Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain
Abstract
:1. Introduction
2. Results and Discussion
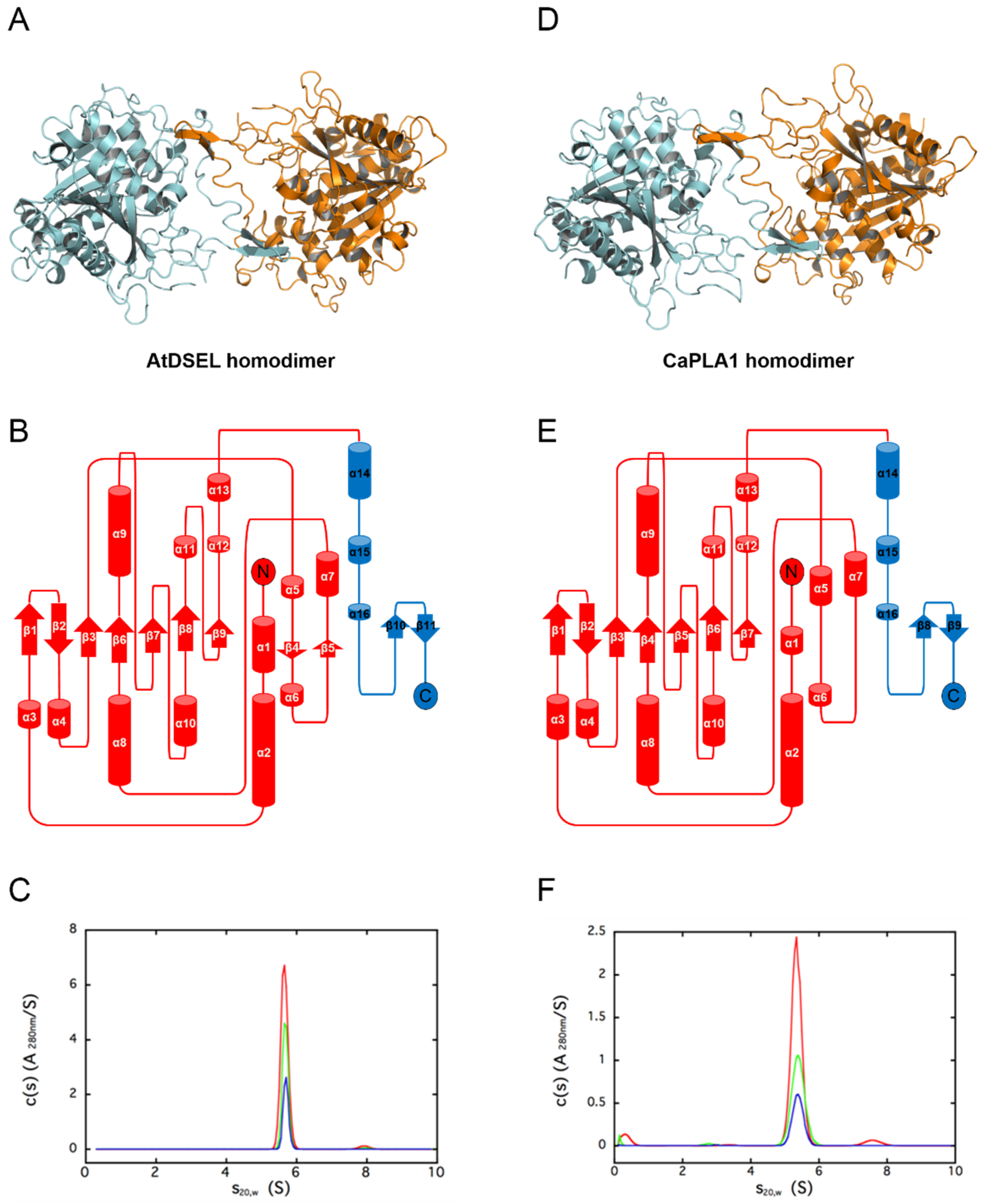
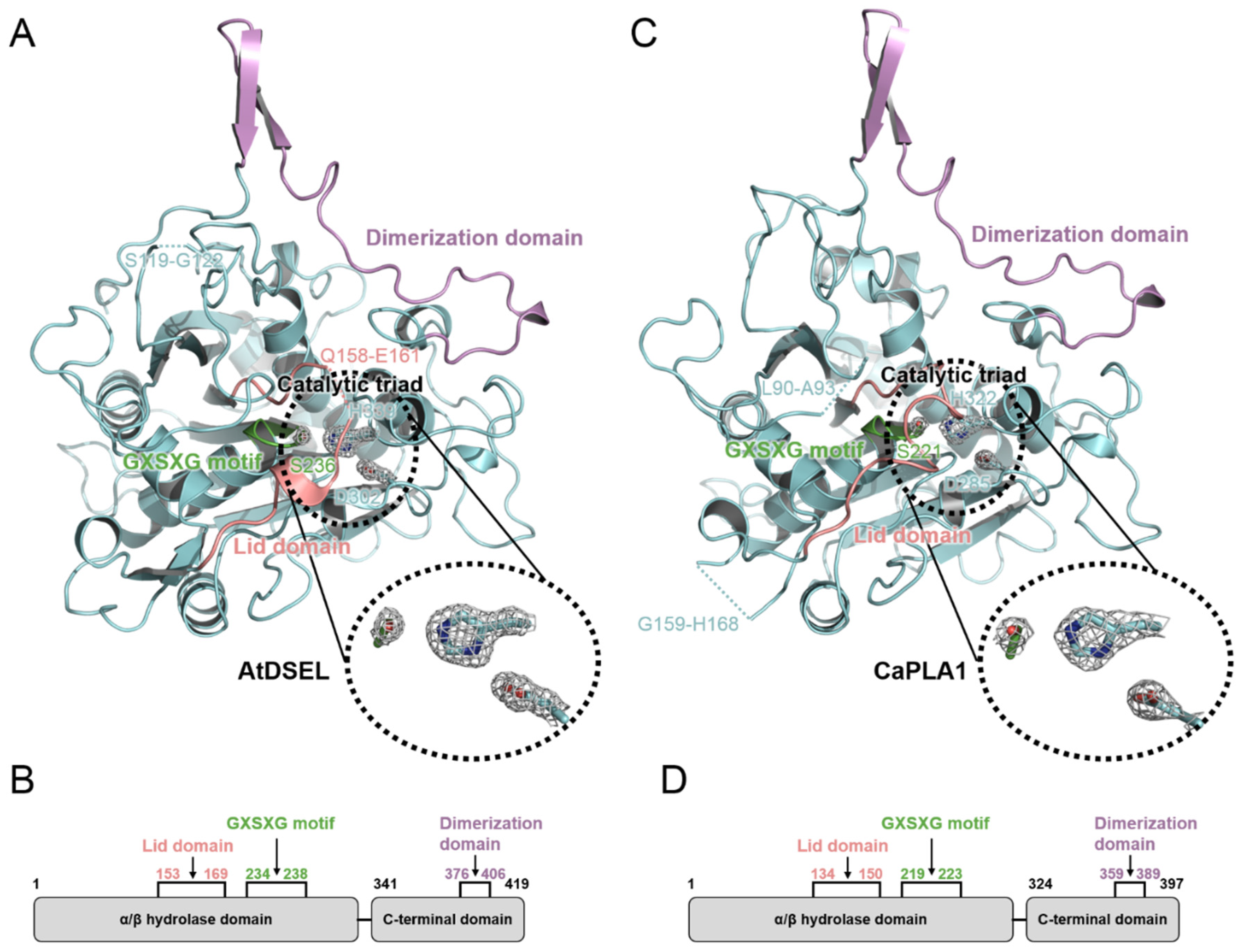
2.1. Structural Features of AtDSEL and CaPLA1
2.2. Conserved Motifs and Domains of AtDSEL and CaPLA1
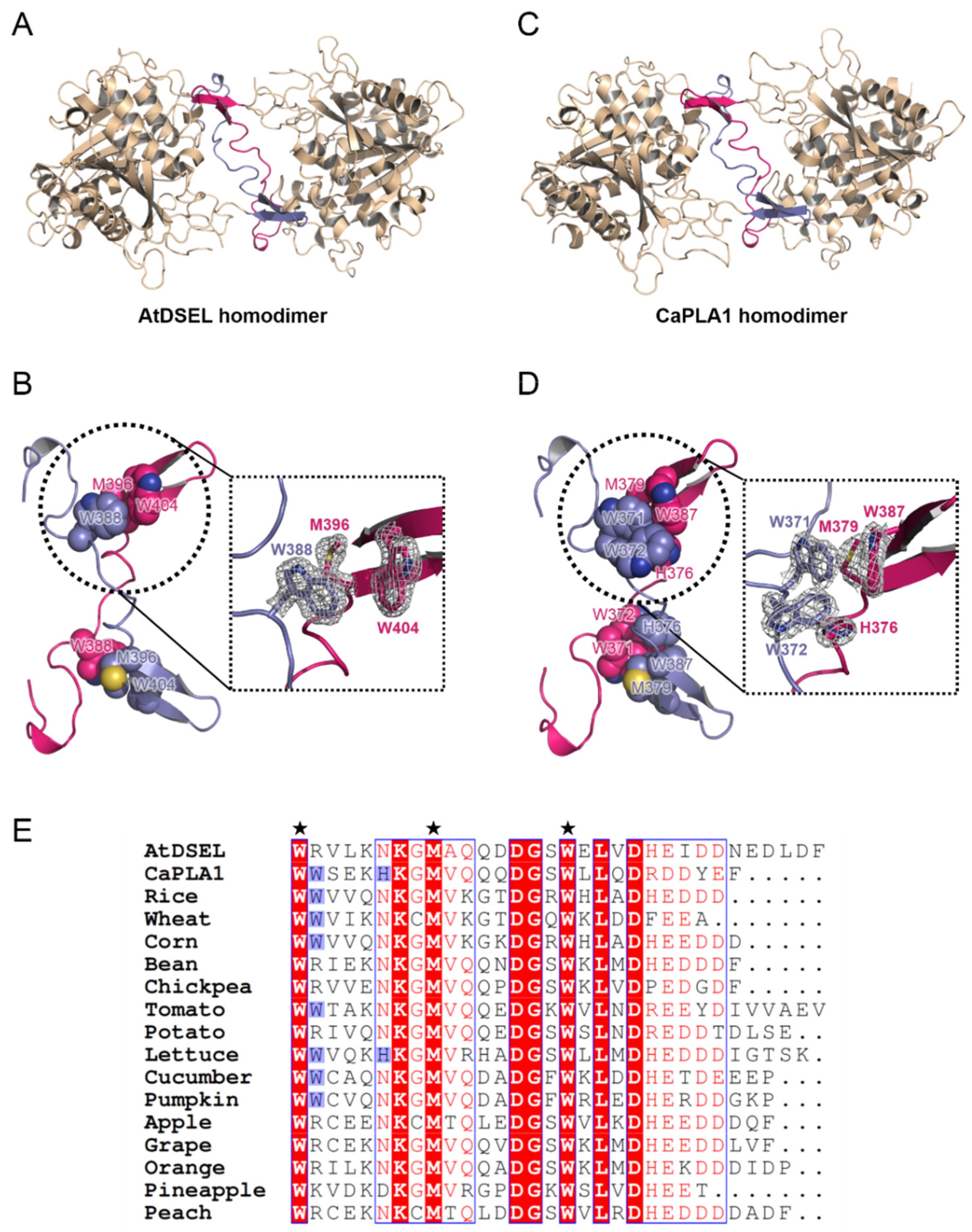
2.3. Interactions between Homodimers of AtDSEL and CaPLA1
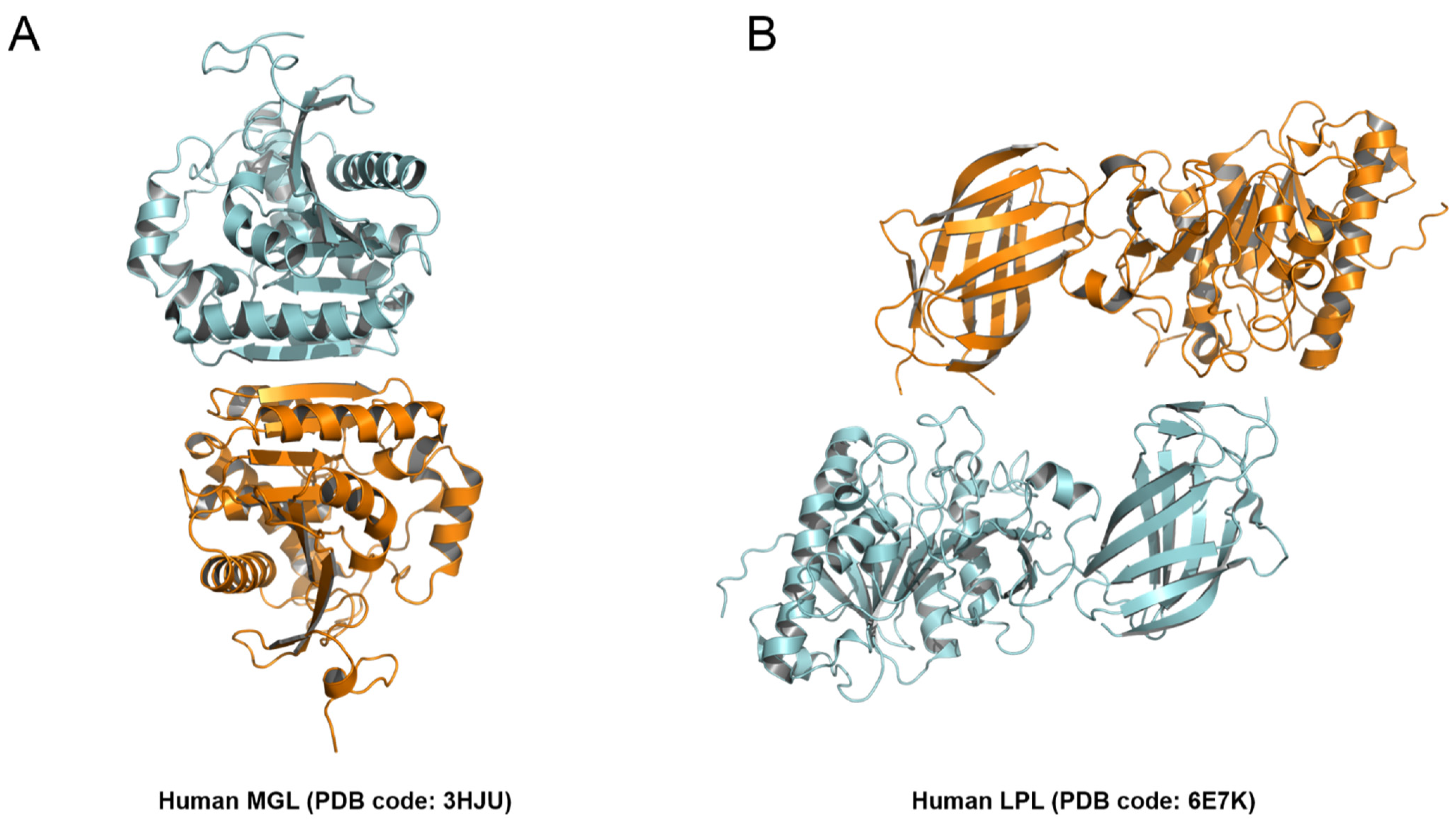
2.4. Structural Comparison among PLAs
3. Conclusions
4. Materials and Methods
4.1. Cloning, Expression, and Purification
4.2. Crystallization, Data Collection, and Structure Determination
4.3. Analytical Ultracentrifugation (AUC) Experiment
4.4. In Vitro Lipase Assay
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Richmond, G.S.; Smith, T.K. Phospholipases A1. Int. J. Mol. Sci. 2011, 12, 588–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filkin, S.Y.; Lipkin, A.V.; Fedorov, A.N. Phospholipase Superfamily: Structure, Functions, and Biotechnological Applications. Biochemistry 2020, 85, S177–S195. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.; Inoue, A.; Makide, K.; Saiki, N.; Arai, H. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie 2007, 89, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.-H.; Chuang, C.-Y.; Ko, T.-P.; Hu, N.-J.; Chou, C.-C.; Shih, Y.-P.; Ho, C.-L.; Wang, A.H.-J. Crystal structure of vespid phospholipase A(1) reveals insights into the mechanism for cause of membrane dysfunction. Insect Biochem. Mol. Biol. 2016, 68, 79–88. [Google Scholar] [CrossRef]
- Skjold-Jørgensen, J.; Vind, J.; Moroz, O.V.; Blagova, E.; Bhatia, V.K.; Swendsen, A.; Wilson, K.S.; Bjerrum, M.J. Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 20–27. [Google Scholar] [CrossRef]
- Zhang, M.; Yu, X.-W.; Xu, Y.; Guo, R.-T.; Swapna, G.V.T.; Szyperski, T.; Hunt, J.F.; Montelione, G.T. Structural Basis by Which the N-Terminal Polypeptide Segment of Rhizopus chinensis Lipase Regulates Its Substrate Binding Affinity. Biochemistry 2019, 58, 3943–3954. [Google Scholar] [CrossRef]
- Birrane, G.; Beigneux, A.P.; Dwyer, B.; Strack-Logue, B.; Kristensen, K.K.; Francone, O.L.; Fong, L.G.; Mertens, H.D.T.; Pan, C.Q.; Ploug, M.; et al. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc. Natl. Acad. Sci. USA 2019, 116, 1723–1732. [Google Scholar] [CrossRef] [Green Version]
- Arora, R.; Nimonkar, A.V.; Baird, D.; Wang, C.; Chiu, C.-H.; Horton, P.A.; Hanrahan, S.; Cubbon, R.; Weldon, S.; Tschantz, W.R.; et al. Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. USA 2019, 116, 10360–10365. [Google Scholar] [CrossRef] [Green Version]
- Tilbeurgh, H.v.; Sarda, L.; Verger, R.; Cambillau, C. Structure of the pancreatic lipase-procolipase complex. Nature 1992, 359, 159–162. [Google Scholar] [CrossRef]
- Labar, G.; Bauvois, C.; Borel, F.; Ferror, J.-L.; Wouters, J.; Lambert, D.M. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem 2010, 11, 218–227. [Google Scholar] [CrossRef]
- Bian, C.; Yuan, C.; Chen, L.; Meehan, E.J.; Jiang, L.; Huang, Z.; Lin, L.; Huang, M. Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD. Proteins 2010, 78, 1601–1605. [Google Scholar] [PubMed]
- Noble, M.E.; Cleasby, A.; Johnson, L.N.; Egmond, M.R.; Frenken, L.G. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 1993, 331, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Derewenda, U.; Swenson, L.; Wei, Y.; Green, R.; Kobos, P.M.; Joerger, R.; Haas, M.J.; Derewenda, Z.S. Conformational lability of lipases observed in the absence of an oil-water interface: Crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J. Lipid Res. 1994, 35, 524–534. [Google Scholar] [CrossRef]
- Yapoudjian, S.; Ivanova, M.G.; Brzozowski, A.M.; Patkar, S.A.; Vind, J.; Svendsen, A.; Verger, R. Binding of Thermomyces (Humicola) lanuginosa lipase to the mixed micelles of cis-parinaric acid/NaTDC. Eur. J. Biochem. 2002, 269, 1613–1621. [Google Scholar] [CrossRef]
- Kohno, M.; Funatsu, J.; Mikami, B.; Kugimiya, W.; Matsuo, T.; Morita, Y. The crystal structure of lipase II from Rhizopus niveus at 2.2 A resolution. J. Biochem. 1996, 120, 505–510. [Google Scholar] [CrossRef]
- Wang, G.; Ryu, S.; Wang, X. Plant phospholipases: An overview. Methods Mol. Biol. 2012, 861, 123–137. [Google Scholar]
- Wang, X. Plant Phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 211–231. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The defective in anther dehiscience gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.Y.; Seo, Y.S.; Kim, W.T. AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. J. Plant Physiol. 2011, 168, 1705–1709. [Google Scholar] [CrossRef]
- Seo, Y.S.; Kim, E.Y.; Mang, H.G.; Kim, W.T. Heterologous expression, and biochemical and cellular characterization of CaPLA1 encoding a hot pepper phospholipase A1 homolog. Plant J. 2008, 53, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Derewenda, U. Relationships among serine hydrolases: Evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem. Cell Biol. 1991, 69, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Ploier, B.; Scharwey, M.; Koch, B.; Schmidt, C.; Schatte, J.; Rechberger, G.; Kollroser, M.; Hermetter, A.; Daum, G. Screening for hydrolytic enzymes reveals Ayr1p as a novel triacylglycerol lipase in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 36061–36072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, R.J.; Suh, M.C. The GxSxG motif of Arabidopsis monoacylglycerol lipase (MAGL6 and MAGL8) is essential for their enzyme activities. Appl. Biol. Chem. 2016, 59, 833–840. [Google Scholar] [CrossRef]
- Xu, T.; Liu, L.; Hou, S.; Xu, J.; Yang, B.; Wang, Y.; Liu, J. Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity. J. Struct. Biol. 2012, 178, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Lowe, M.E. The catalytic site residues and interfacial binding of human pancreatic lipase. J. Biol. Chem. 1992, 267, 17069–17073. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Struct. Biol. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Seo, Y.S.; Kim, E.Y.; Kim, J.H.; Kim, W.T. Enzymatic characterization of class I DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Lett. 2009, 583, 2301–2307. [Google Scholar] [CrossRef] [Green Version]

| Protein | AtDSEL | CaPLA1 | ||||
|---|---|---|---|---|---|---|
| PDB Code | 7X0C | 7X0D | ||||
| Data collection | ||||||
| Space group | P1211 | P1211 | ||||
| Cell dimensions | ||||||
| a, b, c (Å) | 73.977 | 95.343 | 79.278 | 102.313 | 59.568 | 147.319 |
| α, β, γ (°) | 90.000 | 105.144 | 90.000 | 90.000 | 90.139 | 90.000 |
| Resolution (Å) | 50–1.80 (1.86–1.80) | 50–2.40 (2.44–2.40) | ||||
| Rmerge | 0.086 (0.400) | 0.119 (0.589) | ||||
| I/σ (I) | 35.29 (4.70) | 13.97 (2.79) | ||||
| Completeness (%) | 99.8 (100.0) | 96.1 (95.5) | ||||
| Multiplicity | 7.5 (7.5) | 3.5 (3.0) | ||||
| Refinement | ||||||
| Resolution (Å) | 33.29–1.80 | 49.11–2.40 | ||||
| No. reflections | 98,085 | 67,351 | ||||
| Rwork/Rfree | 0.1783/0.2117 | 0.2065/0.2602 | ||||
| No. atoms | ||||||
| Protein | 6409 | 12,431 | ||||
| Water | 423 | 508 | ||||
| B-factors (Å2) | ||||||
| Protein | 24.68 | 26.50 | ||||
| Water | 28.83 | 28.53 | ||||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.012 | 0.002 | ||||
| Bond angles (°) | 1.127 | 0.545 | ||||
| Ramachandran plot (%) | ||||||
| Favored | 98.35 | 97.29 | ||||
| Allowed | 1.65 | 2.45 | ||||
| Disallowed | 0.00 | 0.26 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heo, Y.; Lee, I.; Moon, S.; Yun, J.-H.; Kim, E.Y.; Park, S.-Y.; Park, J.-H.; Kim, W.T.; Lee, W. Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain. Molecules 2022, 27, 2317. https://doi.org/10.3390/molecules27072317
Heo Y, Lee I, Moon S, Yun J-H, Kim EY, Park S-Y, Park J-H, Kim WT, Lee W. Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain. Molecules. 2022; 27(7):2317. https://doi.org/10.3390/molecules27072317
Chicago/Turabian StyleHeo, Yunseok, Inhwan Lee, Sunjin Moon, Ji-Hye Yun, Eun Yu Kim, Sam-Yong Park, Jae-Hyun Park, Woo Taek Kim, and Weontae Lee. 2022. "Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain" Molecules 27, no. 7: 2317. https://doi.org/10.3390/molecules27072317
APA StyleHeo, Y., Lee, I., Moon, S., Yun, J.-H., Kim, E. Y., Park, S.-Y., Park, J.-H., Kim, W. T., & Lee, W. (2022). Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain. Molecules, 27(7), 2317. https://doi.org/10.3390/molecules27072317







