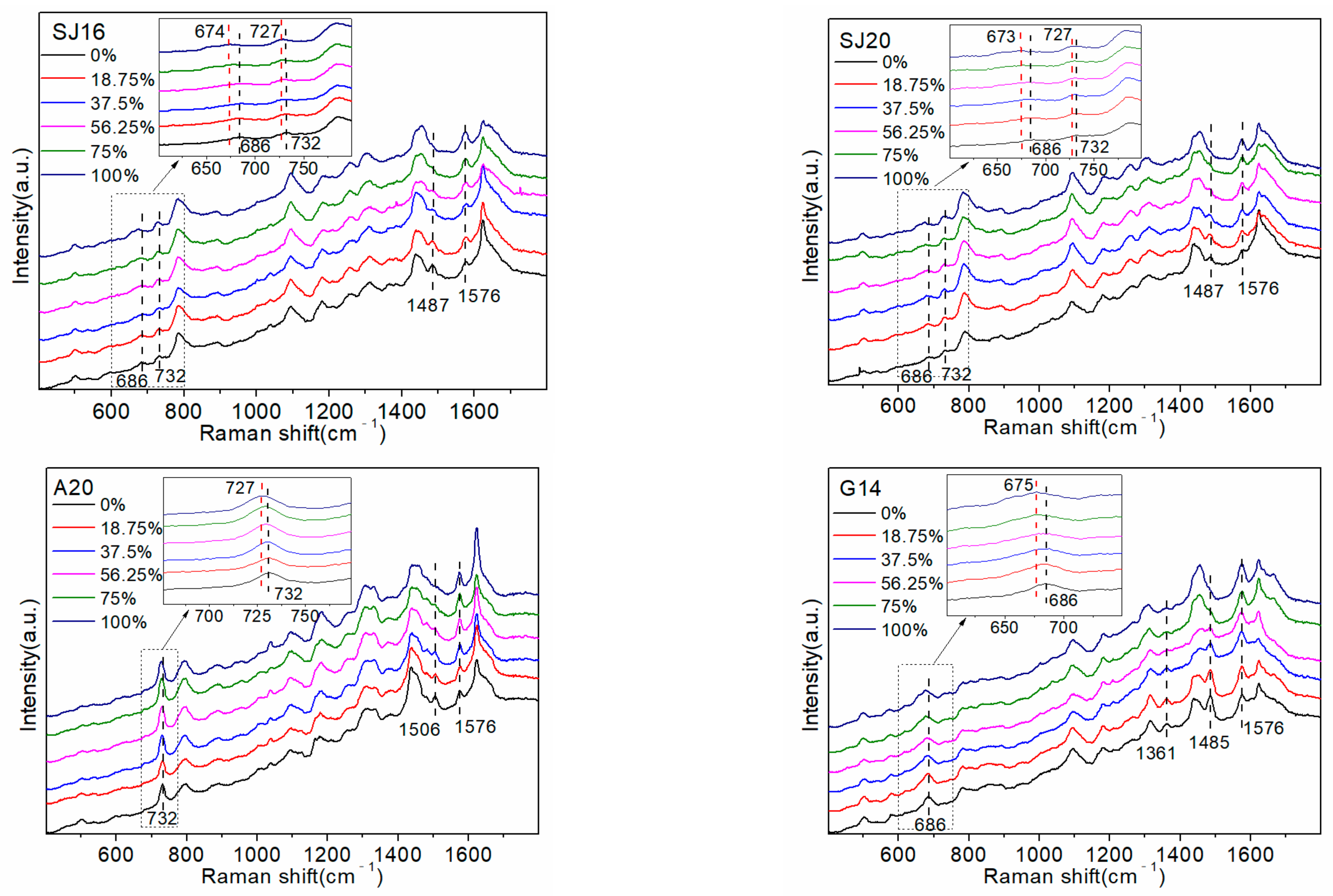
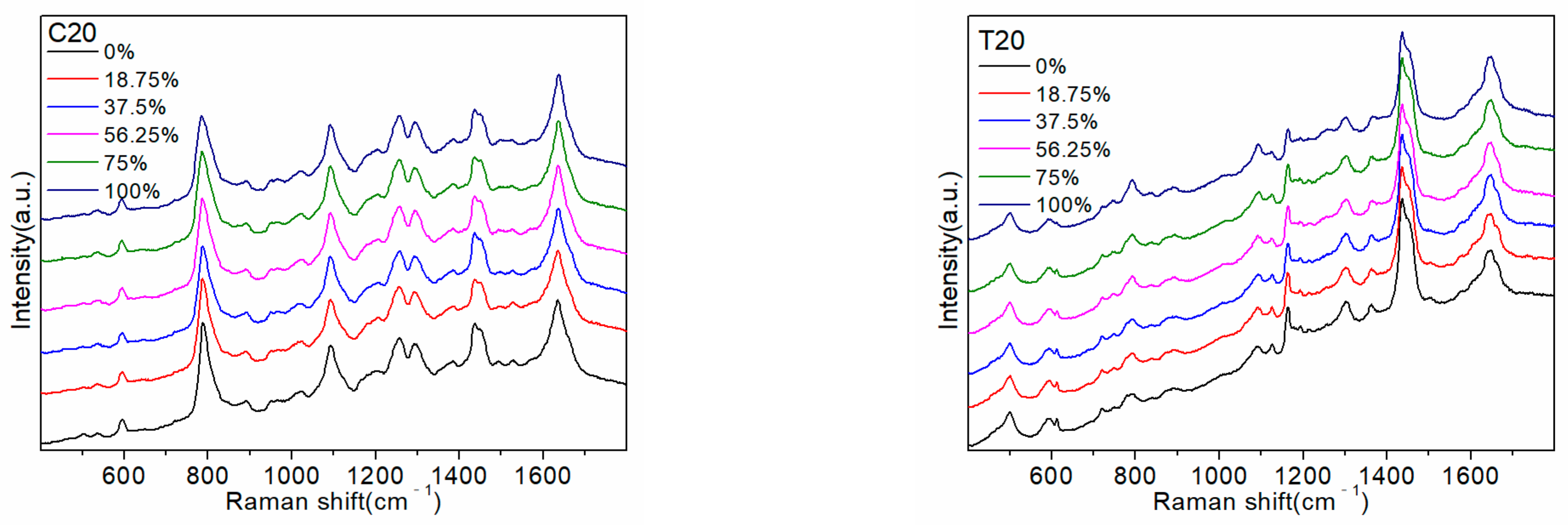
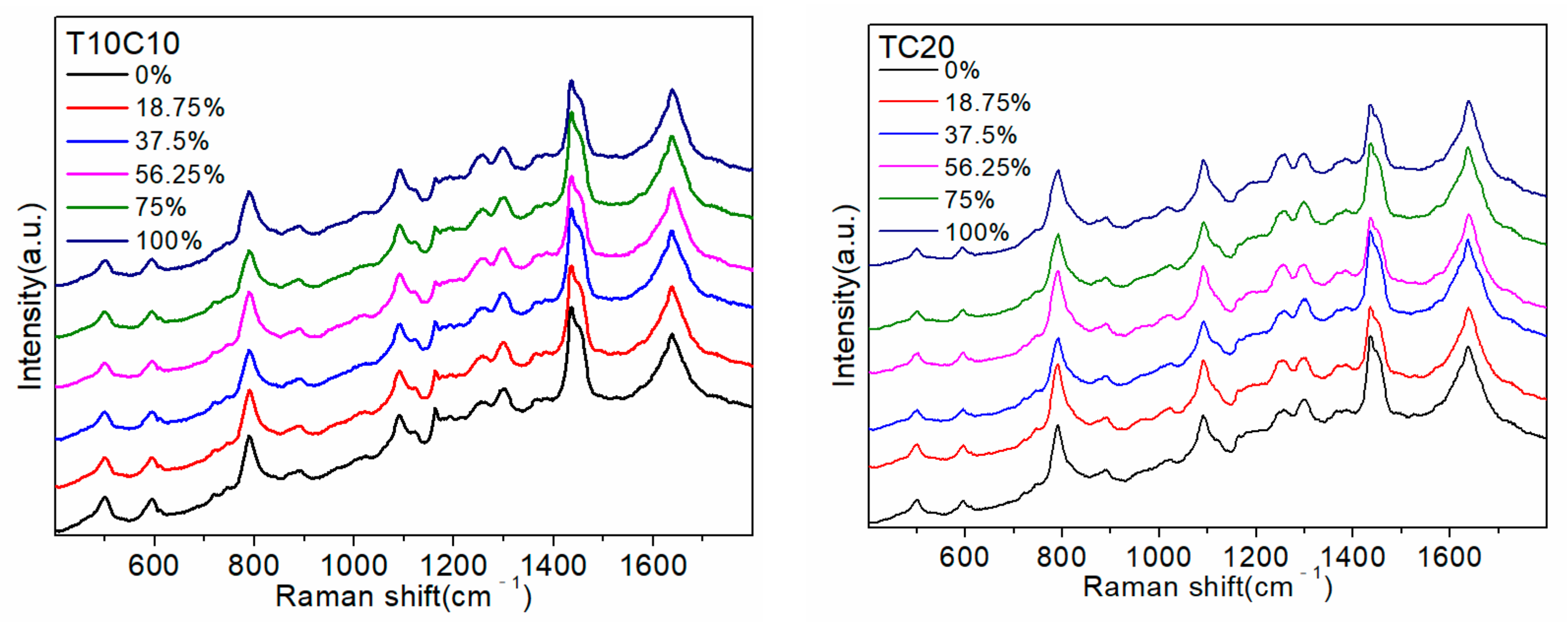
2.1. Detection of Random Sequences and Pure Base Sequences
To explore the interaction between D
2O and ssDNA, two random sequences, SJ16 and SJ20, were detected first. The specific sequences SJ16 and SJ20 represent are shown in
Table 1. Subsequently, pure base sequences A20, G14 (the number of consecutive G bases did not exceed 14), C20, and T20 were detected to attribute the SERS peaks and determine the common active sites between them. The results are shown in
Figure 1.
Replacing the atoms of the molecular vibrating group with their isotopes shifted the vibration frequency and changed the spectral intensity.
Figure 1 shows the 686 cm
−1 signal belonging to the G base ring respiratory vibration peak [
19,
22,
23], which gradually shifted to 675 cm
−1 as the D
2O proportion increased. This likely occurred because some hydrogen atoms in the G base were replaced by deuterium atoms, which increased the overall mass, reduced the vibration frequency, and caused a red shift in the peak. The peak at 732 cm
−1 was attributed to the A base ring respiratory vibration peak [
19,
23,
24], which gradually decreased with the increase in the proportion of D
2O, and the frequency shifted to 727 cm
−1 (100% D
2O).
The decrease in the intensity of the peak at 732 cm
−1 occurred because D
2O has a higher viscosity than ordinary water, making it more conducive to nucleic acid folding [
8]. Therefore, the A base is squeezed by the surrounding groups, its ring breathing vibration is affected, and the peak intensity is weakened. The frequency shift is affected by the H/D isotope exchange, which increases the overall mass and decreases the vibration frequency. Therefore, there was a slight red shift in the peak position. In the pure A base, the peak at 732 cm
−1 did not weaken because all sequences were A bases, which would be difficult to fold. The respiratory vibration of the A base ring was only slightly affected by the environmental D
2O, and therefore, the peak at 732 cm
−1 did not change markedly.
The peak at 1485 cm
−1 was attributed to the hydrogen bond formed by N7 on the G base and the surrounding H
2O/D
2O [
20,
22]. Gradually decreasing the surrounding H
2O concentration (i.e., gradually increasing the D
2O concentration) increases the number of deuterium–hydrogen bonds formed by N7 and D
2O. This action causes the isotopic effect to weaken the vibration tendency of the deuterium–hydrogen bonds and the peak intensity. As the hydrogen bonds formed between both molecules are not tetrahedral, there is no peak frequency shift [
25]. The peak intensity at 1576 cm
−1 increased with an increasing proportion of D
2O in both the A20 and G14 sequences, but the change in the G14 sequence was more obvious.
Therefore, the change in the peak at 1576 cm−1 was caused by the interaction between the NH2 group on the A and G bases in the sequence and D2O. Furthermore, deuterium atoms do not directly participate in the formation of hydrogen bonds, which are still NH…O, and the O-D bond in D2O has higher energy and a shorter bond length than the O-H bond in H2O. Therefore, the NH…O bond polarizability, Raman activity, and Raman strength increase.
The following two interesting phenomena were observed: the peaks at 1506 cm
−1 and 1361 cm
−1 changed with increasing D
2O proportion in pure base sequences A20 and G14. However, these changes were not evident in the SERS spectra of random sequences, i.e., SJ16 and SJ20. The 1506 cm
−1 peak in the SERS spectrum of the A20 sequence was attributed to the A base. As the number of A bases in the sequences SJ16 and SJ20 (three and four, respectively) and the Raman intensity of this peak was relatively low, it did not appear in the SERS spectra of SJ16 and SJ20. Increasing the number of A bases in the sequence, such as with the sequence A12GTC in 3.3 (including 12 A bases), caused the peak at 1506 cm
−1 to appear in the SERS spectra, and the intensity increased with the increasing proportion of D
2O in the environment. A comparison of the SERS spectra of SJ16, SJ20, and the four pure base sequences showed that the 1361 cm
−1 peak was attributable to the G and T bases [
24]. Increasing the proportion of D
2O did not change the peak of the pure T base at 1361 cm
−1, whereas the peak intensity of the pure G base changed significantly, but the change was very weak in the SJ16 and SJ20 sequences. This likely occurred because the number of G bases in the sequence was relatively low (seven and eight, respectively). Although a total of eight G bases is small, it accounts for 40% of all bases, and the intensity of the peak in G14 was not high. Therefore, the change caused by eight G bases would not be very obvious. In the G12ATC sequence of the follow-up experiment, there were 12 G bases, accounting for 60% of the total number of bases, and the peak intensity change of 1361 cm
−1 was more visible than that of SJ16 and SJ20. However, the apparent degree was still less than that of G14 (100% G base) at the 1361 cm
−1 peak.
In summary, the Raman peaks that changed with an increasing proportion of D
2O in the random sequence SERS spectrum were almost all related to the A and G bases. Therefore, we preliminarily inferred that the interaction site of D
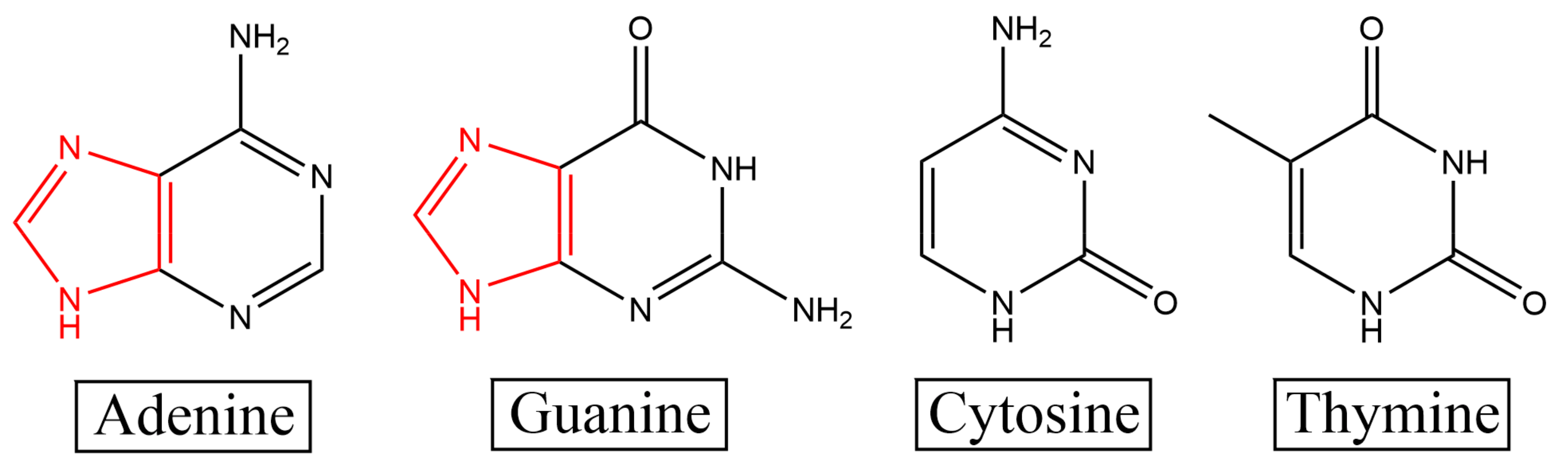
2O and DNA was mainly at the A and G bases. A comparison of the structures of the four bases determined that the A and G bases had a unique five-membered ring structure (as shown in
Figure 2), which increased the site of interaction with water. Owing to their hydrophobicity, the bases in DNA gather in water to reduce contact with water [
26]. As the number of sites on the A and G bases that interact with water is higher than that on the C and T bases, the A and G bases are less hydrophobic (more hydrophilic) than the C and T bases, and their ability to bind to H
2O is stronger. C and T bases have fewer interaction sites with H
2O than A and G bases; therefore, they are more hydrophobic, which is not conducive to interaction with H
2O. When the concentration of D
2O in the environment increases, more sites of action not only increase the number of hydrogen–deuterium exchanges between A and G bases and D
2O and the number of deuterium–hydrogen bonds formed, but also reduce the total energy of the formed complex and thus improve its stability. Simultaneously, they increase the influence of D
2O on the group vibration in the A and G bases, which is conducive to the capture of the SERS instrument. As the C and T bases and D
2O interact with fewer sites, it not only reduces the number of hydrogen–deuterium exchanges and the formation of deuterium–hydrogen bonds between the two but also makes the total energy of the complex higher and lowers its stability. This leads to a reduction in the influence of D
2O on the group vibration of C and T bases, which is not conducive to the capturing of changes by the SERS instrument. Therefore, the SERS peaks related to the C and T bases barely changed, except the 1170 cm
−1 peak belonging to T bases was weakened.
2.3. Effect of Number and Position of G Base in Sequence
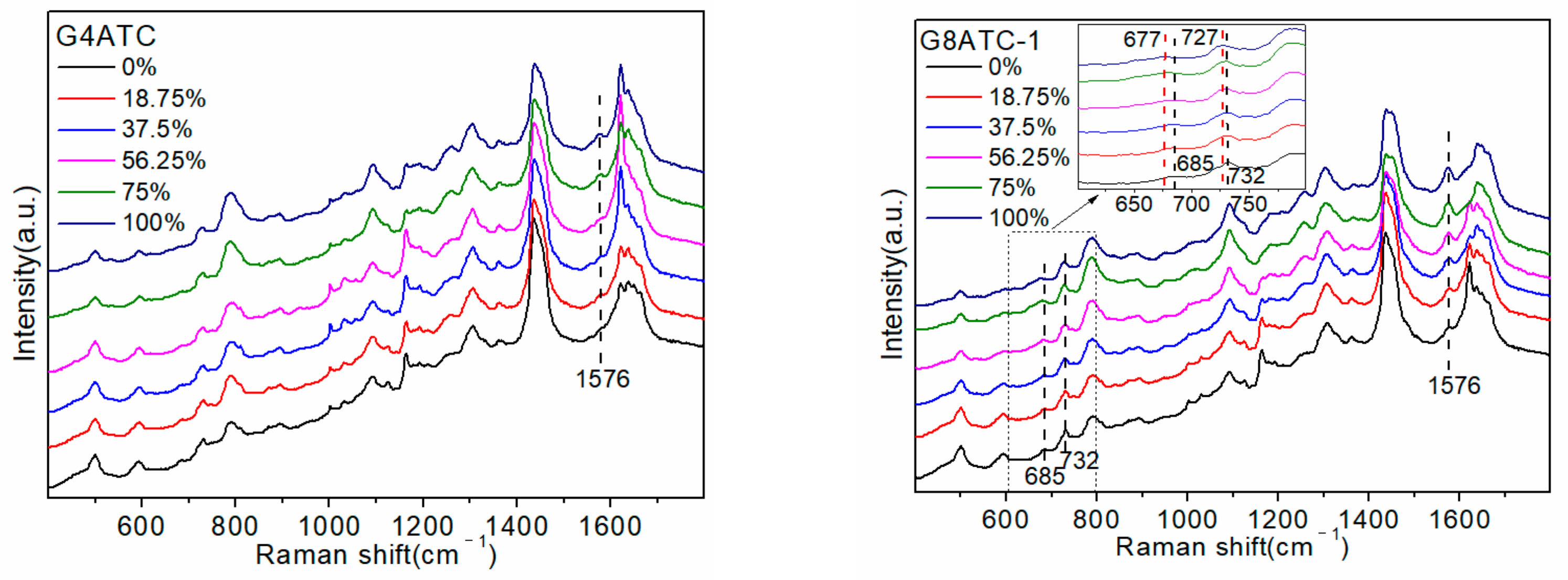
To explore the effects of the number and position of G bases in the sequence, the following five sequences were designed: G4ATC, G8ATC-1, G8ATC-2, G8ATC-3, and G12ATC, as shown in
Table 1. All sequences contained 20 bases, and the number of G bases was 4, 8, 8, 8, and 12, respectively. The four G and eight G bases replaced the C bases that did not interact with D
2O. The three G8ATC (-1, -2, and -3) sequences contained the same number and types of bases, but the order of arrangement was different. The five ssDNAs detected are shown in
Figure 4a.
The increase in the proportion of D
2O produced a Raman peak in the spectrum that was consistent with that of SJ16 and SJ20. Specifically, the 686 cm
−1 and 732 cm
−1 peaks had a red shift, and the intensities of the 732 cm
−1 and 1485 cm
−1 peaks gradually weakened, whereas that of the peak at 1576 cm
−1 gradually increased. The largest change was in the intensity of the 1576 cm
−1 peak. Using the peak intensity as a reference, we explored the influence of the number and position of G bases in the sequence on their interaction with D
2O. The change in the peak intensity with the increasing proportion of D
2O in the different sequences and the linear relationship between the peak intensity and the proportion of D
2O are shown in
Figure 4b.
The G4ATC, G8ATC-1, and G12ATC sequences are all 20 bases long. The extra G bases of the latter two were replaced by C bases that did not interact with D
2O; therefore, the changes in the three maps were mostly caused by the increase in the number of G bases. In the SERS spectrum, when the proportion of D
2O was 0%, the difference in the SERS spectra of the three sequences was mainly reflected in the peaks at 1485 cm
−1 and 1576 cm
−1, which were both attributed to the G base. In contrast, the peak at 732 cm
−1 attributed to the A base showed almost no difference.
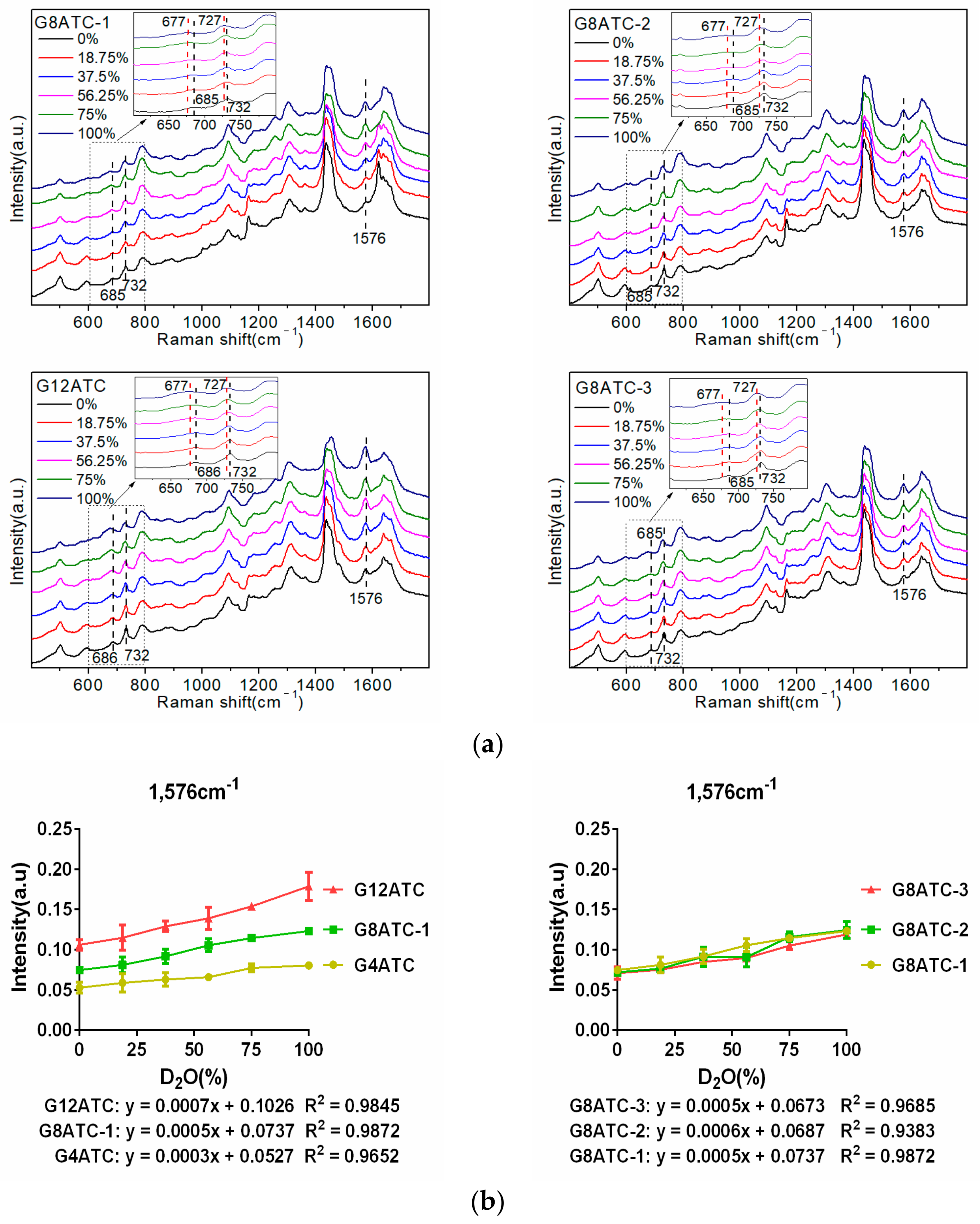
Figure 4b shows that the 1576 cm
−1 peak intensity of the three sequences was positively correlated with the number of G bases, and the slopes of the linear equations of the three sequences were 0.0003, 0.0005, and 0.0007, respectively, which showed an upward trend. Specifically, increasing the number of G bases also increased the amplitude of the intensity of the 1576 cm
−1 peak with an increase in the proportion of D
2O. Therefore, the greater the number of G bases, the stronger the interaction between the sequence and D
2O.
Differences in the arrangement of sequences produced slight variations in the intensities of some peaks in the SERS spectra of the three G8ATC sequences. For example, for the 732 cm
−1 peak, a D
2O proportion of 0% produced an A8GTC-2 sequence that had a stronger intensity than the A8GTC-1 sequence. However, under D
2O conditions, the trend of peak intensity and displacement in the three sequences was the same as the proportion of the D
2O increase.
Figure 4b shows that the 1576 cm
−1 peak intensity of the three G8ATC sequences was between 0.075 and 0.135, and the slopes of the linear equation were almost equal (0.0005–0.0006). Therefore, when the number of G bases in the sequence was constant, the difference in the amplitude of the 1576 cm
−1 peak intensity of the three sequences was very small and was smaller than the difference caused by the difference in the number of G bases. We believe that this phenomenon occurred because the volume of the D
2O molecules was small relative to the base and ssDNA sequences in the system. Regardless of the position of the base in the sequence, the active site of both molecules was always “exposed”, and the small D
2O molecule always localized to and interacted with the active site. Therefore, the sequence interaction with D
2O was not affected by the positions of the A and G bases in the sequence.
2.4. Effect of Number and Position of A Base in Sequence
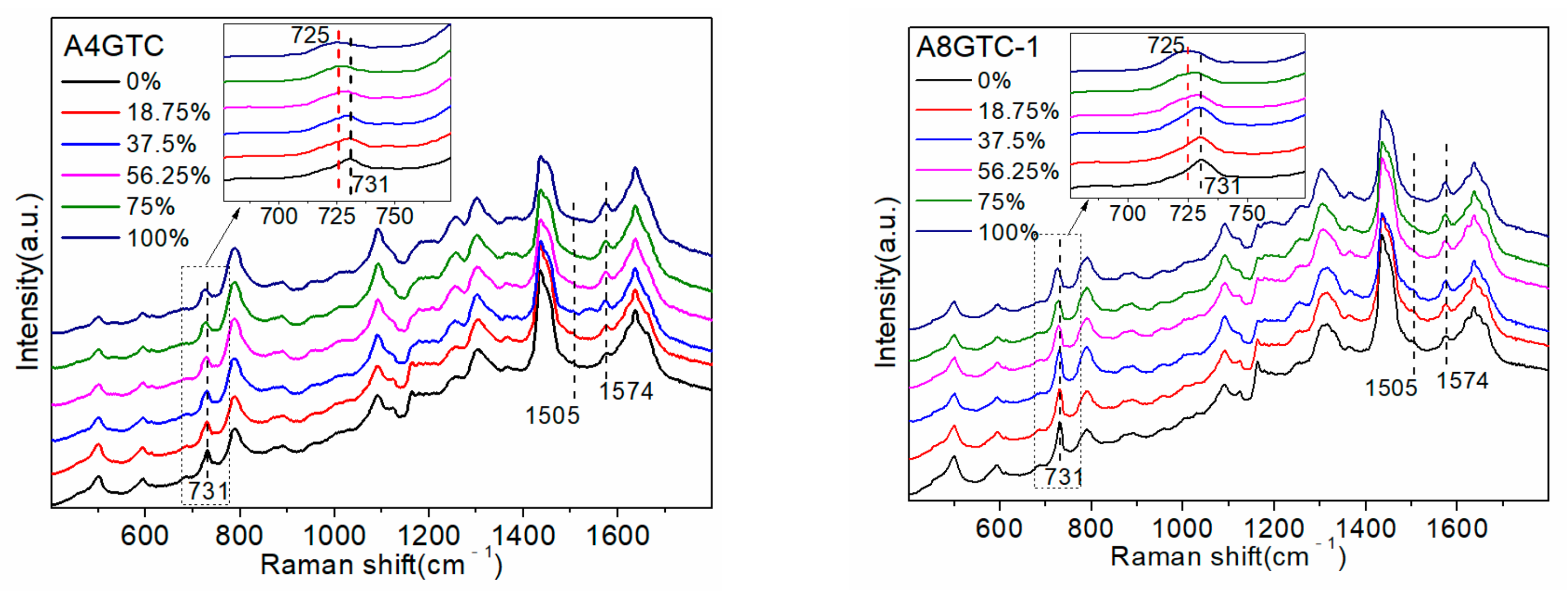
In addition, to explore the influence of the number and position of the A bases, the following five sequences were designed: A4GTC, A8GTC (-1, -2, and -3), and A12GTC; the specific sequences they represent are shown in
Table 1. All sequences contained 20 bases, and the number of A bases was 4, 8, 8, 8, and 12, respectively. The increase in four and eight A bases replaced the C bases that did not interact with D
2O. Three A8GTC (-1, -2, and -3) sequences contained the same number and types of bases, but they were arranged in a different order. The five ssDNAs detected are shown in
Figure 5a.
As all sequences had the same number of G bases, their position did not affect the intensity of the 1574 cm
−1 peak, which was only altered by a change in the A base. Increasing the proportion of D
2O resulted in changes in the Raman peak spectrum that were consistent with those observed in the SJ16 and SJ20 spectra. Specifically, the peak at 731 cm
−1 had a red shift, and the intensities of the 731 cm
−1 and 1505 cm
−1 peaks gradually decreased, whereas that of the 1574 cm
−1 peak gradually increased. We used the peaks at 731 cm
−1 and 1574 cm
−1, which had the largest intensity changes, as references to explore the influence of the number of A bases and their positions in the sequence on the interaction between the sequence and D
2O. The trend of change in the peak intensities at 731 cm
−1 and 1574 cm
−1 with increasing D
2O proportion and the linear equation of the peak intensity with respect to the proportion of D
2O are shown in
Figure 5b.
The A4GTC, A8GTC-2, and A12GTC sequences were all 20 bases long. The extra A bases in the A8GTC-2 and A12GTC sequences were included by replacing the C bases that do not interact with D
2O. Therefore, the changes in the three SERS maps could almost all be attributed to an increase in the number of A bases.
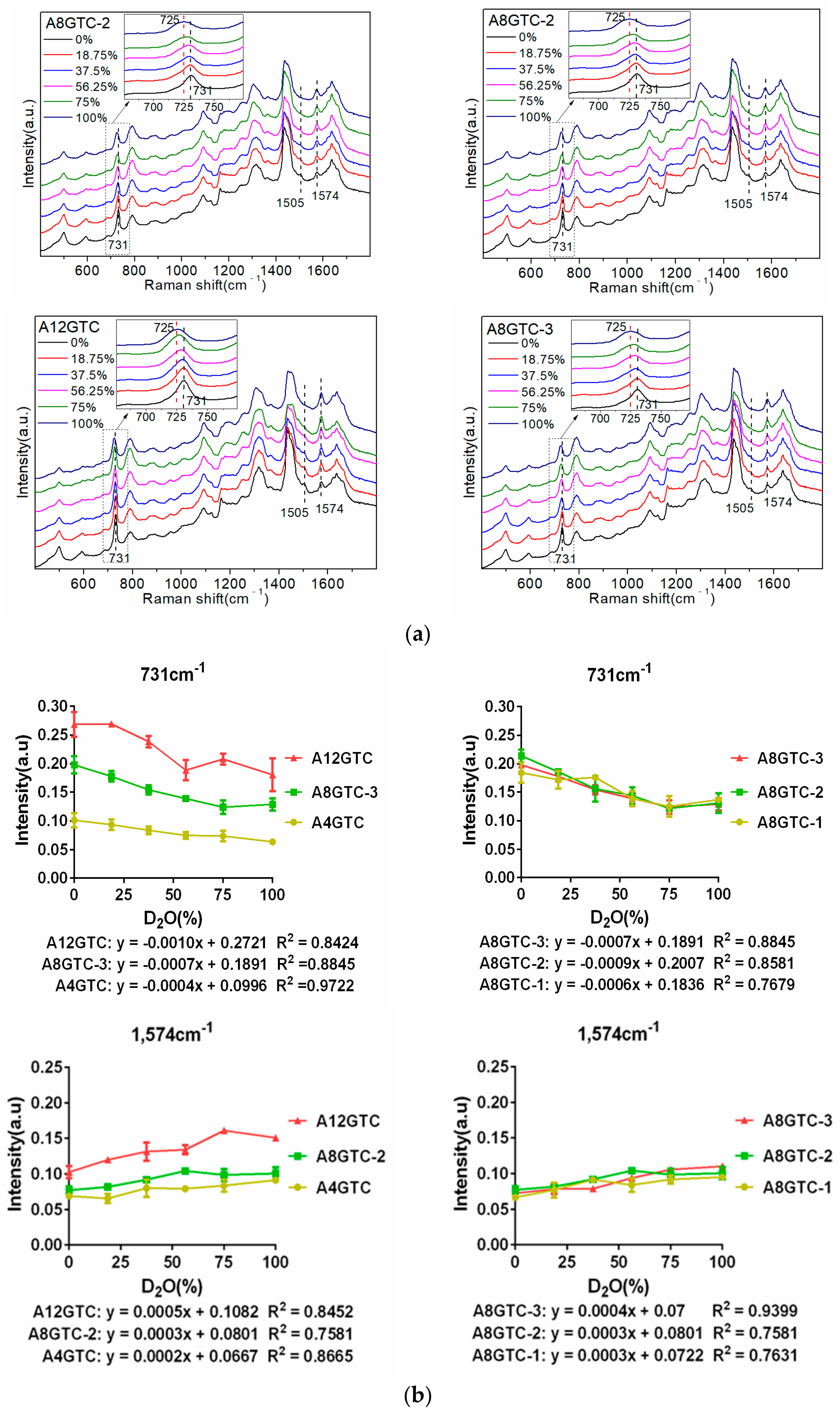
Figure 5b shows that the 731 cm
−1 peak intensity of the three sequences was positively correlated with the number of A bases and decreased with an increase in the D
2O proportion. The linear equation slopes of the peak intensity with respect to the proportion of D
2O were −0.0004, −0.0007, and −0.0010, respectively, and the absolute value showed an increasing trend. Moreover, as the number of A bases increased, the reduction in the intensity of the 731 cm
−1 peak increased with an increase in the proportion of D
2O. The 1574 cm
−1 peak intensity of the three sequences was positively correlated with the number of A bases and increased with an increase in the proportion of D
2O. The linear equation slopes of the peak intensity with respect to the proportion of D
2O were 0.0002, 0.0004, and 0.0005, respectively: i.e., they showed an upward trend. Specifically, as the number of A bases increased, the magnitude of enhancement of the intensity of the 1574 cm
−1 peak increased with an increase in the proportion of D
2O. Therefore, the greater the number of A bases in the sequence, the stronger the interaction between the sequence and D
2O. In
Section 2.3, it was shown that the linear equation slopes of the 1576 cm
−1 peak intensity of the G4ATC, G8ATC, and G12ATC sequences increased with the proportion of D
2O, i.e., 0.0003, 0.0005, and 0.0007, respectively. Importantly, these values were greater than the linear equation slopes of the 1574 cm
−1 peak intensity of the A4GTC, A8GTC-2, and A12GTC sequences and the proportion of D
2O. Therefore, the interaction between the G base and D
2O was stronger, causing a greater change in the intensity of the 1576 cm
−1 peak than that caused by the A base.
Differences in the order of arrangement induced slight differences in the intensities of some peaks of the SERS spectra of the three A8GTC sequences. However, the peaks and trends of changes for the three sequences with respect to the proportion of D2O were consistent. Furthermore, the 731 cm−1 and 1574 cm−1 peaks with pronounced changes were similarly analyzed. The intensity of the 731 cm−1 peak of the three sequences was between 0.11 and 0.21 (linear equation slopes: −0.0006, −0.0009, and −0.0007), and that of the 1574 cm−1 peak was between 0.06 and 0.11 (linear equation slopes: 0.0003, 0.0003, and 0.0004). The absolute slopes of the linear equation were not significantly different, and the slopes of the 1574 cm−1 peak were almost equal. Therefore, when the number of A bases in the sequence was constant, the difference in the peak intensities of the three sequences at 731 cm−1 and 1574 cm−1 was very small (smaller than the difference induced in response to variations in the number of A bases).
Investigation of the number and position of the A and G bases in the sequence suggests that the interaction between D2O and sequences with the same length was affected by the change in the number of A and G bases. In contrast, changes in the position of the A and G bases had less pronounced effects. Owing to the volatility of SERS, for sequences that cannot be folded into specific secondary structures, such as G-quadruplexes and i-motifs, their interaction with D2O is unaffected by the position of A and G bases.
2.5. Interaction between G-Quadruplex Sequence and D2O
We concluded that D2O mainly interacts with the A and G bases in the sequence and is mainly affected by their number rather than location, provided that the sequence does not form specific secondary structures. Therefore, we investigated the outcome when the sequence formed a secondary structure.
We selected the G-quadruplex as the paradigm for investigating the interaction between D
2O and the ssDNA secondary structure. G-quadruplexes are unique secondary structures formed by guanine-rich nucleic acids. Considerable in vitro biophysical and structural evidence supports the formation of a G-quadruplex [
27,
28]. Computer research and sequencing methods have revealed the ubiquity of G4 sequences in the gene regulatory regions of different genomes, including human genomes [
29,
30]. Experiments using chemical, molecular, and cellular biological methods have demonstrated the presence of G4 sequences in chromatin, DNA, and RNA, and the formation of G-quadruplexes is very important for key biological processes, such as transcription, translation, genome instability, and cancer development [
31,
32,
33].
In addition, G-quadruplexes can also serve as potential therapeutic targets for human diseases, thereby improving the therapeutic effect on diseases such as cancer and genetic diseases [
34]. Therefore, it is necessary to explore the interaction between D
2O and G4 sequences, which would contribute to enhancing our understanding of the structure and properties of G-quadruplexes. Furthermore, such studies would provide partial theoretical guidance for further studies of various biological processes that require the simultaneous involvement of D
2O and G-quadruplexes. We synthesized six sequences that form G-quadruplexes, i.e., (G
3T)
4, TG
7T, Tel21, 93del [
20], GO18, and GO18T [
35] (
Table 1 shows the specific sequences they represent) and explored their interaction with D
2O. The results are shown in
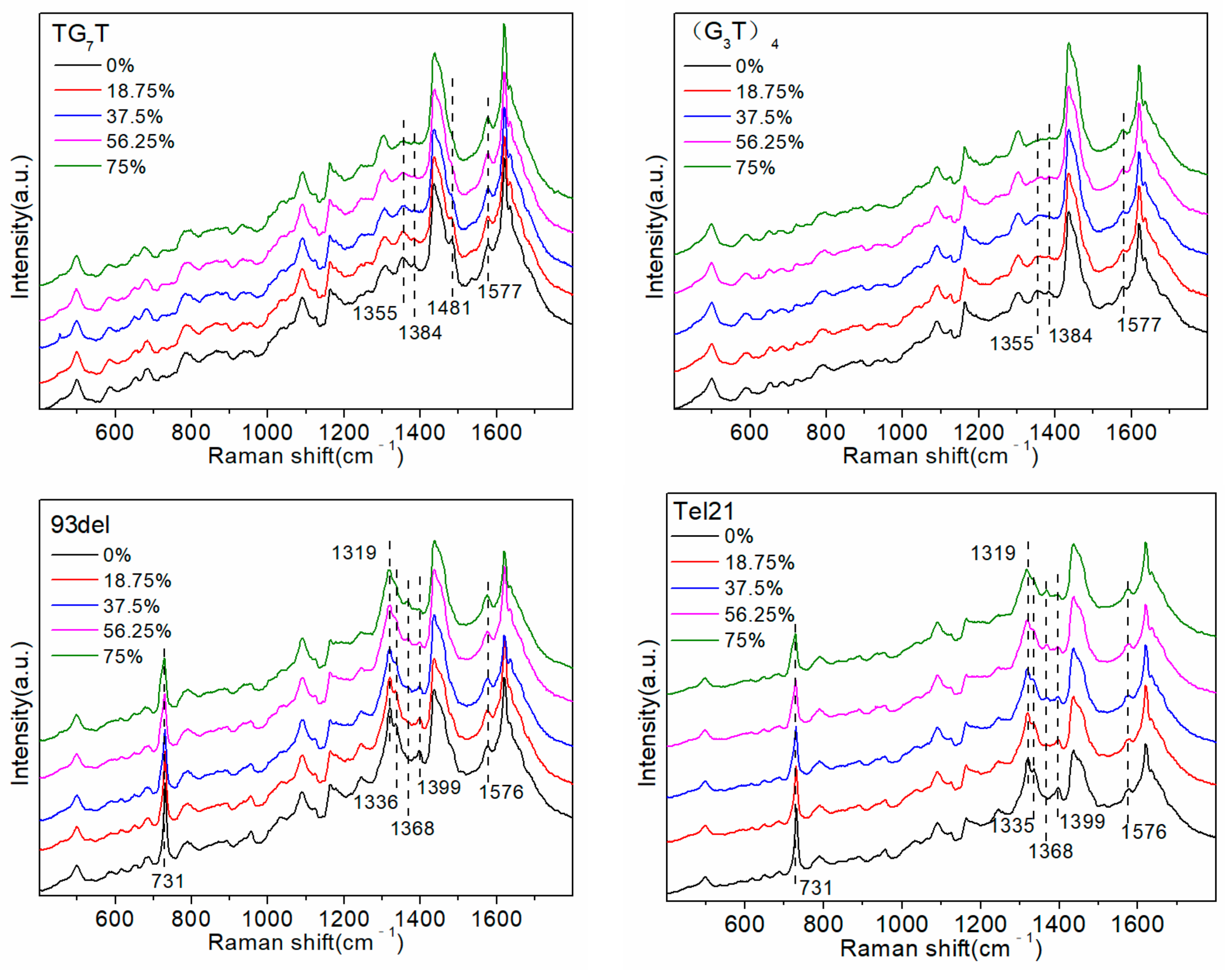
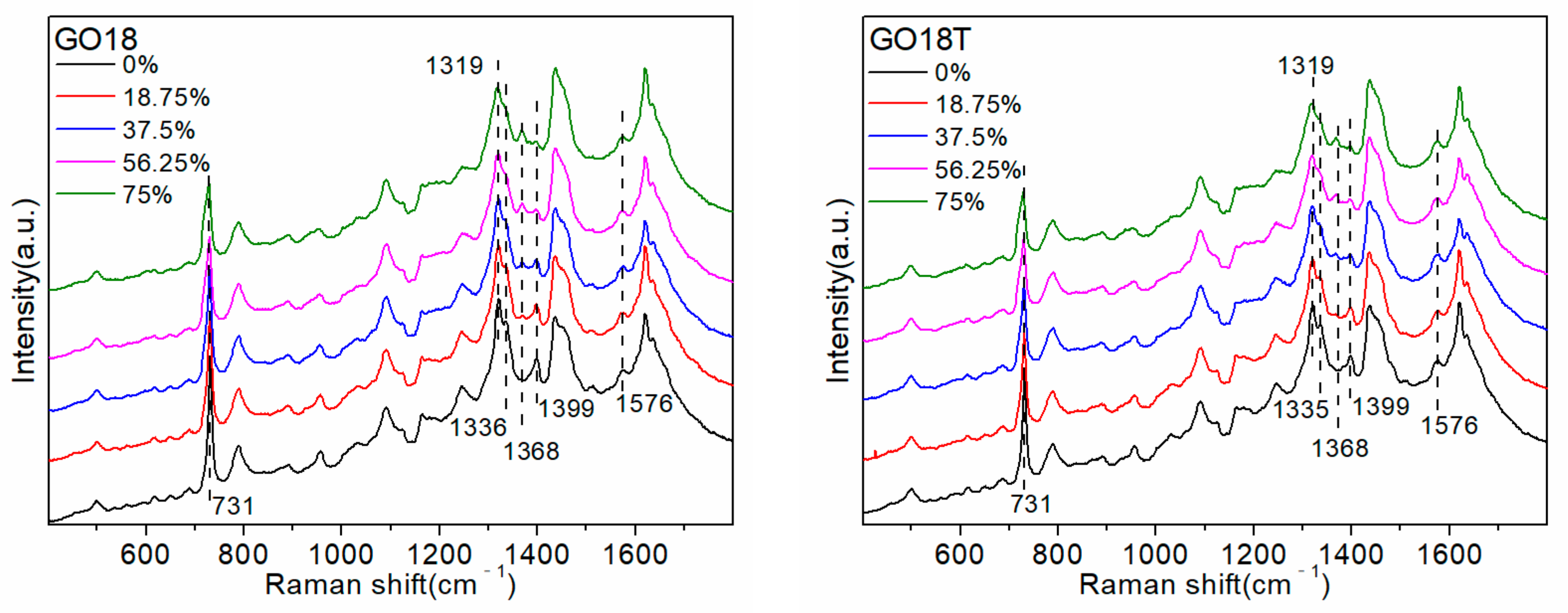
Figure 6.
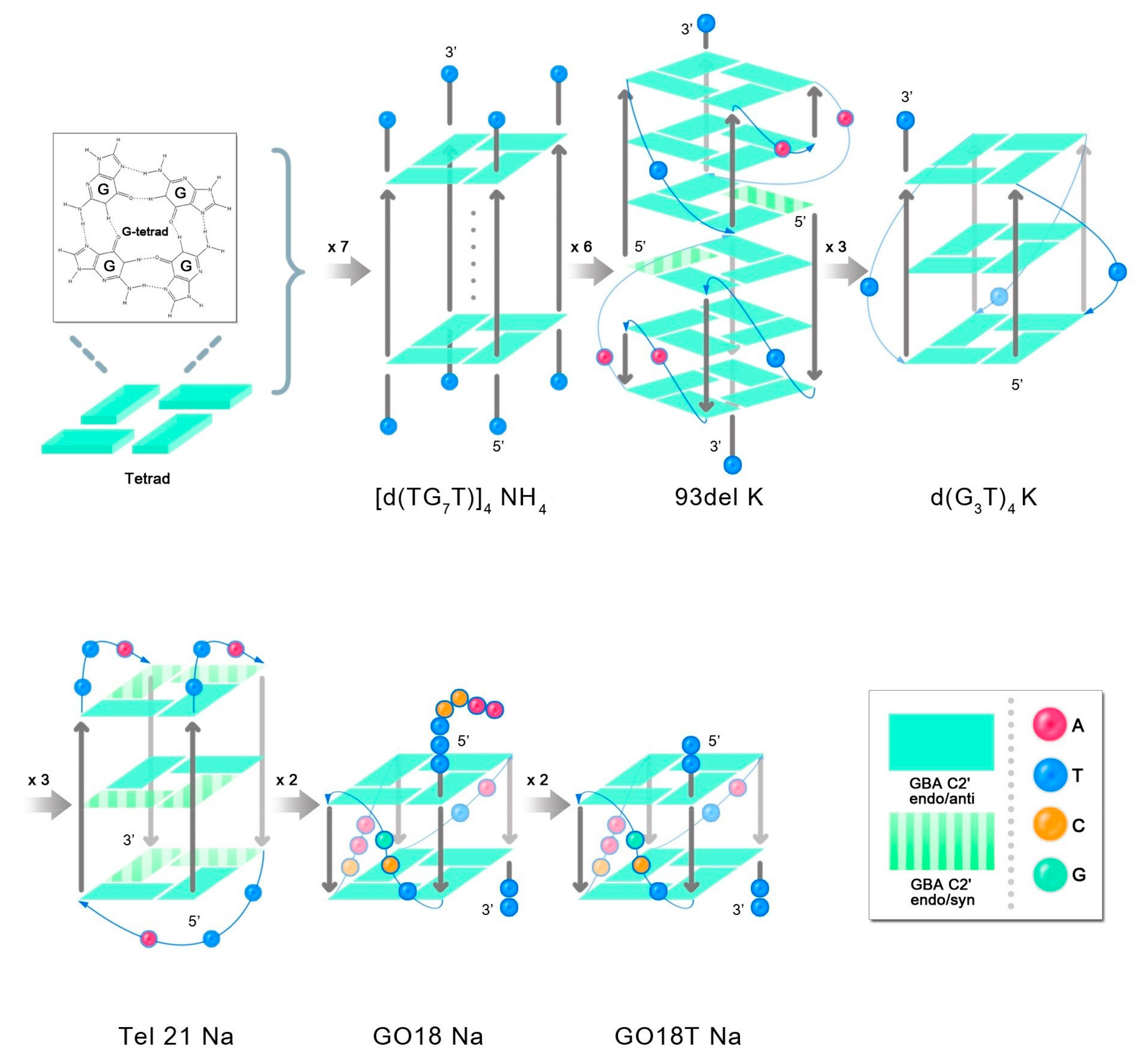
After adding a cationic acetate buffer (pH 4.5), the ssDNA formed a G-quadruplex structure [
20,
23], which is shown in
Figure 7, thereby inducing partial changes in the sites where it interacted with D
2O. First, the intensity of the 1576 cm
−1 peak changed very slightly with a change in the D
2O proportion, as the sites that originally interacted with D
2O were involved in the formation of intramolecular hydrogen bonds, forming a G-quadruplex structure, and showed no intermolecular hydrogen bonding interactions with D
2O. Although the A base also interacts with D
2O to induce an alteration in the intensity of the 1574 cm
−1 peak, the number of A bases in the G4 sequence is often very small. The smaller the number of A bases, the weaker the interaction with D
2O. Therefore, there was almost no change in the intensity of the 1576 cm
−1 peak with a change in the proportion of D
2O. The 1399 cm
−1 peak, attributable to the T base [
20,
23], only appeared in the G-quadruplex sequence, and its intensity decreased with the increasing proportion of D
2O. This was because after the G-quadruplex structure was formed, T bases were located outside the G-tetrad or even in the G-quadruplex, which enabled them to move closer to the surface of the silver nanoparticles. Furthermore, the SERS signal of the T bases was enhanced more strongly, and therefore, the signal was revealed in the SERS spectra. Moreover, compared to that of the sequence that did not form the G-quadruplex structure, the intensity of the 731 cm
−1 peak of the G-quadruplex sequence was exceptionally strong ([G
3T]
4 and TG
7T did not contain A bases, and this shift was not associated with any peak). This observation was also likely due to the fact that after the G-quadruplex structure is formed, more A bases are located outside the G-tetrad or even the G-quadruplex. Consequently, they are located closer to the surface of the silver nanoparticles, and the A base SERS signal is more strongly enhanced. Therefore, in the G-quadruplex sequence containing A and T bases, the intensities of the 1399 cm
−1 and 731 cm
−1 peaks reflected the number and stability of G-quadruplex structures. The greater the number and stability of G-quadruplexes, the greater the strength of these two peaks.
Figure 6 shows that as the proportion of D
2O increased, the intensities of the 1399 cm
−1 and 730 cm
−1 peaks were gradually reduced, indicating that D
2O reduced the number and stability of G-quadruplex structures.
In addition, increasing the proportion of D
2O in the solution gradually resulted in reduced intensities of the 1319 cm
−1 and 1336 cm
−1 peaks, whereas that of the 1368 cm
−1 peak gradually increased. The positions and intensities of the Raman bands in the ranges 550–700 cm
−1 and 1300–1380 cm
−1 have been reported to be related to the glycosidic bond angle (GBA) conformation of all G, A, and T bases [
36]. The number and type of GBAs contained in different G-quadruplexes are different, resulting in differences in the Raman shifts attributed to the GBA structure in the Raman signal of the G4 sequence. In the literature, the 1319 cm
−1 and 1336 cm
−1 peaks are assigned to dG C2′-endo/anti, and the 1368 cm
−1 peak is assigned to dG C2′-endo/syn; (G
3T)
4 and TG
7T does not contain the C2′-endo/syn structure, and therefore, the 1355 cm
−1 peak was assigned to the C2′-endo/anti-structure [
20,
22]. The changes in the peak intensities at 1319 cm
−1, 1336 cm
−1, 1355 cm
−1, and 1368 cm
−1 indicated alterations in the C2′-endo/anti and C2′-endo/syn structures of the G base, which further reflected changes in the syn/anti-GBA conformation of the G-quadruplex.
In the G-quadruplex structure formed by (G3T)4 and TG7T, all sequences were arranged 5′-3′, and all GBAs adopted a trans conformation. Increasing the proportion of D2O gradually resulted in the reduced intensity of the 1355 cm−1 peak, indicating that the number of GBAs in the trans conformation gradually decreased, and the number or stability of G-quadruplexes decreased. In the G-quadruplex structure formed by the sequences of 93del, Tel21, GO18, and GO18T, the arrangement direction of the DNA strands was not uniform, and a few cis conformations were formed in the GBAs. (In the G-quadruplex structure formed by GO18 and GO18T, GBA is a trans conformation, but the cis-GBA signal also appears in the spectrum. Our guess is that their two-layer G-quadruplex structure is very close due to π-π stacking, which squeezes the three bases on the connecting chain, presenting a conformation quite similar to cis-GBA.) The SERS signal of the trans conformation GBAs was dominant in the SERS spectrum of the four G-quadruplex sequences. As the proportion of D2O increased, the intensities of the 1319 cm−1 and 1336 cm−1 peaks greatly decreased, indicating that the number of GBAs in the trans conformation decreased, whereas the intensity of the 1368 cm−1 peak, representing the cis conformation of GBAs, only slightly increased, indicating that the quantity or stability of the G-quadruplexes decreased.
In summary, D
2O was not conducive to the formation and stability of the G-quadruplex structure, which is inconsistent with the conclusion that D
2O stabilizes the folded structure of nucleic acids, as reported in the literature [
8]. We believe that because of the increased viscosity, D
2O induces DNA to fold, but this fold is not necessarily fixed and only randomly forms a part of the intramolecular hydrogen bonds. In the presence of the cationic buffer, the G-quadruplex structure formed by a specific sequence was fixed. However, D
2O negatively affected this “fixation”, as it increased the tendency of DNA to fold into other random structures.
Therefore, for ssDNA that cannot form a specific structure, D2O promotes its random folding, whereas for ssDNA that does form a specific structure, D2O inhibits the process of folding into specific structures. This discovery has important implications for certain biological processes involving G-quadruplexes. For example, the human chromosome telomere sequence contains numerous G-quadruplex structures, and this is of particular importance, as telomeres have a significant impact on the occurrence of cancer, cell senescence, and apoptosis.
D
2O reduces the number and stability of G-quadruplex structures, which provides a new demonstration to prove that D
2O is not conducive to the survival, growth, and reproduction of organisms [
37,
38,
39] but can lead to the occurrence of genetic mutations [
40] and other deleterious physiological effects. In addition, D
2O can also be used as a “regulatory switch” for G-quadruplexes, which could reduce the D
2O content around the sequence when more G-quadruplexes are needed. Furthermore, the amount of D
2O around the sequence would be decreased when fewer G-quadruplexes are needed, which would have potential applicability in research, including genetics, in vitro studies, and nucleotide medicine.
The above conclusion highlights the significance of studies on the interaction between D2O and ssDNA. Here are six conclusions that summarize the interactions between D2O and ssDNA. 1. D2O may have the following effects on DNA transcription and replications: (i) A small exchange of hydrogen and deuterium alters the bond energy and molecular activity of some bonds. (ii) The deuterium bond formed between the DNA molecule and D2O is stronger than the hydrogen bond, and the binding strength between DNA molecules, and consequently, the surface water, changes. (iii) D2O could alter the number of secondary structures such as G-quadruplexes that form part of the DNA, and these changes could affect DNA transcription, replication, and other processes. 2. The dsDNA forms numerous intramolecular hydrogen bonds through base pair complementation, but numerous groups can form intermolecular deuterium bonds with environmental D2O. Both RNA and ssDNA are single-stranded structures, and their interactions with D2O should be similar. Therefore, this study could enhance our understanding of the interactions between dsDNA, RNA, and H2O/D2O. 3. Oligonucleotide drugs are relatively stable, but it is also important to ensure stability during long-term storage and transportation. The interaction between D2O and ssDNA could change the activity of some groups. For some ssDNAs that can form a specific secondary structure, the number of secondary structures can also be changed. This can affect the interaction of ssDNA with other substances in the environment, thereby ensuring its stability. 4. Nucleic acid aptamers can bind to ligands with high affinity and strong specificity, which makes the slight change in the aptamer have a remarkable influence on binding. The effect of D2O on nucleic acids is almost constant, where there is a small hydrogen–deuterium exchange and numerous intermolecular hydrogen bonds, and the number of secondary structures is affected, which can serve as a means to appropriately alter the aptamer. This would increase the power and specificity of nucleic acid aptamers and ligand recognition, improving the sensitivity and specificity of aptamer-based biosensing. 5. The number of active H atoms or the number and type of groups capable of forming H bonds in different compounds vary; thus, the strength of the interaction between the bonds and D2O is theoretically different. By determining the intensity of the SERS peak or the degree of shift change, the active H of the compound and the number or type of groups that can form H bonds can be compared to identify the compound and determine whether the structure of the compound has changed. 6. Biological macromolecules contain numerous active H groups capable of forming hydrogen bonds, which are D2O active sites. As D2O and these active sites basically exhibit the same activity, an understanding of the interaction between D2O and ssDNA could provide a reference when studying the interaction between D2O and other biological macromolecules.