, anti-tick, repellency and anti-cholinesterase activities of Cupressus sempervirens L. and Mentha pulegium L. combinations against Hyalomma scupense (Acari: Ixodidae). Vet. Parasitol. 2022, 303, 109665. [Google Scholar] [CrossRef]
- He, Y.L.; Shi, J.Y.; Peng, C.; Hu, L.J.; Liu, J.; Zhou, Q.M.; Guo, L.; Xiong, L. Angiogenic effect of motherwort (Leonurus japonicus) alkaloids and toxicity of motherwort essential oil on zebrafish embryos. Fitoterapia 2018, 128, 36–42. [Google Scholar] [CrossRef]
- Haddad, J.G.; Picard, M.; Bénard, S.; Desvignes, C.; Desprès, P.; Diotel, N.; El Kalamouni, C. Ayapana triplinervis essential oil and its main component thymohydroquinone dimethyl ether inhibit Zika virus at doses devoid of toxicity in zebrafish. Molecules 2019, 24, 3447. [Google Scholar] [CrossRef] [Green Version]
- Thitinarongwate, W.; Mektrirat, R.; Nimlamool, W.; Khonsung, P.; Pikulkaew, S.; Okonogi, S.; Kunanusorn, P. Phytochemical and Safety Evaluations of Zingiber ottensii Valeton Essential Oil in Zebrafish Embryos and Rats. Toxics 2021, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.X.; He, X.; Wu, Y.Q.; Liu, J.Z. Enhancing Production of Pinene in Escherichia coli by Using a Combination of Tolerance, Evolution, and Modular Co-culture Engineering. Front. Microbiol. 2018, 9, 1623. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Carneiro, M.R.; Viana, M.E.; Felzenszwalb, I.; Paumgartten, F.J. Evaluation of beta-myrcene, alpha-terpinene and (+)- and (−)-alpha-pinene in the Salmonella/microsome assay. Food Chem. Toxicol. 2005, 43, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Kamal, E.H.; Al-Ajmi, M.F.; Abdullah, M.B. Some cardiovascular effects of the dethymoquinonated Nigella sativa volatile oil and its major components α-pinene and p-cymene in rats. Saudi Pharm. J. 2003, 11, 104–110. [Google Scholar]
- Him, A.; Ozbek, H.; Turel, I.; Oner, A.C. Antinociceptive activity of alphapinene and fenchone. Pharmacologyonline 2008, 3, 363–369. [Google Scholar]
- Orhan, I.; Küpeli, E.; Aslan, M.; Kartal, M.; Yesilada, E. Bioassay-guided evaluation of anti-inflammatory and antinociceptive activities of pistachio, Pistacia vera L. J. Ethnopharmacol. 2006, 105, 235–240. [Google Scholar] [CrossRef]
- FDA. Code of Federal Regulations Title 21; Food and Drug Administration: Washington, DC, USA, 2015.
- Cavaleiro, C.; Pinto, E.; Gonçalves, M.J.; Salgueiro, L. Antifungal activity of Juniperus essential oils against dermatophyte, Aspergillus and Candida strains. J. Appl. Microbiol. 2006, 100, 1333–1338. [Google Scholar]
- Niccolini, P.; Pasotti, V.; Caliari, W. New Pharmacological Properties of Delta-3-Carene. Antibacterial and Expectorant Effects. Boll. Chim. Farm. 1964, 103, 598–608. [Google Scholar]
- Gil, M.L.; Jimenez, J.; Ocete, M.A.; Zarzuelo, A.; Cabo, M.M. Comparative study of different essential oils of Bupleurum gibraltaricum Lamarck. Pharmazie 1989, 44, 284–287. [Google Scholar]
- Ravichandran, C.; Badgujar, P.; Gundev, P.; Ashutosh Upadhyay, A. Review of toxicological assessment of d -limonene, a food and cosmetics additive. Food Chem. Toxicol. 2018, 120, 668–680. [Google Scholar] [CrossRef]
- Chebet, J.J.; Ehiri, J.E.; McClelland, D.J.; Taren, D.; Hakim, I.A. Effect of d-limonene and its derivatives on breast cancer in human trials: A scoping review and narrative synthesis. BMC Cancer 2021, 21, 902. [Google Scholar] [CrossRef] [PubMed]
- Quintans-Junior, L.J.; Souza, T.T.; Leite, B.S.; Lessa, N.M.N.; Bonjardim, L.R.; Santos, M.R.V.; Alves, P.B.; Blank, A.F.; Antoniolli, A.R. Phythochemical screening and anticonvulsant activity of Cymbopogon winterianus Jowitt (Poaceae) leaf essential oil in rodents. Phytomed 2008, 15, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.S.; Guimarães, A.G.; Santana, M.F.; Siqueira, R.S.; De Lima, A.C.; Dias, A.S.; Santos, M.R.V.; Onofre, A.S.C.; Quintans, J.S.S.; De Sousa Almeida, D.P.J.; et al. Anti-inflammatory and redox-protective activities of citronellal. Biol. Res. 2011, 44, 363–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amri, I.; Hamrouni, L.; Hanana, M.; Gargouri, S.; Jamoussi, B. Chemical composition, bio-herbicidal and antifungal activities of essential oils isolated from Tunisian common cypress (Cupressus sempervirens L.). J. Med. Plant Res. 2013, 7, 1070–1080. [Google Scholar]
- Moo, C.L.; Osman, M.A.; Yang, S.K.; Yap, W.S.; Ismail, S.; Lim, S.H.E.; Chong, C.M.; Lai, K.S. Antimicrobial activity and mode of action of 1,8-cineol against carbapenemase-producing Klebsiella pneumoniae. Sci. Rep. 2021, 11, 20824. [Google Scholar] [CrossRef]
- Alamoti, M.P.; Gilani, B.B.; Mahmoudi, R.; Reale, A.; Pakbin, B.; Di Renzo, T.; Ata, A. Essential Oils from Indigenous Iranian Plants: A Natural Weapon vs. Multidrug-Resistant Escherichia coli. Microorganisms 2022, 10, 109. [Google Scholar] [CrossRef]
- O’Sullivana, M.E.; Songa, Y.; Greenhousea, R.; Lina, R.; Pereza, A.; Atkinsona, P.J.; MacDonaldb, J.P.; Siddiquib, Z.; Lagascab, D.; Comstockc, K.; et al. Dissociating antibacterial from ototoxic effects ofgentamicin C-subtypes. Proc. Natl. Acad. Sci. USA 2020, 117, 32423–32432. [Google Scholar] [CrossRef]
- Damayanti, S.; Permana, J.; Tjahjono, D.H. The use of computational chemistry to predict toxicity of antioxidants food additives and its metabolites as a reference for food safety regulation. Pharma Chem J. 2015, 7, 174–181. [Google Scholar]
- Valente, M.J.; Araújo, A.M.; Bastos, M.D.L.; Fernandes, E.; Carvalho, F.; Guedes de Pinho, P.; Carvalho, M. Editor’s highlight: Characterization of hepatotoxicity mechanisms triggered by designer cathinone drugs (β-Keto amphetamines). Toxicol 2016, 153, 89–102. [Google Scholar] [CrossRef] [Green Version]
- Eftekhari, A.; Ahmadian, E.; Azarmi, Y.; Parvizpur, A.; Fard, J.K.; Eghbal, M.A. The effects of cimetidine, N-acetylcysteine, and taurine on thioridazine metabolic activation and induction of oxidative stress in isolated rat hepatocytes. Pharm. Chem. J. 2018, 51, 965–969. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.L.; Fu, B.M.; Zhang, Z.J. Borneol, a novel agent that improves central nervous system drug delivery by enhancing blood–brain barrier permeability. Drug Deliv. 2017, 24, 1037–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samreen Qais, F.A.; Ahmad, I. In silico screening and in vitro validation of phytocompounds as multidrug efflux pump inhibitor against E. coli. J. Biomol. Struct. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hoyberghs, J.; Bars, C.; Pype, C.; Foubert, K.; Hernando, M.A.; Van Ginneken, C.; Van Cruchten, S. Refinement of the zebrafish embryo developmental toxicity assay. MethodsX 2020, 7, 101087. [Google Scholar] [CrossRef] [PubMed]
- Wang-Kan, X.; Blair, J.M.; Chirullo, B.; Betts, J.; La Ragione, R.M.; Ivens, A.; Ricci, V.; Opperman, T.J.; Piddock, L.J.V. Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar Typhimurium. MBio 2017, 8, e00968-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsuka, Y.; Chen, C.; Nikaido, H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 6559–6565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumaraswami, M.; Schuman, J.T.; Seo, S.M.; Kaatz, G.W.; Brennan, R.G. Structural and biochemical characterization of MepR, a multidrug binding transcription regulator of the Staphylococcus aureus multidrug efflux pump MepA. Nucleic Acids Res. 2009, 37, 1211–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Kaizaki, A.; Tanaka, S.; Numazawa, S. New recreational drug 1-phenyl-2-(1-pyrrolidinyl)-1-pentanone (alpha-PVP) activates central nervous system via dopaminergic neuron. J. Toxicol. Sci. 2014, 39, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Roymahapatra, G.; Paul, D.; Mandal, S.M. Theoretical analysis of bacterial efflux pumps inhibitors: Strategies in-search of competent molecules and develop next. Compul. Biol. Chem. 2020, 87, 107275. [Google Scholar] [CrossRef]
- Kovač, J.; Šimunović, K.; Wu, Z.; Klančnik, A.; Bucar, F.; Zhang, Q.; Možina, S.S. Antibiotic resistance modulation and modes of action of (-)-α-pinene in Campylobacter jejuni. PLoS ONE 2015, 10, e0122871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nidhi, P.; Rolta, R.; Kumar, V.; Dev, K.; Sourirajan, A. Synergistic potential of Citrus aurantium L. essential oil with antibiotics against Candida albicans. J. Ethnopharmacol. 2020, 262, 113135. [Google Scholar] [CrossRef] [PubMed]
- Fadli, M.; Chevalier, J.; Hassani, L.; Mezrioui, N.E.; Pagès, J.M. Natural extracts stimulate membrane-associated mechanisms of resistance in Gram-negative bacteria. Lett. Appl. Microbiol. 2014, 58, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Thorsing, M.; Klitgaard, J.K.; Atilano, M.L.; Skov, M.N.; Kolmos, H.J.; Filipe, S.R.; Kallipolitis, B.H. Thioridazine induces major changes in global gene expression and cell wall composition in methicillin-resistant Staphylococcus aureus USA300. PLoS ONE 2013, 8, e64518. [Google Scholar] [CrossRef] [Green Version]
- Rana, T.; Singh, S.; Kaur, N.; Pathania, K.; Farooq, U. A review on efflux pump inhibitors of medically important bacteria from plant sources. Int. J. Pharm. Sci. Rev. Res. 2014, 26, 101–111. [Google Scholar]
- Ruth, M.M.; Pennings, L.J.; Koeken, V.A.; Schildkraut, J.A.; Hashemi, A.; Wertheim, H.F.L.; Hoefsloot, W.; van Ingen, J. Thioridazine is an efflux pump inhibitor in Mycobacterium avium complex but of limited clinical relevance. Antimicrob. Agents Chemother. 2020, 64, e00181-20. [Google Scholar] [CrossRef]
- Dabul, A.N.G.; Avaca-Crusca, J.S.; Van Tyne, D.; Gilmore, M.S.; Camargo, I.L.B.C. Resistance in in vitro selected Tigecycline-resistant methicillin-resistant Staphylococcus aureus sequence type 5 is driven by mutations in mepR and mepA genes. Microb. Drug Resist. 2018, 24, 519–526. [Google Scholar] [CrossRef] [Green Version]
- de Medeiros, V.M.; do Nascimento, Y.M.; Souto, A.L.; Madeiro, S.A.L.; de Oliveira Costa, V.C.; Silva, S.M.P.; dos SantosFalcão, V.; de Fátima Agra, M.; de Siqueira-Júnio, J.P.; Tavares, J.F. Chemical composition and modulation of bacterial drug resistance of the essential oil from leaves of Croton grewioides. Microb. Pathog. 2017, 111, 468–471. [Google Scholar] [CrossRef]
- Coêlho, M.L.; Ferreira, J.H.L.; de Siqueira Júnior, J.P.; Kaatz, G.W.; Barreto, H.M.; Cavalcante, A.A.D.C.M. Inhibition of the NorA multi-drug transporter by oxygenated monoterpenees. Microb. Pathog. 2016, 99, 173–177. [Google Scholar] [CrossRef]
- Agreles, M.A.A.; Cavalcanti, I.D.L.; Cavalcanti, I.M.F. The Role of Essential Oils in the Inhibition of Efflux Pumps and Reversion of Bacterial Resistance to Antimicrobials. Curr. Microbiol. 2021, 78, 3609–3619. [Google Scholar] [CrossRef]
- Limaverde, P.W.; Campina, F.F.; da Cunha, F.A.; Crispim, F.D.; Figueredo, F.G.; Lima, L.F.; de, M. Oliveira-Tintino, C.D.; de Matos, Y.M.L.S.; Siqueira-Júnior, J.P. et al. Inhibition of the TetK efflux-pump by the essential oil of Chenopodium ambrosioides L. and α-terpinene against Staphylococcus aureus IS-58. Food Chem. Toxicol. 2017, 109, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z. Advances in homology protein structure modeling. Curr. Protein Pept. Sci. 2006, 7, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacoppo, J.D.O.; Carregal, J.B.; Junior, M.C.; Cunha, E.F.D.; Ramalho, T.C. Towards the understanding of tetrahydroquinolines action in Aedes aegypti: Larvicide or adulticide? Mol. Simul. 2017, 43, 121–133. [Google Scholar] [CrossRef]
- Robinson, A.; Van Oijen, A.M. Bacterial replication, transcription and translation: Mechanistic insights from single-molecule biochemical studies. Nat. Rev. Microbiol. 2013, 11, 303–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Bai, M.; Sun, Y.; Abdel-Samie, M.A.S.; Lin, L. Antibacterial activity and mechanism of Chuzhou chrysanthemum essential oil. J. Funct. Foods 2018, 48, 159–166. [Google Scholar] [CrossRef]
- Utegenova, G.A.; Pallister, K.B.; Kushnarenko, S.V.; Özek, G.; Özek, T.; Abidkulova, K.T.; Voyich, J.M. Chemical composition and antibacterial activity of essential oils from Ferula L. species against methicillin-resistant Staphylococcus aureus. Molecules 2018, 23, 1679. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Hu, D.H.; Feng, Y. Antibacterial activity and mode of action of the Artemisia capillaris essential oil and its constituents against respiratory tract infection-causing pathogens. Mol. Med. Rep. 2015, 11, 2852–2860. [Google Scholar] [CrossRef] [Green Version]
- Melkina, O.E.; Plyuta, V.A.; Khmel, I.A.; Zavilgelsky, G.B. The Mode of Action of Cyclic Monoterpenees (−)-Limoneneand (+)-α-Pinene on Bacterial Cells. BioMolecules 2021, 11, 806. [Google Scholar] [CrossRef]
- Luís, Â.; Ramos, A.; Domingues, F. Pullulan films containing rockrose essential oil for potential food packaging applications. Antibiotics 2020, 9, 681. [Google Scholar] [CrossRef]
- Aghoutane, Y.; Moufid, M.; Motia, S.; Padzys, G.S.; Omouendze, L.P.; Llobet, E.; El Bari, N. Characterization and analysis of Okoume and aiele essential oils from Gabon by gc-ms, electronic nose, and their antibacterial activity assessment. Sensors 2020, 20, 6750. [Google Scholar] [CrossRef]
- Anka, L.; Rammal, H.; Kobeissi, A.; Saab, H.B. Chemical composition and biological potentials of Lebanese Cupressus sempervirensL. leaves extracts. J. Med. Plant Res. 2020, 14, 292–299. [Google Scholar]
- Lee, S.Y.; Kim, S.H.; Hong, C.Y.; Park, M.J.; Choi, I.G. Effects of (−)-borneol on the growth and morphology of Aspergillus fumigatus and Epidermophyton floccosom. Flavour Fragr. J. 2013, 28, 129–134. [Google Scholar] [CrossRef]
- Xiong, Z.Y.; Xiao, F.M.; Xu, X.; Wu, Y.F.; Jiang, X.M. Studies on pharmacological activity of borneol. China J. Chin. Mater. Med. 2013, 38, 786–790. [Google Scholar]
- Yang, L.; Zhan, C.; Huang, X.; Hong, L.; Fang, L.; Wang, W.; Su, J. Durable Antibacterial Cotton Fabrics Based on Natural Borneol-Derived Anti-MRSA Agents. Adv. Healthc. Mater. 2020, 9, 2000186. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Li, G.; Luan, D.; Yuan, Q.; Wei, Y.; Wang, X. Antibacterial adhesion of borneol-based polymer via surface chiral stereochemistry. ACS Appl. Mater. Interfaces 2014, 6, 19371–19377. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Qian, Z.; Luo, L.; Yuan, Q.; Guo, X.; Tao, L.; Wei, Y.; Wang, X. Antibacterial adhesion of poly (methyl methacrylate) modified by borneol acrylate. ACS Appl. Mater. Interfaces 2016, 8, 28522–28528. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ren, Z.; Wang, L.; Cai, Y.; Ma, H.; Fang, L.; Su, J. Nanoparticle-stabilized encapsulation of borneol and citral: Physicochemical characteristics, storage stability, and enhanced antibacterial activities. J. Food Sci. 2021, 86, 4554–4565. [Google Scholar] [CrossRef]
- Dai, J.; Li, C.; Cui, H.; Lin, L. Unraveling the anti-bacterial mechanism of Litsea cubeba essential oil against E. coli O157: H7 and its application in vegetable juices. Int. J. Food Microbiol. 2021, 338, 108989. [Google Scholar] [CrossRef]
- De Souza Moura, W.; de Souza, S.R.; Campos, F.S.; Cangussu, A.S.R.; Santos, E.M.S.; Andrade, B.S.; Gomes, C.H.B.; Viana, K.F.; Haddi, K.; Oliveira, E.E.; et al. Antibacterial activity of Siparuna guianensis essential oil mediated by impairment of membrane permeability and replication of pathogenic bacteria. Ind Crops Prod. 2020, 146, 112142. [Google Scholar] [CrossRef]
- Sellem, I.; Chakchouk-Mtibaa, A.; Zaghden, H.; Smaoui, S.; Ennouri, K.; Mellouli, L. Harvesting season dependent variation in chemical composition and biological activities of the essential oil obtained from Inula graveolens (L.) grown in Chebba (Tunisia) salt marsh. Arab. J. Chem. 2020, 13, 4835–4845. [Google Scholar] [CrossRef]
- Güven, K.; Yücel, E.; Cetintaş, F. Antimicrobial activities of fruits of Crataegus and Pyrus species. Pharm. Biol. 2006, 44, 79–83. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Venkatesalu, V. Antibacterial and antifungal activity of Syzygium jambolanum seeds. J. Ethnopharmacol. 2004, 91, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Lennerz, B.S.; Vafai, S.B.; Delaney, N.F.; Clish, C.B.; Deik, A.A.; Pierce, K.A.; Ludwig, D.S.; Mootha, V.K. Effects of sodium benzoate, a widely used food preservative, on glucose homeostasis and metabolic profiles in humans. Mol. Genet. Metab. 2015, 114, 73–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosaei, H.; Zenkin, N. Inhibition of RNA polymerase by Rifampicin and Rifamycin-like Molecules. EcoSal Plus 2020, 9, 1–16. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Amer, A.A.; Marzouk, A.A.; Gomaa, H.A.; Youssif, B.G.; Abdelhamid, A.A. Design, synthesis, and antibacterial screening of some novel heteroaryl-based ciprofloxacin derivatives as DNA gyrase and topoisomerase IV inhibitors. Pharmaceuticals 2021, 14, 399. [Google Scholar] [CrossRef]
- Ferrari, T.; Gini, G. An open source multistep model to predict mutagenicity from statistical analysis and relevant structural alerts. Chem. Cent. J. 2010, 4, S2. [Google Scholar] [CrossRef] [Green Version]
- Fjodorova, N.; Vračko, M.; Tušar, M.; Jezierska, A.; Novič, M.; Kühne, R.; Schüürmann, G. Quantitative and qualitative models for carcinogenicity prediction for non-congeneric chemicals using CP ANN method for regulatory uses. Mol. Divers. 2010, 14, 581–594. [Google Scholar] [CrossRef]
- Benfenati, E.; Chaudhry, Q.; Gini, G.; Dorne, J.L. Integrating in silico models and read-across methods for predicting toxicity of chemicals: A step-wise strategy. Environ. Int. 2019, 131, 105060. [Google Scholar] [CrossRef]
- Roncaglioni, A.; Piclin, N.; Pintore, M.; Benfenati, E. Binary classification models for endocrine disrupter effects mediated through the estrogen receptor. SAR QSAR Environ. Res. 2008, 19, 697–733. [Google Scholar] [CrossRef]
- Bruning, J.B.; Parent, A.A.; Gil, G.; Zhao, M.; Nowak, J.; Pace, M.C.; Smith, C.L.; Afonine, P.V.; Adams, P.D.; Katzenellenbogen, J.A.; et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 2010, 6, 837–843. [Google Scholar] [CrossRef] [Green Version]
- Baderna, D.; Gadaleta, D.; Lostaglio, E.; Selvestrel, G.; Raitano, G.; Golbamaki, A.; Lombardo, A.; Benfenati, E. New in silico models to predict in vitro micronucleus induction as marker of genotoxicity. J. Hazard. Mater. 2020, 385, 121638. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.vegahub.eu/ (accessed on 27 January 2022).
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://tox-new.charite.de/protox_II/ (accessed on 27 January 2022).
- No, O.T. 236: Fish embryo acute toxicity (FET) test. OECD Guidel. Test. Chem. 2013, 2, 1–22. [Google Scholar]
- De Guzman, M.C.; Chua, P.A.P.; Sedano, F.S. Embryotoxic and teratogenic effects of polyethylene microbeads found in facial wash products in Zebrafish (Danio rerio) using the Fish Embryo Acute Toxicity Test. bioRxiv 2020. [Google Scholar] [CrossRef]
- Available online: https://swissmodel.expasy.org (accessed on 28 January 2022).
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.uniprot.org (accessed on 28 January 2022).
- UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://www.ebi.ac.uk/thornton-srv/databases/ProFunc (accessed on 29 January 2022).
- Sharma, A.D.; Kaur, G.R.I.; Kocher, G.S. Molecular modeling and in-silico characterization of alkaline protease from bacillus circulans MTCC 7906. Online J. Bioinform. 2015, 16, 61–87. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.drugbank.ca/ (accessed on 26 January 2022).
- Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 26 January 2022).
- Available online: http://www.molecular-networks.com/online_demos/corina_demo (accessed on 26 January 2022).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Pawar, S.S.; Rohane, S.H. Review on discovery studio: An important tool for molecular docking. Asian J. Res. Chem. 2021, 14, 86–88. [Google Scholar] [CrossRef]




| Compound | Molar Mass (g/mol) | Molecular Formula | Retention Time (min) | EO (%) |
|---|---|---|---|---|
| β-terpinene | 136.23 | C10H16 | 4.80 | 0.11 |
| Tricyclene | 136.23 | C10H16 | 5.25 | 0.22 |
| α-pinene | 136.23 | C10H16 | 5.76 | 38.47 |
| α-fenchene | 136.23 | C10H16 | 5.95 | 1.36 |
| Sabinene | 136.23 | C10H16 | 6.67 | 1.18 |
| β-pinene | 136.23 | C10H16 | 6.75 | 2.04 |
| β-myrcene | 136.23 | C10H16 | 7.22 | 2.78 |
| δ-3-carene | 136.23 | C10H16 | 7.91 | 25.14 |
| D-limonene | 136.23 | C10H16 | 7.99 | 5.84 |
| P-cymene | 134.22 | C10H14 | 8.23 | 0.86 |
| Linalool | 154.25 | C10H18O | 10.62 | 0.38 |
| Isopulegol | 154.25 | C10H18O | 11.96 | 0.88 |
| Citronellal | 154.25 | C10H18O | 12.23 | 5.33 |
| Borneol | 154.25 | C10H18O | 12.58 | 1.37 |
| Terpinen-4-ol | 154.25 | C10H18O | 12.92 | 1.55 |
| α-terpineol | 154.25 | C10H18O | 13.39 | 0.54 |
| β-citronellol | 156.26 | C10H20O | 14.54 | 0.21 |
| α-fenchyl acetate | 196.29 | C12H20O2 | 15.95 | 1.28 |
| Camphene | 136.23 | C10H16 | 16.29 | 0.29 |
| α-terpinyl acetate | 196.29 | C12H20O2 | 17.73 | 2.82 |
| α-zingibirene | 204.35 | C15H24 | 19.31 | 0.52 |
| α-carophyllene | 204.35 | C15H24 | 19.49 | 0.83 |
| α-humulene | 204.35 | C15H24 | 20.36 | 0.18 |
| Germacrene D | 204.35 | C15H24 | 20.60 | 0.83 |
| α-amorphene | 204.35 | C15H24 | 20.94 | 0.18 |
| δ-cadinene | 204.35 | C15H24 | 20.09 | 0.30 |
| Cedrol | 222.37 | C15H26O | 24.02 | 2.24 |
| Monoterpenes hydrocarbons | 78.29 (%) | |||
| Oxygenated monoterpenes | 17.28 (%) | |||
| Sesquiterpens | 2.84 (%) | |||
| Total | 98.41 (%) | |||
| Bacterial Strains | Inhibition Zones Diameters (mm) | MIC (µg/mL) | ||
|---|---|---|---|---|
| (CSEO) | Gentamicin | (CSEO) | Gentamicin | |
| S. aureus ATCC 6538 | 21 ± 1.00 a | 20 ± 0.83 a | 6.25 ± 0.00 a | 12.5 ± 0.00 b |
| L. monocytogenes ATCCC 19117 | 21 ± 0.83 a | 20 ± 0.66 a | 6.25 ± 0.00 a | 12.5 ± 0.00 b |
| S. Typhimurium ATCC 14028 | 15 ± 0.5 a | 25 ± 1.00 b | 12.5 ± 0.00 b | 2.5 ± 0.00 a |
| E. coli ATCC 8739 | 15 ± 0.5 a | 25 ± 1.25 b | 12.5 ± 0.00 b | 2.5 ± 0.00 a |
| Toxicity Measurements | Mutagenicity (Ames Test) Model (CAESAR) 2.1.13 | Carcinogenicity Model (CAESAR) 2.1.9 | Developmental Toxicity Model (CAESAR) 2.1.7 | Developmental/Reproductive Toxicity Library (PG) 1.1.0 | Estrogen Receptor Relative Binding Affinity Model (IRFMN) | Androgen Receptor-Mediated Effect (IRFMN/COMPARA) 1.0.0 | Thyroid Receptor Alpha Effect (NRMEA) 1.0.0 | Thyroid Receptor Beta Effect (NRMEA) 1.0.0 | In Vitro Micronucleus Activity (IRFMN/VERMEER) 1.0.0 |
|---|---|---|---|---|---|---|---|---|---|
| Compound | |||||||||
| β-terpinene | - | - | - | - | - | - | - | - | - |
| Tricyclene | - | - | - | - | + | - | - | - | - |
| α-pinene | - | - | + | - | - | - | - | - | - |
| α-fenchene | - | - | - | - | - | - | - | - | - |
| Sabinene | - | - | - | - | - | - | - | - | - |
| β-pinene | - | - | + | - | - | - | - | - | - |
| β-myrcene | - | + | - | - | - | - | - | - | + |
| δ-3-carene | - | - | - | - | - | - | - | - | - |
| D-limonene | - | + | - | - | - | - | - | - | - |
| P-cymene | - | - | - | + | - | - | - | - | - |
| Linalool | - | - | - | - | - | - | - | - | + |
| Isopulegol | - | + | + | + | - | - | - | - | - |
| Citronellal | - | - | - | - | - | - | - | - | - |
| Borneol | - | - | - | - | - | - | - | - | - |
| Terpinen-4-ol | - | - | + | - | - | - | - | - | - |
| α-terpineol | - | - | + | - | - | - | - | - | - |
| β-citronellol | - | - | - | - | - | - | - | - | - |
| α-fenchyl acetate | - | - | - | - | - | - | - | - | - |
| Camphene | - | - | - | - | - | - | - | - | - |
| α-terpinyl acetate | - | - | - | - | - | - | - | - | - |
| α-zingibirene | - | - | - | - | - | - | - | - | + |
| α-carophyllene | - | - | - | - | - | - | - | - | + |
| α-humulene | - | - | - | - | - | - | - | - | + |
| Germacrene D | - | - | - | - | - | - | - | - | + |
| α-amorphene | - | + | + | - | - | - | - | - | + |
| δ-cadinene | - | + | + | - | - | - | - | - | - |
| Cedrol | - | - | + | - | - | - | - | - | - |
| Ciprofloxacin | + | - | + | + | - | - | - | - | + |
| Rifamycin SV | + | - | + | - | - | - | - | - | + |
| Toxicity Measurements | Citric Acid | Butylated Hydroxyanisol (BHA) | Ascorbic Acid | Propionic Acid | Benzoic Acid | Cathinone | Thioridazine |
|---|---|---|---|---|---|---|---|
| Mutagenicity (Ames test) model (CAESAR) 2.1.13 | - | - | - | - | - | - | - |
| Carcinogenicity model (CAESAR) 2.1.9 | - | + | - | - | - | - | - |
| Developmental Toxicity model (CAESAR) 2.1.7 | - | - | - | + | + | + | + |
| Developmental/Reproductive Toxicity library (PG) 1.1.0 | - | - | - | + | - | + | + |
| Estrogen Receptor Relative Binding Affinity model (IRFMN) | - | - | - | - | - | - | + |
| Androgen Receptor-mediated effect (IRFMN/COMPARA) 1.0.0 | - | - | - | - | - | - | - |
| Thyroid Receptor Alpha effect (NRMEA) 1.0.0 | - | - | - | - | - | - | - |
| Thyroid Receptor Beta effect (NRMEA) 1.0.0 | - | - | - | - | - | - | - |
| In vitro Micronucleus activity (IRFMN/VERMEER) 1.0.0 | + | - | + | Not predicted | - | + | + |
| Compound | Cytotoxicity | Probability | LD 50 (mg/kg) | Toxicity Class |
|---|---|---|---|---|
| β-terpinene | Inactive | 0.80 | 4400 | 5 |
| Tricyclene | Inactive | 0.77 | 15,380 | 6 |
| α-pinene | Inactive | 0.75 | 3700 | 5 |
| α-fenchene | Inactive | 0.74 | 5000 | 5 |
| Sabinene | Inactive | 0.71 | 5000 | 5 |
| β-pinene | Inactive | 0.71 | 4700 | 5 |
| β-myrcene | Inactive | 0.75 | 5000 | 5 |
| δ-3-carene | Inactive | 0.71 | 4800 | 5 |
| D-limonene | Inactive | 0.82 | 4400 | 5 |
| P-cymene | Inactive | 0.89 | 3 | 1 |
| Linalool | Inactive | 0.82 | 2200 | 5 |
| Isopulegol | Inactive | 0.93 | 5000 | 5 |
| Citronellal | Inactive | 0.82 | 2420 | 5 |
| Borneol | Inactive | 0.88 | 500 | 4 |
| Terpinen-4-ol | Inactive | 0.88 | 1016 | 4 |
| α-terpineol | Inactive | 0.64 | 2830 | 5 |
| β-citronellol | Inactive | 0.86 | 3450 | 5 |
| α-fenchyl acetate | Inactive | 0.73 | 3100 | 5 |
| Camphene | Inactive | 0.76 | 5000 | 5 |
| α-terpinyl acetate | Inactive | 0.80 | 4800 | 5 |
| α-zingibirene | Inactive | 0.82 | 1680 | 4 |
| α-caryophyllene | Inactive | 0.79 | 3650 | 5 |
| α-humulene | Inactive | 0.79 | 3650 | 5 |
| Germacrene D | Inactive | 0.83 | 5300 | 5 |
| α-amorphene | Inactive | 0.76 | 4400 | 5 |
| δ-cadinene | Inactive | 0.69 | 4390 | 5 |
| Cedrol | Inactive | 0.87 | 2000 | 4 |
| Rifamycin SV | Inactive | 0.60 | 2120 | 5 |
| Ciprofloxacin | Inactive | 0.92 | 2000 | 4 |
| Citric acid | Inactive | 0.73 | 80 | 3 |
| BHA | Inactive | 0.83 | 700 | 4 |
| L-ascorbic acid | Inactive | 0.65 | 3367 | 5 |
| Propionic acid | Inactive | 0.75 | 300 | 3 |
| Benzoic acid | Inactive | 0.86 | 235 | 3 |
| Cathinone | Inactive | 0.82 | 400 | 4 |
| Thioridazine | Inactive | 0.68 | 360 | 4 |
| Compound | AcrsB Efflux Pump | MepR |
|---|---|---|
| β-terpinene | −5.1 | −5.5 |
| Tricyclene | −5.8 | −4.7 |
| α-pinene | −6.7 | −6.5 |
| α-fenchene | −5.3 | −4.8 |
| Sabinene | −5.0 | −5.2 |
| β-pinene | −5.0 | −4.9 |
| β-myrcene | −4.8 | −4.5 |
| δ-3-carene | −6.4 | −6.2 |
| D-limonene | −5.3 | −5.3 |
| P-cymene | −6.2 | −5.6 |
| Linalool | −4.8 | −4.4 |
| Isopulegol | −5.2 | −5.1 |
| Citronellal | −4.2 | −3.9 |
| Borneol | −7.9 | −7.7 |
| Terpinen-4-ol | −5.3 | −5.0 |
| α-terpineol | −5.3 | −5.3 |
| β-citronellol | −4.4 | −4.3 |
| α-fenchyl acetate | −6.4 | −4.3 |
| Camphene | −5.4 | −4.8 |
| α-terpinyl acetate | −6.2 | −5.2 |
| α-zingibirene | −5.4 | −5.6 |
| α-carophyllene | −6.0 | −6.3 |
| α-humulene | −6.5 | −6.3 |
| Germacrene D | −7.0 | −6.4 |
| α-amorphene | −6.5 | −6.6 |
| δ-cadinene | −6.6 | −6.7 |
| Cedrol | −6.3 | −6.5 |
| Cathinone | −4.8 | - |
| Thioridazine | - | −6.7 |
| S. aureus (Strain Mu50/ATCC 700699) | S. Typhimurium (strain LT2/SGSC1412/ATCC 700720) | |||||
|---|---|---|---|---|---|---|
| Compound | DNA Polymerase | RNA Polymerase | Topoisomerase II | DNA Polymerase | RNA Polymerase | Topoisomerase II |
| β-terpinene | −5.2 | −5.8 | −5.5 | −5.3 | −6 | −5.4 |
| Tricyclene | −5.2 | −5 | −5.4 | −5.2 | −5 | −5.2 |
| α-pinene | −5.2 | −5.1 | −5.3 | −5.4 | −5.4 | −5.2 |
| α-fenchene | −5.4 | −5.6 | −5.2 | −5.2 | −5.1 | −5.5 |
| Sabinene | −5.2 | −5.4 | −5.4 | −5.2 | −5.2 | −5.1 |
| β-pinene | −5.5 | −5.1 | −5.3 | −5.4 | −5.4 | −5.2 |
| β-myrcene | −4.6 | −4.3 | −5.3 | −4.7 | −4.3 | −4.3 |
| δ-3-carene | −5.1 | −6.1 | −5.2 | −5.6 | −5.4 | −5.6 |
| D-limonene | −4.9 | −4.6 | −5.4 | −4.9 | −5.2 | −5.1 |
| P-cymene | −5.4 | −5.2 | −5.6 | −5.4 | −5.5 | −5.2 |
| Linalool | −4.8 | −4.8 | −5.2 | −5.1 | −4.5 | −4.2 |
| Isopulegol | −5.3 | −5.8 | −5.2 | −5.4 | −5.4 | −5 |
| Citronellal | −4.5 | −4.4 | −4.8 | −4.6 | −4.8 | −4.2 |
| Borneol | −7.7 | −8.2 | −8.8 | −7.7 | −7.3 | −7.4 |
| Terpinen-4-ol | −5.2 | −5.3 | −5.7 | −5.7 | −5.7 | −5.3 |
| α-terpineol | −5.3 | −5.9 | −5.6 | −5.7 | −5.7 | −5.2 |
| β-citronellol | −4.8 | −4.3 | −5 | −5.1 | −4.6 | −4.7 |
| α-fenchyl acetate | −5.7 | −5.4 | −5.6 | −5.2 | −5.3 | −5.1 |
| Camphene | −5.2 | −5.4 | −5.1 | −5.5 | −5.5 | −5.2 |
| α-terpinyl acetate | −5.4 | −5.1 | −6.4 | −6 | −6 | −5.3 |
| α-zingibirene | −5.8 | −5.1 | −6.1 | −6.1 | −5.2 | −5.8 |
| α-carophyllene | −6.2 | −5.9 | −6.8 | −6.1 | −6.1 | −6.3 |
| α-humulene | −6.1 | −6.3 | −6.8 | −6.1 | −6.1 | −6.3 |
| Germacrene D | −6.5 | −7.1 | −6.7 | −6.2 | −6.2 | −6.5 |
| α-amorphene | −6.8 | −6.2 | −6.9 | −6.8 | −6.2 | −6 |
| δ-cadinene | −6.3 | −6.7 | −6.7 | −6.5 | −6.7 | −6.1 |
| Cedrol | −6.6 | −6.7 | −6.8 | −6.9 | −6.2 | −6.7 |
| Rifamycin SV | −9.2 | −9.8 | - | −8.4 | −8.6 | - |
| Ciprofloxacin | - | - | −6.7 | - | - | −6.3 |
| Bacteria | Compound | Targets | Number of Residues Interacting | Residues with H-Bond |
|---|---|---|---|---|
| S. aureus (strain Mu50/ATCC 700699) | α-pinene | DNA polymerase | 6 | - |
| RNA polymerase | 6 | - | ||
| Topoisomerase II | 6 | - | ||
| δ-3-carene | DNA polymerase | 8 | - | |
| RNA polymerase | 4 | - | ||
| Topoisomerase II | 4 | - | ||
| Borneol | DNA polymerase | 4 | Ser 901, Lys 901 | |
| RNA polymerase | 11 | Glu 79, Ala 672, Gly 670, Gln 725 | ||
| Topoisomerase II | 5 | Thr 194 | ||
| S. Typhimurium (strain LT2/SGSC1412/ATCC 700720) | α-pinene | DNA polymerase | 3 | - |
| RNA polymerase | 9 | - | ||
| Topoisomerase II | 8 | - | ||
| δ-3-carene | DNA polymerase | 7 | - | |
| RNA polymerase | 6 | - | ||
| Topoisomerase II | 2 | - | ||
| Borneol | DNA polymerase | 8 | Gly 640 | |
| RNA polymerase | 8 | Asp81, Glu 963 | ||
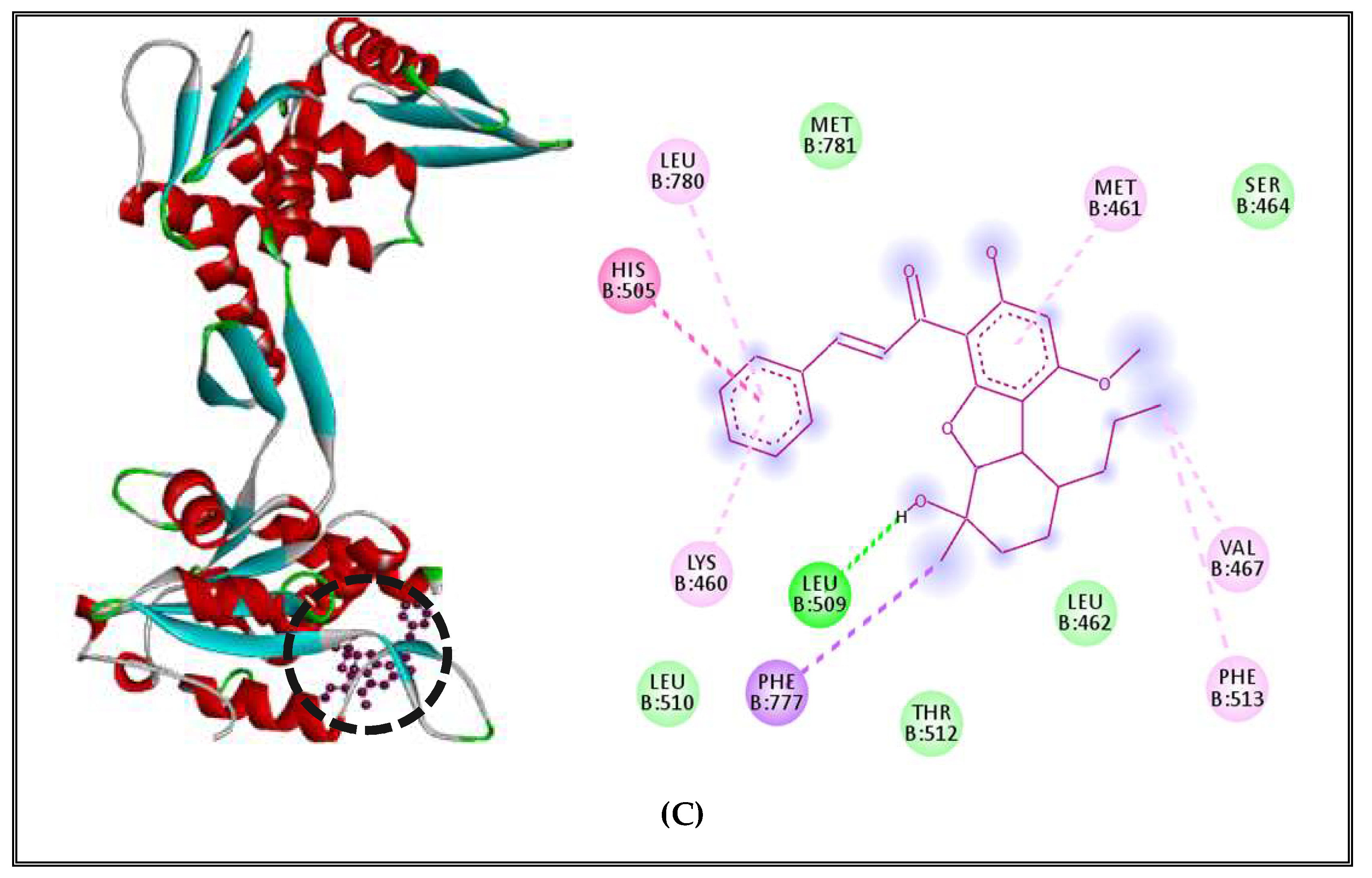
| Topoisomerase II | 8 | Leu 509 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akermi, S.; Smaoui, S.; Elhadef, K.; Fourati, M.; Louhichi, N.; Chaari, M.; Chakchouk Mtibaa, A.; Baanannou, A.; Masmoudi, S.; Mellouli, L. Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos. Molecules 2022, 27, 2630. https://doi.org/10.3390/molecules27092630
Akermi S, Smaoui S, Elhadef K, Fourati M, Louhichi N, Chaari M, Chakchouk Mtibaa A, Baanannou A, Masmoudi S, Mellouli L. Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos. Molecules. 2022; 27(9):2630. https://doi.org/10.3390/molecules27092630
Chicago/Turabian StyleAkermi, Sarra, Slim Smaoui, Khaoula Elhadef, Mariam Fourati, Nacim Louhichi, Moufida Chaari, Ahlem Chakchouk Mtibaa, Aissette Baanannou, Saber Masmoudi, and Lotfi Mellouli. 2022. "Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos" Molecules 27, no. 9: 2630. https://doi.org/10.3390/molecules27092630
APA StyleAkermi, S., Smaoui, S., Elhadef, K., Fourati, M., Louhichi, N., Chaari, M., Chakchouk Mtibaa, A., Baanannou, A., Masmoudi, S., & Mellouli, L. (2022). Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos. Molecules, 27(9), 2630. https://doi.org/10.3390/molecules27092630







