Development of a Genus-Universal Nucleotide Signature for the Identification and Supervision of Ephedra-Containing Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Materials
2.2. DNA Extraction
2.3. Primer Design, PCR Amplification and Sequencing
2.4. Sequence Analysis
3. Results
3.1. Development of the Genus-Universal Nucleotide Signature for Ephedra L.
3.2. Verification of the Nucleotide Signature in the Species and Varieties within Ephedra L.
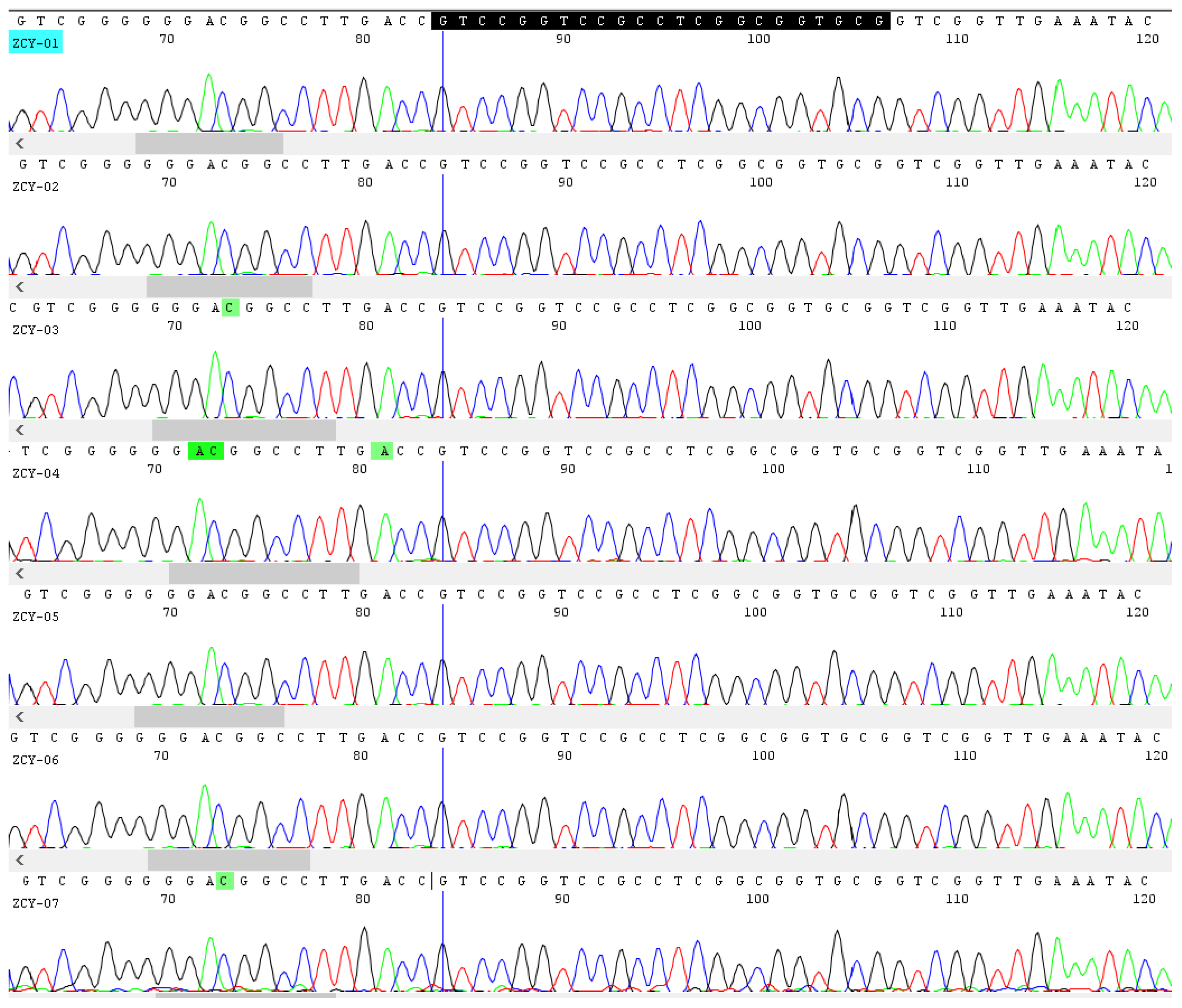
3.3. Application of the Nucleotide Signature on the Detection of Ephedra Herb in Compound Preparations
4. Discussion
4.1. Significance of the Development of a Nucleotide Signature for Ephedra
4.2. Supervision of Ephedra-Containing Products with the Genus-Universal Nucleotide Signature
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bánki, O.; Roskov, Y.; Vandepitte, L.; DeWalt, R.E.; Remsen, D.; Schalk, P.; Orrell, T.; Keping, M.; Miller, J.; Aalbu, R.; et al. Catalogue of Life Checklist; Catalogue of Life: Leiden, The Netherlands, 2021. [Google Scholar] [CrossRef]
- González-Juárez, D.E.; Escobedo-Moratilla, A.; Flores, J.; Hidalgo-Figueroa, S.; Martínez-Tagüeña, N.; Morales-Jiménez, J.; Muñiz-Ramírez, A.; Pastor-Palacios, G.; Pérez-Miranda, S.; Ramírez-Hernández, A.; et al. A Review of the Ephedra genus: Distribution, Ecology, Ethnobotany, Phytochemistry and Pharmacological Properties. Molecules 2020, 25, 3283. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.-M.; Zhang, Q.; Bi, X.-B.; Cui, J.-L.; Wang, M.-L. A review of the phytochemistry and pharmacological activities of Ephedra herb. Chin. J. Nat. Med. 2020, 18, 321–344. [Google Scholar] [CrossRef]
- Seif, M.; Deabes, M.; El-Askary, A.; El-Kott, A.F.; Albadrani, G.M.; Seif, A.; Wang, Z. Ephedra sinica mitigates hepatic oxidative stress and inflammation via suppressing the TLR4/MyD88/NF-κB pathway in fipronil-treated rats. Environ. Sci. Pollut. Res. 2021, 28, 62943–62958. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Khalifeh, H.; Saad, F.; Serale, N.; Salis, A.; Damonte, G.; Lupidi, G.; Daher, A.; Vergani, L. Protective effects of extracts from Ephedra foeminea Forssk fruits against oxidative injury in human endothelial cells. J. Ethnopharmacol. 2020, 260, 112976. [Google Scholar] [CrossRef]
- Palamar, J. How ephedrine escaped regulation in the United States: A historical review of misuse and associated policy. Health Policy 2011, 99, 1–9. [Google Scholar] [CrossRef]
- Kim, B.-S.; Song, M.-Y.; Kim, H. The anti-obesity effect of Ephedra sinica through modulation of gut microbiota in obese Korean women. J. Ethnopharmacol. 2014, 152, 532–539. [Google Scholar] [CrossRef]
- Munafò, A.; Frara, S.; Perico, N.; Di Mauro, R.; Cortinovis, M.; Burgaletto, C.; Cantarella, G.; Remuzzi, G.; Giustina, A.; Bernardini, R. In search of an ideal drug for safer treatment of obesity: The false promise of pseudoephedrine. Rev. Endocr. Metab. Disord. 2021, 22, 1013–1025. [Google Scholar] [CrossRef]
- Duan, S.; Xie, L.; Zheng, L.; Huang, J.; Guo, R.; Sun, Z.; Xie, Y.; Lv, J.; Lin, Z.; Ma, S. Long-term exposure to ephedrine leads to neurotoxicity and neurobehavioral disorders accompanied by up-regulation of CRF in prefrontal cortex and hippocampus in rhesus macaques. Behav. Brain Res. 2020, 393, 112796. [Google Scholar] [CrossRef]
- Takemoto, H.; Takahashi, J.; Hyuga, S.; Odaguchi, H.; Uchiyama, N.; Maruyama, T.; Yamashita, T.; Hyuga, M.; Oshima, N.; Amakura, Y.; et al. Ephedrine Alkaloids-Free Ephedra Herb Extract, EFE, Has No Adverse Effects Such as Excitation, Insomnia, and Arrhythmias. Biol. Pharm. Bull. 2018, 41, 247–253. [Google Scholar] [CrossRef] [Green Version]
- Zell-Kanter, M.; Quigley, M.A.; Leikin, J.B. Reduction in Ephedra Poisonings after FDA Ban. N. Engl. J. Med. 2015, 372, 2172–2174. [Google Scholar] [CrossRef]
- Food and Drug Administration. Final Rule Declaring Dietary Supplements Containing Ephedrine Alkaloids Adulterated Because They Present an Unreasonable Risk; Final Rule. Fed. Regist. 2004, 69, 6788–6854. Available online: https://www.govinfo.gov/content/pkg/FR-2004-02-11/pdf/04-2912.pdf (accessed on 5 February 2022).
- Lai, S.; Yu, C.; Dennehy, C.E.; Tsourounis, C.; Lee, K.P. Online Marketing of Ephedra Weight Loss Supplements: Labeling and Marketing Compliance with the U.S. Food and Drug Administration Ban on Ephedra. J. Altern. Complement. Med. 2021, 27, 796–802. [Google Scholar] [CrossRef]
- Głowacka, K.; Wiela-Hojeńska, A. Pseudoephedrine—Benefits and Risks. Int. J. Mol. Sci. 2021, 22, 5146. [Google Scholar] [CrossRef]
- China National Narcotics Control Commission. Drug Situation in China (2019). Available online: http://www.nncc626.com/2020-06/25/c_1210675877.htm (accessed on 5 February 2022).
- Yu, W.-N.; Wang, L.-H.; Cheng, H.-W. Regulatory analysis on the medical use of ephedrine-related products in Taiwan. J. Food Drug Anal. 2018, 26, 481–486. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Wang, Z.; Xin, Z.; Ren, Z. Problems in Supervision of Ephedrine Compound Prescription Preparation. Chin. J. Pharmacovigil. 2010, 7, 673–675. [Google Scholar]
- Zhu, B.; Conlan, X.; Cao, J.; Meng, L.; Zheng, K.; Yang, D.; Yang, W. Case studies on illegal production of ephedrine/pseudoephedrine within Fujian China. Forensic Sci. Int. 2020, 312, 110326. [Google Scholar] [CrossRef]
- CBOL Plant Working Group; Hollingsworth, P. M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef]
- Lo, Y.-T.; Li, M.; Shaw, P.-C. Identification of constituent herbs in ginseng decoctions by DNA markers. Chin. Med. 2015, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Meusnier, I.; Singer, G.A.; Landry, J.-F.; Hickey, D.A.; Hebert, P.D.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 214. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, X.; Wang, L.; Chen, X.; Pang, X.; Han, J. A Nucleotide Signature for the Identification of American Ginseng and Its Products. Front. Plant Sci. 2016, 7, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, Y.; Wang, L.; Han, J.; Chen, S. A Nucleotide Signature for the Identification of Angelicae Sinensis Radix (Danggui) and Its Products. Sci. Rep. 2016, 6, 34940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Jiang, W.; Yu, J.; Pang, X. Investigating the authenticity of Ophiopogonis Radix and its Chinese patent medicines by using a nucleotide signature. J. Ethnopharmacol. 2020, 261, 113134. [Google Scholar] [CrossRef] [PubMed]
- De Boer, H.J.; Ichim, M.C.; Newmaster, S.G. DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf. 2015, 38, 611–620. [Google Scholar] [CrossRef]
- Matthes, N.; Pietsch, K.; Rullmann, A.; Näumann, G.; Popping, B.; Szabo, K. The Barcoding Table of Animal Species (BaTAnS): A new tool to select appropriate methods for animal species identification using DNA barcoding. Mol. Biol. Rep. 2020, 47, 6457–6461. [Google Scholar] [CrossRef]
- Tnah, L.; Lee, S.; Tan, A.; Lee, C.T.; Ng, K.K.S.; Ng, C.; Farhanah, Z.N. DNA barcode database of common herbal plants in the tropics: A resource for herbal product authentication. Food Control 2018, 95, 318–326. [Google Scholar] [CrossRef]
- Han, J.; Pang, X.; Liao, B.; Yao, H.; Song, J.; Chen, S. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 2016, 6, 18723. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, X.; Wang, P.; Huang, B.; Sun, W.; Xiong, C.; Hu, Z.; Chen, S. Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding. Molecules 2018, 23, 967. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Dong, G.-Q.; Zhang, Y.-Q.; Liu, X.; Sun, W. Identification of processed Chinese medicinal materials using DNA mini-barcoding. Chin. J. Nat. Med. 2017, 15, 481–486. [Google Scholar] [CrossRef]
- Ibragic, S.; Sofic, E. Chemical composition of various Ephedra species. Bosn. J. Basic Med. Sci. 2015, 15, 21. [Google Scholar] [CrossRef] [Green Version]
- Yun, N.; Kim, H.J.; Park, S.C.; Park, G.; Kim, M.K.; Choi, Y.H.; Jang, Y.P. Localization of Major Ephedra Alkaloids in Whole Aerial Parts of Ephedrae Herba Using Direct Analysis in Real Time-Time of Flight-Mass Spectrometry. Molecules 2021, 26, 580. [Google Scholar] [CrossRef]
- Pawar, R.S.; Grundel, E. Overview of regulation of dietary supplements in the USA and issues of adulteration with phenethylamines (PEAs). Drug Test. Anal. 2017, 9, 500–517. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific Opinion on safety evaluation of Ephedra species for use in food. EFSA J. 2013, 11, 3467. [Google Scholar]
- Gardner, S.F.; Franks, A.; Gurley, B.J.; Haller, C.A.; Singh, B.K.; Mehta, J.L. Effect of a multicomponent, ephedra-containing dietary supplement (Metabolife 356) on Holter monitoring and hemostatic parameters in healthy volunteers. Am. J. Cardiol. 2003, 91, 1510–1513. [Google Scholar] [CrossRef]
- Al-Tayyib, A.; Koester, S.; Langegger, S.; Raville, L. Heroin and Methamphetamine Injection: An Emerging Drug Use Pattern. Subst. Use Misuse 2017, 52, 1051–1058. [Google Scholar] [CrossRef] [Green Version]

| No. | Species | Voucher No. | Collection Site | GenBank Accession |
|---|---|---|---|---|
| 1 | E. equisetina | IMD0001954 | Hebei | OL456774 |
| 2 | E. equisetina | IMD0001953 | Shaanxi | OL456770 |
| 3 | E. equisetina | IMD0001955 | Shanxi | OL456771 |
| 4 | E. equisetina | IMD0001952 | Hebei | OL456772 |
| 5 | E. equisetina | IMD0001951 | Xinjiang | OL456773 |
| 6 | E. equisetina | IMD0001950 | Xinjiang | OL456775 |
| 7 | E. equisetina | IMD0001949 | Xinjiang | OL456776 |
| 8 | E. equisetina | IMD0001946 | Xinjiang | OL456777 |
| 9 | E. equisetina | IMD0001964 | Beijing | OL456763 |
| 10 | E. equisetina | IMD0001963 | Beijing | OL456764 |
| 11 | E. equisetina | IMD0001966 | Xinjiang | OL456765 |
| 12 | E. equisetina | IMD0001961 | Hebei | OL456766 |
| 13 | E. equisetina | IMD0001958 | Jiangsu | OL456767 |
| 14 | E. equisetina | IMD0001957 | Beijing | OL456768 |
| 15 | E. equisetina | IMD0001956 | Beijing | OL456769 |
| 16 | E. fedtschenkoae | IMD0001967 | Qinghai | OL456778 |
| 17 | E. gerardiana | IMD0001970 | Xizang | OL456779 |
| 18 | E. gerardiana | IMD0001969 | Xizang | OL456780 |
| 19 | E. gerardiana | IMD0001968 | Xizang | OL456781 |
| 20 | E. gerardiana | IMD0001971 | Xizang | OL456782 |
| 21 | E. gerardiana | IMD0001972 | Xizang | OL456783 |
| 22 | E. intermedia | IMD0001980 | Qinghai | OL456784 |
| 23 | E. intermedia | IMD0001978 | Qinghai | OL456785 |
| 24 | E. intermedia | IMD0001976 | Xinjiang | OL456786 |
| 25 | E. intermedia | IMD0001975 | Qinghai | OL456787 |
| 26 | E. intermedia | IMD0001974 | Xinjiang | OL456788 |
| 27 | E. intermedia | IMD0001995 | Sichuan | OL456789 |
| 28 | E. intermedia | IMD0001994 | Neimenggu | OL456790 |
| 29 | E. intermedia | IMD0001992 | Xinjiang | OL456791 |
| 30 | E. intermedia | IMD0001991 | Xinjiang | OL456792 |
| 31 | E. intermedia | IMD0001989 | Xinjiang | OL456793 |
| 32 | E. intermedia | IMD0001988 | Hebei | OL456794 |
| 33 | E. intermedia | IMD0001996 | Neimenggu | OL456795 |
| 34 | E. intermedia | IMD0001986 | Xinjiang | OL456796 |
| 35 | E. intermedia | IMD0001984 | Gansu | OL456797 |
| 36 | E. intermedia | IMD0001983 | Xinjiang | OL456798 |
| 37 | E. intermedia | IMD0001981 | Gansu | OL456799 |
| 38 | E. intermedia | IMD0001985 | Sichuan | OL456800 |
| 39 | E. intermedia | IMD0001987 | Xinjiang | OL456801 |
| 40 | E. intermedia var. tibetica | IMD0001982 | Xizang | OL456802 |
| 41 | E. intermedia var. tibetica | IMD0002003 | Xizang | OL456803 |
| 42 | E. intermedia var. tibetica | IMD0002002 | Xizang | OL456804 |
| 43 | E. intermedia var. tibetica | IMD0002001 | Xizang | OL456805 |
| 44 | E. intermedia var. tibetica | IMD0002000 | Xizang | OL456806 |
| 45 | E. intermedia var. tibetica | IMD0001999 | Xizang | OL456807 |
| 46 | E. intermedia var. tibetica | IMD0001998 | Xizang | OL456808 |
| 47 | E. intermedia var. tibetica | IMD0001997 | Xizang | OL456809 |
| 48 | E. intermedia var. tibetica | IMD0002016 | Xizang | OL456810 |
| 49 | E. intermedia var. tibetica | IMD0002014 | Xizang | OL456811 |
| 50 | E. intermedia var. tibetica | IMD0002013 | Xizang | OL456812 |
| 51 | E. intermedia var. tibetica | IMD0002012 | Xizang | OL456813 |
| 52 | E. intermedia var. tibetica | IMD0002011 | Xizang | OL456814 |
| 53 | E. intermedia var. tibetica | IMD0002010 | Xizang | OL456815 |
| 54 | E. intermedia var. tibetica | IMD0002009 | Xizang | OL456816 |
| 55 | E. intermedia var. tibetica | IMD0002007 | Xizang | OL456817 |
| 56 | E. intermedia var. tibetica | IMD0002006 | Xizang | OL456818 |
| 57 | E. intermedia var. tibetica | IMD0002005 | Xizang | OL456819 |
| 58 | E. intermedia var. tibetica | IMD0002008 | Xizang | OL456820 |
| 59 | E. intermedia var. tibetica | IMD0002021 | Xizang | OL456821 |
| 60 | E. intermedia var. tibetica | IMD0002017 | Xizang | OL456822 |
| 61 | E. intermedia var. tibetica | IMD0002019 | Xizang | OL456823 |
| 62 | E. intermedia var. tibetica | IMD0002020 | Xizang | OL456824 |
| 63 | E. intermedia var. tibetica | IMD0002018 | Xizang | OL456825 |
| 64 | E. intermedia var. tibetica | IMD0002024 | Xizang | OL456826 |
| 65 | E. intermedia var. tibetica | IMD0002023 | Xizang | OL456827 |
| 66 | E. intermedia var. tibetica | IMD0002022 | Xizang | OL456828 |
| 67 | E. likiangensis | IMD0002027 | Yunnan | OL456829 |
| 68 | E. likiangensis | IMD0002026 | Sichuan | OL456830 |
| 69 | E. likiangensis | IMD0002025 | Yunnan | OL456831 |
| 70 | E. minuta | IMD0002029 | Xizang | OL456832 |
| 71 | E. minuta | IMD0002030 | Xizang | OL456833 |
| 72 | E. minuta | IMD0002028 | Sichuan | OL456834 |
| 73 | E. monosperma | IMD0002037 | Qinghai | OL456835 |
| 74 | E. monosperma | IMD0002043 | Xizang | OL456836 |
| 75 | E. monosperma | IMD0002044 | Xizang | OL456837 |
| 76 | E. przewalskii | IMD0002060 | Gansu | OL456841 |
| 77 | E. przewalskii | IMD0002059 | Qinghai | OL456842 |
| 78 | E. przewalskii | IMD0002057 | Gansu | OL456843 |
| 79 | E. przewalskii | IMD0002056 | Xinjiang | OL456844 |
| 80 | E. przewalskii | IMD0002055 | Qinghai | OL456845 |
| 81 | E. przewalskii | IMD0002063 | Gansu | OL456846 |
| 82 | E. przewalskii | IMD0002062 | Gansu | OL456838 |
| 83 | E. przewalskii | IMD0002061 | Gansu | OL456839 |
| 84 | E. przewalskii | IMD0002051 | Xinjiang | OL456840 |
| 85 | E. regeliana | IMD0002066 | Qinghai | OL456850 |
| 86 | E. regeliana | IMD0002064 | Xinjiang | OL456847 |
| 87 | E. regeliana | IMD0002067 | Qinghai | OL456848 |
| 88 | E. regeliana | IMD0002070 | Xinjiang | OL456849 |
| 89 | E. saxatilis | IMD0002071 | Xizang | OL456851 |
| 90 | E. saxatilis | IMD0002078 | Xizang | OL456852 |
| 91 | E. saxatilis | IMD0002076 | Xizang | OL456853 |
| 92 | E. saxatilis | IMD0002075 | Xizang | OL456854 |
| 93 | E. saxatilis | IMD0002077 | Sichuan | OL456855 |
| 94 | E. saxatilis | IMD0002081 | Xizang | OL456856 |
| 95 | E. saxatilis | IMD0002080 | Xizang | OL456857 |
| 96 | E. saxatilis | IMD0002079 | Xizang | OL456858 |
| 97 | E. saxatilis | IMD0002090 | Xizang | OL456859 |
| 98 | E. saxatilis | IMD0002089 | Xizang | OL456860 |
| 99 | E. saxatilis | IMD0002088 | Xizang | OL456861 |
| 100 | E. saxatilis | IMD0002087 | Xizang | OL456862 |
| 101 | E. sinica | IMD0002098 | Hebei | OL456887 |
| 102 | E. sinica | IMD0002099 | Hebei | OL456863 |
| 103 | E. sinica | IMD0002097 | Hebei | OL456864 |
| 104 | E. sinica | IMD0002095 | Shanxi | OL456865 |
| 105 | E. sinica | IMD0002106 | Hebei | OL456866 |
| 106 | E. sinica | IMD0002105 | Neimenggu | OL456867 |
| 107 | E. sinica | IMD0002104 | Hebei | OL456868 |
| 108 | E. sinica | IMD0002103 | Hebei | OL456869 |
| 109 | E. sinica | IMD0002101 | Shanxi | OL456870 |
| 110 | E. sinica | IMD0002109 | Neimenggu | OL456871 |
| 111 | E. sinica | IMD0002108 | Hebei | OL456872 |
| 112 | E. sinica | IMD0002118 | Neimenggu | OL456873 |
| 113 | E. sinica | IMD0002117 | Neimenggu | OL456874 |
| 114 | E. sinica | IMD0002115 | Neimenggu | OL456875 |
| 115 | E. sinica | IMD0002112 | Neimenggu | OL456876 |
| 116 | E. sinica | IMD0002111 | Jilin | OL456877 |
| 117 | E. sinica | IMD0002125 | Neimenggu | OL456878 |
| 118 | E. sinica | IMD0002122 | Neimenggu | OL456879 |
| 119 | E. sinica | IMD0002123 | Neimenggu | OL456880 |
| 120 | E. sinica | IMD0002121 | Neimenggu | OL456881 |
| 121 | E. sinica | IMD0002119 | Hebei | OL456882 |
| 122 | E. sinica | IMD0002136 | Neimenggu | OL456883 |
| 123 | E. sinica | IMD0002135 | Neimenggu | OL456884 |
| 124 | E. sinica | IMD0002134 | Neimenggu | OL456885 |
| 125 | E. sinica | IMD0002132 | Neimenggu | OL456886 |
| No. | Sample Name | Type | Collection Site | Number of Ingredients | Ingredients on Label |
|---|---|---|---|---|---|
| ZCY-01 | Mahuang Fuzi Xixin Soup | Concentrated pills | Guangdong | 3 | Ephedrae Herba, Aconiti Lateralis Radix Praeparata, Asari Radix Et Rhizoma |
| ZCY-02 | Maxing Zhike Tablets | Tablets | Jilin | 4 | Ephedrae Herba, Armeniacae Semen Amarum, Gypsum Fibrosum, Glycyrrhizae Radix Et Rhizoma Praeparata Cum Melle |
| ZCY-03 | Mahuang Zhisou Pills | Watered pills | Shandong | 7 | Citri Exocarpium Rubrum, Ephedrae Herba, Platycodonis Radix, Fritillariae Cirrhosae Bulbus, Schisandrae Chinensis Fructus, Poria, Asari Radix Et Rhizoma |
| ZCY-04 | Tongxuan Lifei Pills | Honeyed pills | Beijing | 11 | Perillae Folium, Peucedani Radix, Platycodonis Radix, Armeniacae Semen Amarum, Ephedrae Herba, Glycyrrhizae Radix Et Rhizoma, Citri Reticulatae Pericarpium, Pinelliae Rhizoma, Poria, Aurantii Fructus, Scutellariae Radix |
| ZCY-05 | Xiaoer Feire Kechuan Granules | Granules | Heilongjiang | 11 | Ephedrae Herba, Armeniacae Semen Amarum, Gypsum Fibrosum, Glycyrrhizae Radix Et Rhizoma, Lonicerae Japonicae Flos, Forsythiae Fructus, Anemarrhenae Rhizoma, Scutellariae Radix, Isatidis Radix, Ophiopogonis Radix, Houttuyniae Herba |
| ZCY-06 | Zhengtian Pills | Watered pills | Guangdong | 15 | Uncariae Ramulus Cum Uncis, Paeoniae Radix Alba, Chuanxiong Rhizoma, Angelicae Sinensis Radix, Rehmanniae Radix, Angelicae Dahuricae Radix, Saposhnikoviae Radix, Notopterygii Rhizoma Et Radix, Persicae Semen, Carthami Flos, Asari Radix Et Rhizoma, Angelicae Pubescentis Radix, Ephedrae Herba, Aconiti Lateralis Radix Praeparata, Spatholobi Caulis |
| ZCY-07 | Qiguanyan Pills | Watered pills | Beijing | 31 | Ephedrae Herba, Armeniacae Semen Amarum, Gypsum Fibrosum, Glycyrrhizae Radix Et Rhizoma, Peucedani Radix, Cynanchi Stauntonii Rhizoma Et Radix, Stemonae Radix, Asteris Radix Et Rhizoma, Farfarae Flos, Meretricis Concha Cyclinae Concha, Descurainiae Semen Lepidii Semen, Citri Grandis Exocarpium, Platycodonis Radix, Poria, Pinelliae Rhizoma, Polygalae Radix, Inulae Flos, Pumice, Perillae Fructus, Codonopsis Radix, Jujubae Fructus, Schisandrae Chinensis Fructus, Cinnamomi Ramulus, Allii Macrostemonis Bulbus, Paeoniae Radix Alba, Mori Folium, Belamcandae Rhizoma, Scutellariae Radix, Indigo Naturalis, Taraxaci Herba, Eriobotryae Folium |
| No. | Sequences (5′-3′) | Length/bp | Intergeneric Specificity | Intragenus Conservation |
|---|---|---|---|---|
| S1 | TCGGGGGGACGGCCTTGACCGTCCGGTCCGCCTCGGCGGTGCGGTCGGTTGAAAT | 55 | √ | × |
| S2 | GGGGGACGGCCTTGACCGTCCGGTCCGCCTCGGCGGTGCGGTCGG | 45 | √ | × |
| S3 | GCCTTGACCGTCCGGTCCGCCTCGGCGGTGCGGTC | 35 | √ | × |
| S4 | GACCGTCCGGTCCGCCTCGGCGGTGCGGTC | 30 | √ | × |
| S5 | CCGTCCGGTCCGCCTCGGCGGTGCGGT | 27 | √ | × |
| S6 | GTCCGGTCCGCCTCGGCGGTGCG | 23 | √ | √ |
| S7 | CCGGTCCGCCTCGGCGGTGC | 20 | × | √ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Bai, X.; Chen, X.; Ren, Y.; Han, J. Development of a Genus-Universal Nucleotide Signature for the Identification and Supervision of Ephedra-Containing Products. Molecules 2022, 27, 2342. https://doi.org/10.3390/molecules27072342
Wang G, Bai X, Chen X, Ren Y, Han J. Development of a Genus-Universal Nucleotide Signature for the Identification and Supervision of Ephedra-Containing Products. Molecules. 2022; 27(7):2342. https://doi.org/10.3390/molecules27072342
Chicago/Turabian StyleWang, Gang, Xuanjiao Bai, Xiaochen Chen, Ying Ren, and Jianping Han. 2022. "Development of a Genus-Universal Nucleotide Signature for the Identification and Supervision of Ephedra-Containing Products" Molecules 27, no. 7: 2342. https://doi.org/10.3390/molecules27072342
APA StyleWang, G., Bai, X., Chen, X., Ren, Y., & Han, J. (2022). Development of a Genus-Universal Nucleotide Signature for the Identification and Supervision of Ephedra-Containing Products. Molecules, 27(7), 2342. https://doi.org/10.3390/molecules27072342






