Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability
Abstract
:1. Introduction
2. Results and Discussions
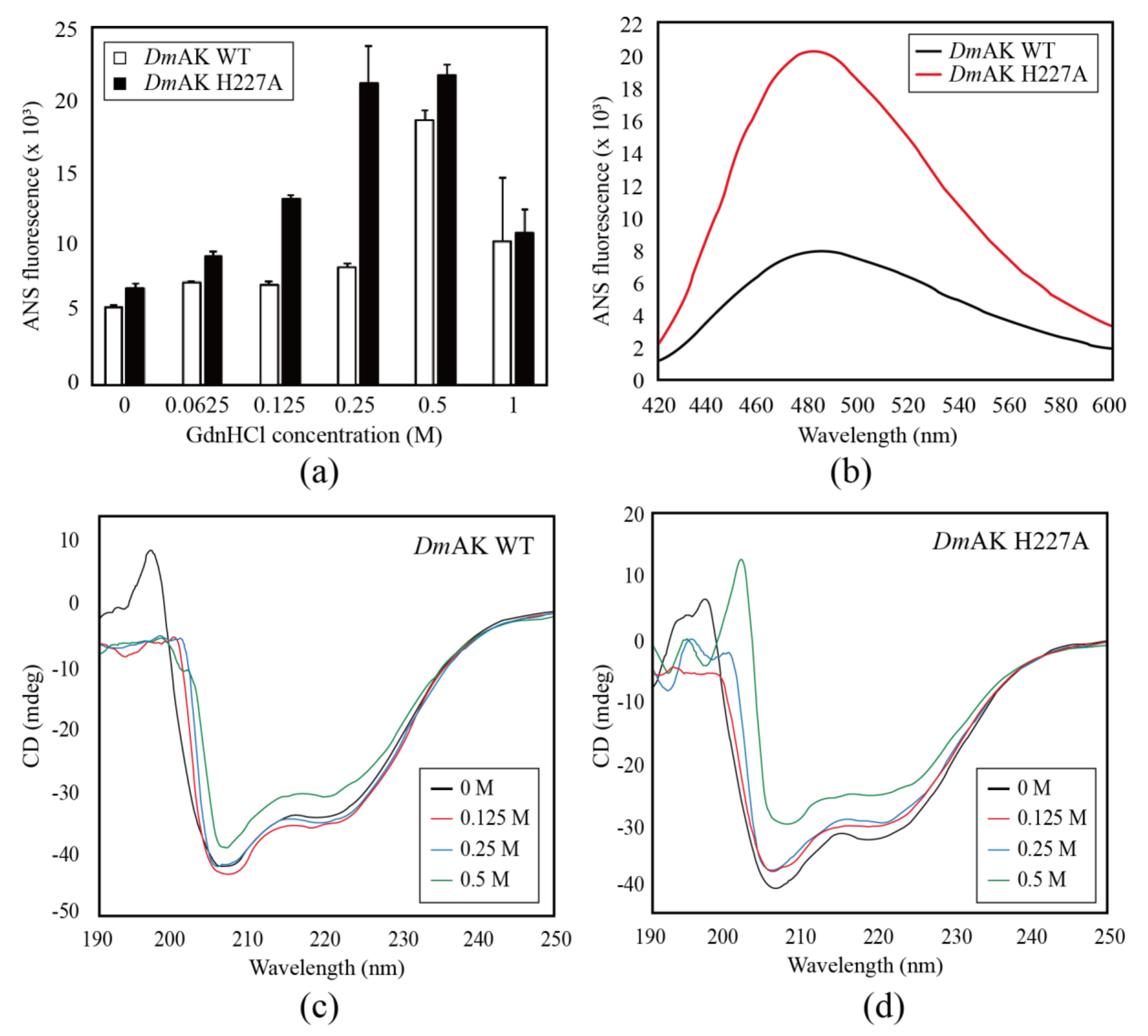
2.1. Assay of Enzyme Activity and Structural Stability
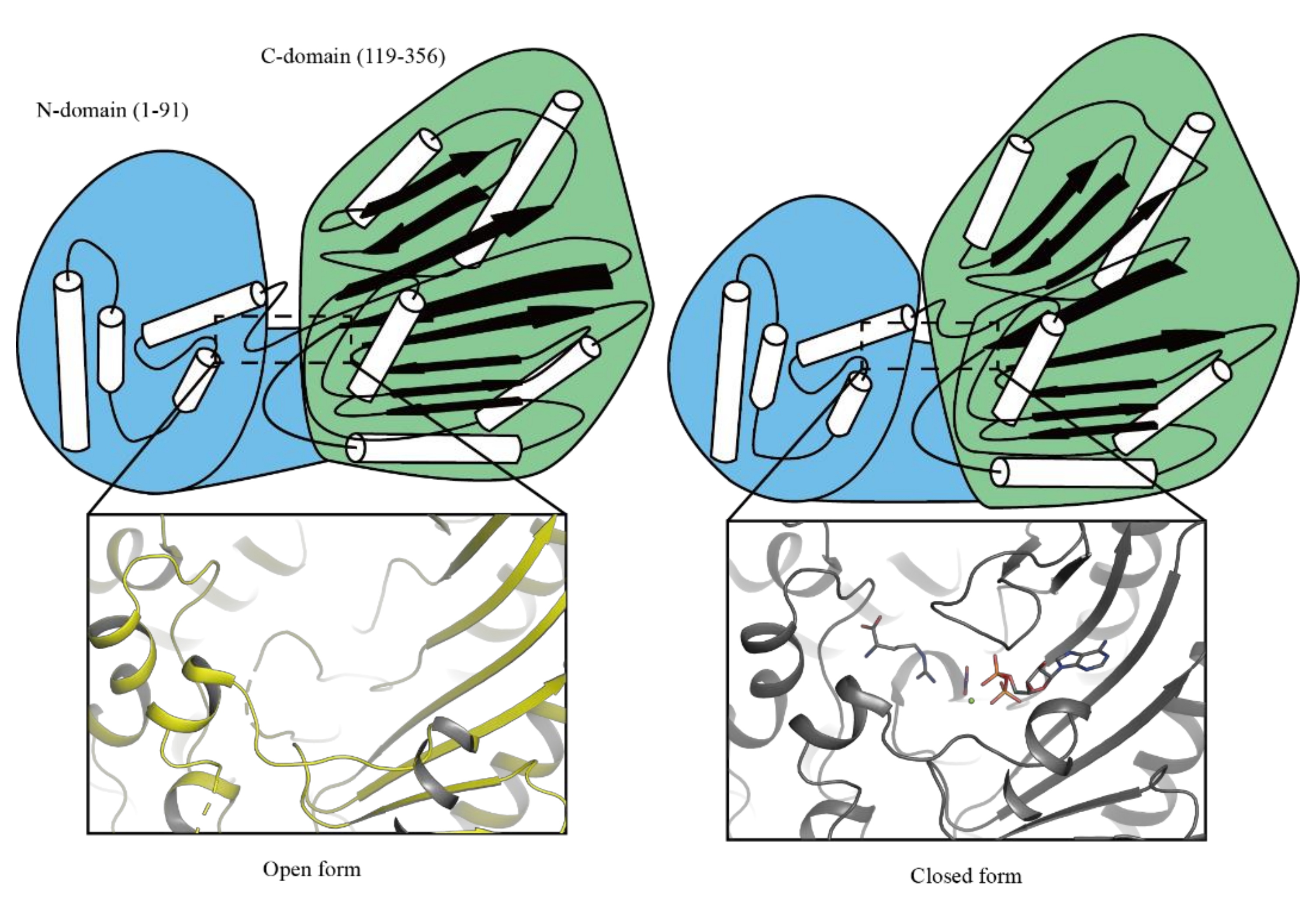
2.2. The Overall Structure of DmAK H227A
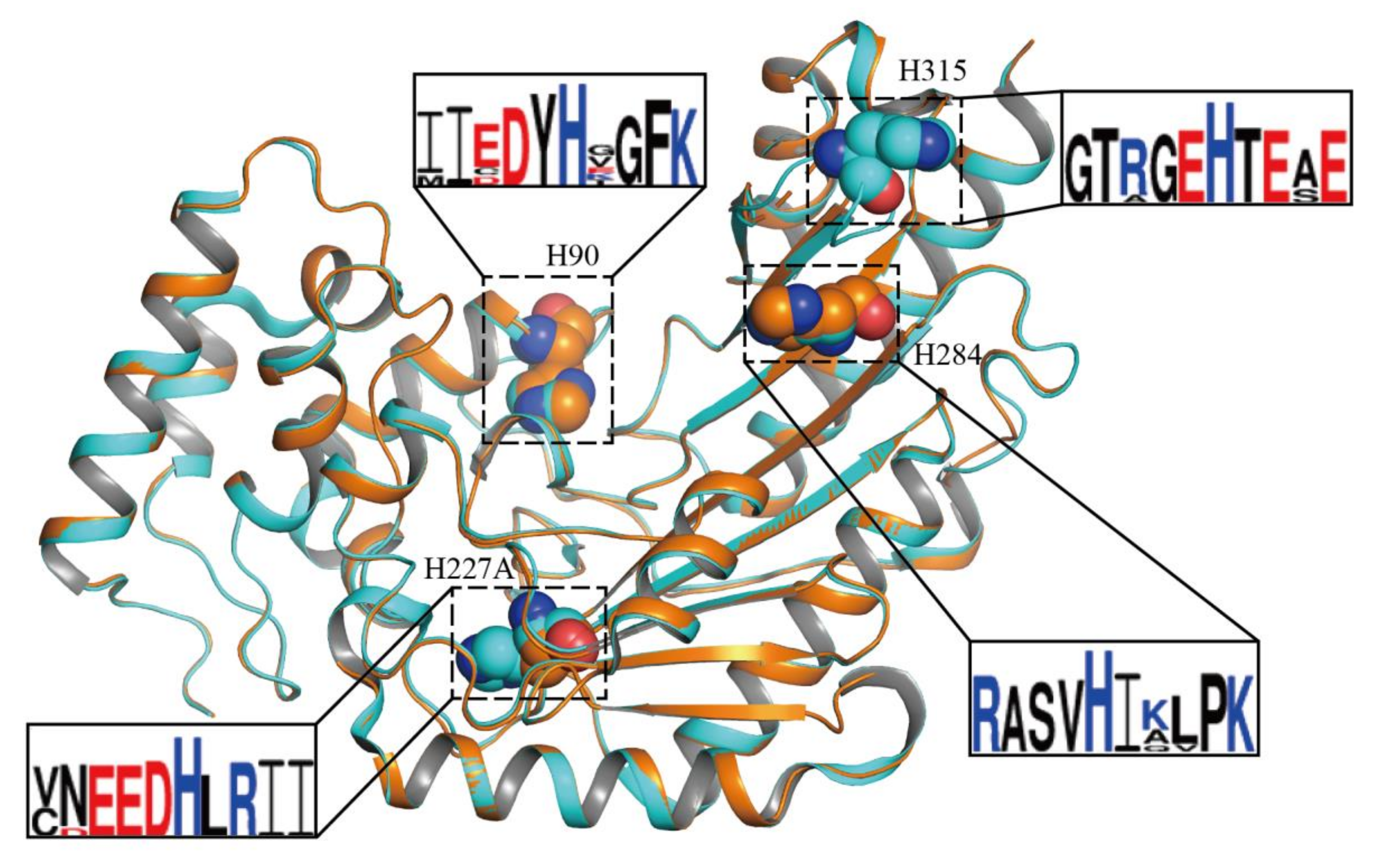
2.3. Structural Features of DmAK H227A
3. Materials and Methods
3.1. Cloning, Expression, and Purification of DmAK WT and Histidine Mutants
3.2. AK Activity Test
3.3. Spectroscopic Measurements
3.4. Crystallization and Structural Determination of DmAK H227A
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbrevations
| ANS | Anilino-1-naphthalenesulfonic acid |
| ATP | Adenosine triphosphate |
| CD | Circular Dichroism |
| DmAK | Daphnia magna Arginine kinase |
| ECL | Enhanced chemiluminescence |
| E. coli | Escherichia coli |
| F | Forward |
| GdnHCl | Guanidine hydrochloride |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TCA | trichloroacetic acid |
| PAL | Pohang accelerator laboratory |
| PCR | Polymerase chain reaction |
| PDR | phosphate determination reagent |
| PVDF | polyvinylidene fluoride |
| R | Reverse |
| WT | Wild type |
References
- Mergeay, J.; Declerck, S.; Verschuren, D.; Meester, L.D. Daphnia community analysis in shallow Kenyan lakes and ponds using dormant eggs in surface sediments. Freshw. Biol. 2006, 51, 399–411. [Google Scholar] [CrossRef]
- Smutna, M.; Babica, P.; Jarque, S.; Hilscherova, K.; Marsalek, B.; Haeba, M.; Blaha, L. Acute, chronic and reproductive toxicity of complex cyanobacterial blooms in Daphnia magna and the role of microcystins. Toxicon 2014, 79, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Lampert, W. Laboratory studies on zooplankton-cyanobacteria interactions. N. Z. J. Mar. Freshw. Res. 1987, 21, 483–490. [Google Scholar] [CrossRef]
- Sarnelle, O.; Wilson, A.E. Local adaptation of Daphnia pulicaria to toxic cyanobacteria. Limnol. Oceanogr. 2005, 50, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Chislock, M.F.; Sarnelle, O.; Olsen, B.K.; Doster, E.; Wilson, A.E. Large effects of consumer offense on ecosystem structure and function. Ecology 2013, 94, 2375–2380. [Google Scholar] [CrossRef] [PubMed]
- Lyu, K.; Zhang, L.; Zhu, X.; Cui, G.; Wilson, A.E.; Yang, Z. Arginine kinase in the cladoceran Daphnia magna: cDNA sequencing and expression is associated with resistance to toxic Microcystis. Aquat. Toxicol. 2015, 160, 13–21. [Google Scholar] [CrossRef]
- Alonso, G.D.; Pereira, C.A.; Remedi, M.S.; Paveto, M.C.; Cochella, L.; Ivaldi, M.S.; Gerez de Burgos, N.M.; Torres, H.N.; Flawia, M.M. Arginine kinase of the flagellate protozoa Trypanosoma cruzi. Regulation of its expression and catalytic activity. FEBS Lett. 2001, 498, 22–25. [Google Scholar] [CrossRef] [Green Version]
- Morris, S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [Google Scholar] [CrossRef]
- Kinsey, S.T.; Lee, B.C. The effects of rapid salinity change on in vivo arginine kinase flux in the juvenile blue crab, Callinectes sapidus. Comp. Biochem. Physiol. Part. B Biochem. Mol. Biol. 2003, 135, 521–531. [Google Scholar] [CrossRef]
- Miranda, M.R.; Canepa, G.E.; Bouvier, L.A.; Pereira, C.A. Trypanosoma cruzi: Oxidative stress induces arginine kinase expression. Exp. Parasitol. 2006, 114, 341–344. [Google Scholar] [CrossRef]
- Silvestre, F.; Dierick, J.F.; Dumont, V.; Dieu, M.; Raes, M.; Devos, P. Differential protein expression profiles in anterior gills of Eriocheir sinensis during acclimation to cadmium. Aquat. Toxicol. 2006, 76, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, P.; Chu, X.; Hao, L.; Guo, X.; Xu, B. Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones 2015, 20, 169–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Huang, X.; Liao, H.; Zhang, Z.; Sun, F.; Kou, S.; Bao, Z. Structure and functional analysis reveal an important regulated role of arginine kinase in Patinopecten yessoensis under low pH stress. Aquat. Toxicol. 2020, 222, 105452. [Google Scholar] [CrossRef]
- Niu, X.; Bruschweiler-Li, L.; Davulcu, O.; Skalicky, J.J.; Bruschweiler, R.; Chapman, M.S. Arginine kinase: Joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility. J. Mol. Biol. 2011, 405, 479–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godsey, M.H.; Davulcu, O.; Nix, J.C.; Skalicky, J.J.; Brüschweiler, R.P.; Chapman, M.S. The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase. Structure 2016, 24, 1658–1667. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, N.; Tanaka, K.; Suzuki, T. Amino acid residues 62 and 193 play the key role in regulating the synergism of substrate binding in oyster arginine kinase. FEBS Lett 2005, 579, 1688–1692. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.Y.; Guo, H.Y.; Geng, H.L.; Ru, B.M.; Cao, J.; Chen, C.; Zeng, L.Y.; Wang, X.Y.; Li, F.; Xu, K.L. T273 plays an important role in the activity and structural stability of arginine kinase. Int. J. Biol. Macromol. 2014, 63, 21–28. [Google Scholar] [CrossRef]
- Geng, H.L.; Bian, M.R.; Liu, Y.; Cao, J.; Chen, C.; Wang, Z.Y.; Li, Z.Y.; Zeng, L.Y.; Wang, X.Y.; Wu, Q.Y.; et al. The D14 and R138 ion pair is involved in dimeric arginine kinase activity, structural stability and folding. Int. J. Biol. Macromol. 2014, 66, 302–310. [Google Scholar] [CrossRef]
- Li, F.; Wu, Q.Y.; Wang, X.Y. The amino acid residue L113 is involved in arginine kinase activity and structural stability. Int. J. Biol. Macromol. 2013, 52, 198–205. [Google Scholar] [CrossRef]
- Wu, Q.Y.; Li, F.; Wang, X.Y. Val65 plays an important role in the substrate synergism, structural stability and activity of arginine kinase. Int. J. Biol. Macromol. 2009, 45, 393–398. [Google Scholar] [CrossRef]
- Wu, Q.Y.; Zhu, Y.Y.; Wei, F.; Tong, Y.X.; Cao, J.; Zhou, P.; Li, Z.Y.; Zeng, L.Y.; Li, F.; Wang, X.Y.; et al. Mutation of the conserved G66 residue in GS region decreased structural stability and activity of arginine kinase. Int. J. Biol. Macromol. 2018, 111, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Somasundaram, T.; Blanc, E.; Chen, Z.; Chapman, M.S. Critical initial real-space refinement in the structure determination of arginine kinase. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 835–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, M.S.; Fabiola, F.; Gattis, J.L.; Somasundaram, T.; Chapman, M.S. Refinement of the arginine kinase transition-state analogue complex at 1.2 A resolution: Mechanistic insights. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, P.; Haouz, A.; Pereira, C.A.; Aguilar, C.; Alzari, P.M. The crystal structure of Trypanosoma cruzi arginine kinase. Proteins 2007, 69, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Rao, Z.; Kim, S.Y.; Li, X.; Kim, D.S.; Kim, Y.J.; Park, J.H. Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase. Mol. Cells 2020, 43, 784–792. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Somasundaram, T.; Blanc, E.; Parthasarathy, G.; Ellington, W.R.; Chapman, M.S. Transition state structure of arginine kinase: Implications for catalysis of bimolecular reactions. Proc. Natl. Acad. Sci. USA 1998, 95, 8449–8454. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Ye, S.; Guo, S.; Yan, W.; Bartlam, M.; Rao, Z. Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity. FASEB J. 2010, 24, 242–252. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, G.Y.; Yang, H.; Hu, M.J.; Cao, M.J.; Su, W.J.; Jin, T.; Liu, G.M. Crystal structure determination of Scylla paramamosain arginine kinase, an allergen that may cause cross-reactivity among invertebrates. Food Chem. 2019, 271, 597–605. [Google Scholar] [CrossRef]
- Laino, A.; Lopez-Zavala, A.A.; Garcia-Orozco, K.D.; Carrasco-Miranda, J.S.; Santana, M.; Stojanoff, V.; Sotelo-Mundo, R.R.; Garcia, C.F. Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus. PeerJ 2017, 5, e3787. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Zavala, A.A.; Garcia-Orozco, K.D.; Carrasco-Miranda, J.S.; Sugich-Miranda, R.; Velazquez-Contreras, E.F.; Criscitiello, M.F.; Brieba, L.G.; Rudino-Pinera, E.; Sotelo-Mundo, R.R. Crystal structure of shrimp arginine kinase in binary complex with arginine-a molecular view of the phosphagen precursor binding to the enzyme. J. Bioenerg. Biomembr. 2013, 45, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Baoyu, C.; Qin, G.; Zhi, G.; Xicheng, W. Improved activity assay method for arginine kinase based on a ternary heteropolyacid system. Tsinghua Sci. Technol. 2003, 8, 422–427. [Google Scholar]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vagin, A.A.; Steiner, R.A.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2184–2195. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Chen, B.; Wang, X. Evidence for proximal cysteine and lysine residues at or near the active site of arginine kinase of Stichopus japonicus. Biochemistry (Mosc.) 2004, 69, 1336–1343. [Google Scholar] [CrossRef]
- Strong, S.J.; Ellington, W.R. Expression of horseshoe crab arginine kinase in Escherichia coli and site-directed mutations of the reactive cysteine peptide. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1996, 113, 809–816. [Google Scholar] [CrossRef]
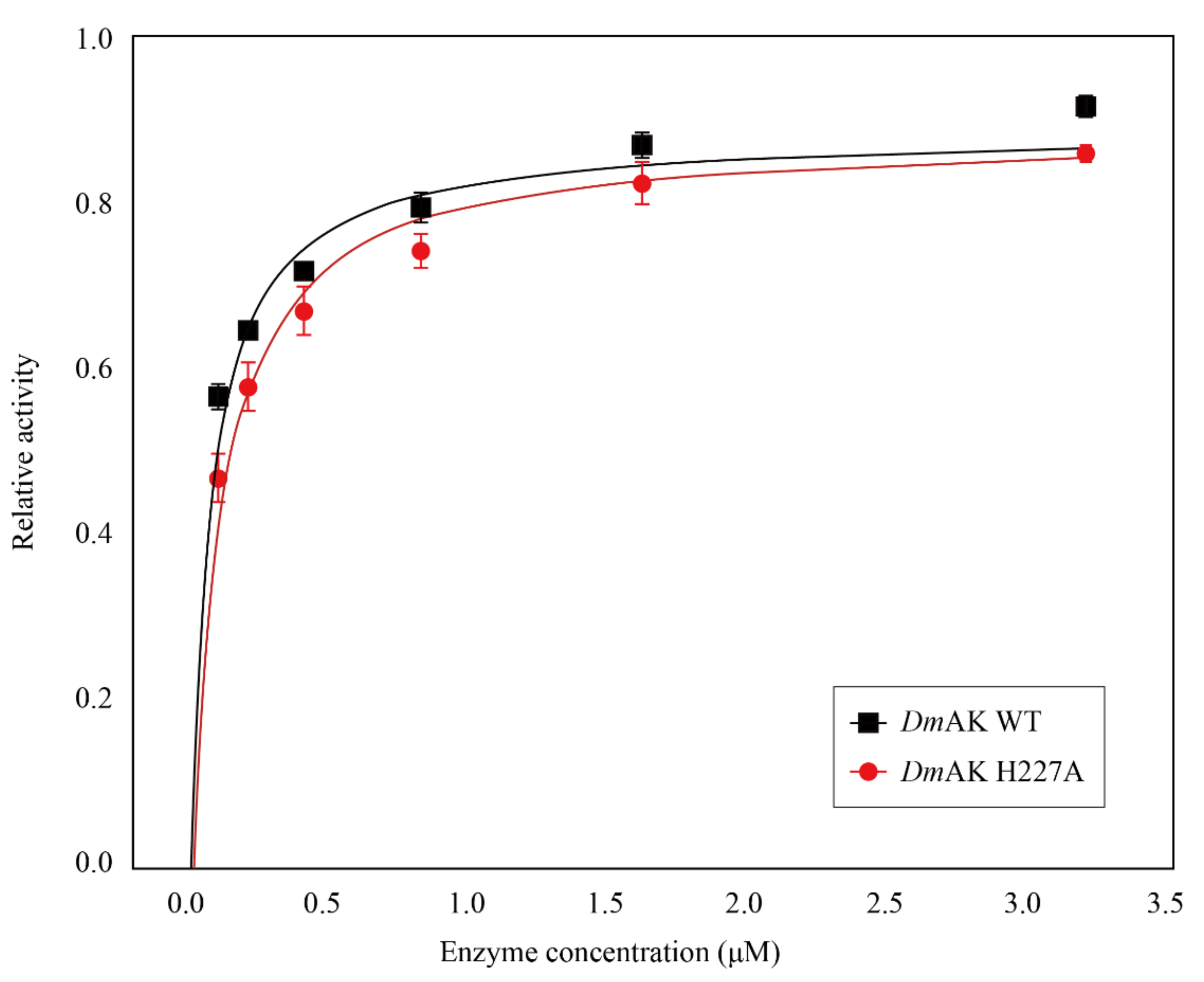

| Vmax (μmolpimin−1 mg−1) | Kcat (s−1) | KmArg (mM) | KmATP (mM) | KdArg (mM) | KdATP (mM) | |
|---|---|---|---|---|---|---|
| DmAK WT | 318.4 ± 7.38 | 3.80 ± 0.09 | 0.22 ± 0.04 | 0.70 ± 0.09 | 2.41 ± 0.34 | 0.80 ± 0.10 |
| DmAK H227A | 261.0 ± 8.61 | 3.12 ± 0.10 | 0.19 ± 0.06 | 0.81 ± 0.13 | 2.74 ± 0.54 | 0.68 ± 0.11 |
| Data Collection | |
|---|---|
| X-ray source (Detector) | PAL 11C µ-MX (Pilatus3 6M) |
| Wavelength (Å) | 0.979490 |
| Space group | C121 |
| Unit cell dimensions a, b, c (Å) | 78.15, 58.21, 75.41 |
| α, β, γ (°) | 90.00, 100.48, 90.00 |
| Resolution range (Å) | 46.40–1.75 (1.78–1.75) |
| Unique reflections | 37,443 (5325) |
| Completeness (%) | 98.4 (98.0) |
| Multiplicity | 3.4 (3.3) |
| <I/σ(I)> | 12.1 (3.3) |
| Rmeas | 0.068 (0.345) |
| Overall B factor from Wilson plot (Å 2) | 13.0 |
| Refinement | |
| Resolution range (Å) | 46.4–1.75 |
| Highest resolution shell (Å) | 1.75 |
| Completeness (%) | 97.9 |
| No. reflections | 33,099 (1621) |
| Rwork/Rfree (%) | 18.1/22.4 |
| No. of atoms / residues | 3123/360 |
| Protein | 2794 |
| Others | 329 |
| PO4 | 5 |
| NO3 | 4 |
| Water | 320 |
| R.m.s. deviations | |
| Bonds length(Å) | 0.008 |
| Bond Angles (°) | 1.038 |
| Ramachandran plot | |
| Most favored (%) | 97.7 |
| Allowed (%) | 2.0 |
| Disallowed (%) | 0.3 |
| Primer Name | Sequence |
|---|---|
| DmAK-NdeI-F | 5′-GCACTCCATATGCATCACCATCATCATCATGTGGAC-3′ |
| DmAK-H90A-F | 5′-GTGATTACGCCACCGGCTTCAAG-3′ |
| DmAK-H227A-F | 5′-ATGAGGAAGATGCCCTGAGAATTATC-3′ |
| DmAK-H284-R | 5′-GCAGTGCAATGGCCACTGAGGCCC-3′ |
| DmAK-H315A-R | 5′-CTGCTTCCGTGGCCTCACCAGCGGTTC-3′ |
| DmAK-NotI-R | 5′-GCACTCGCGGCCGCTTATGCGGCTTC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.S.; Jang, K.; Kim, W.S.; Ryu, M.; Park, J.H.; Kim, Y.J. Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability. Molecules 2022, 27, 884. https://doi.org/10.3390/molecules27030884
Kim DS, Jang K, Kim WS, Ryu M, Park JH, Kim YJ. Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability. Molecules. 2022; 27(3):884. https://doi.org/10.3390/molecules27030884
Chicago/Turabian StyleKim, Da Som, Kiyoung Jang, Wan Seo Kim, Moonhee Ryu, Jung Hee Park, and Yong Ju Kim. 2022. "Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability" Molecules 27, no. 3: 884. https://doi.org/10.3390/molecules27030884
APA StyleKim, D. S., Jang, K., Kim, W. S., Ryu, M., Park, J. H., & Kim, Y. J. (2022). Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability. Molecules, 27(3), 884. https://doi.org/10.3390/molecules27030884






