ISOC1 Modulates Inflammatory Responses in Macrophages through the AKT1/PEX11B/Peroxisome Pathway
Abstract
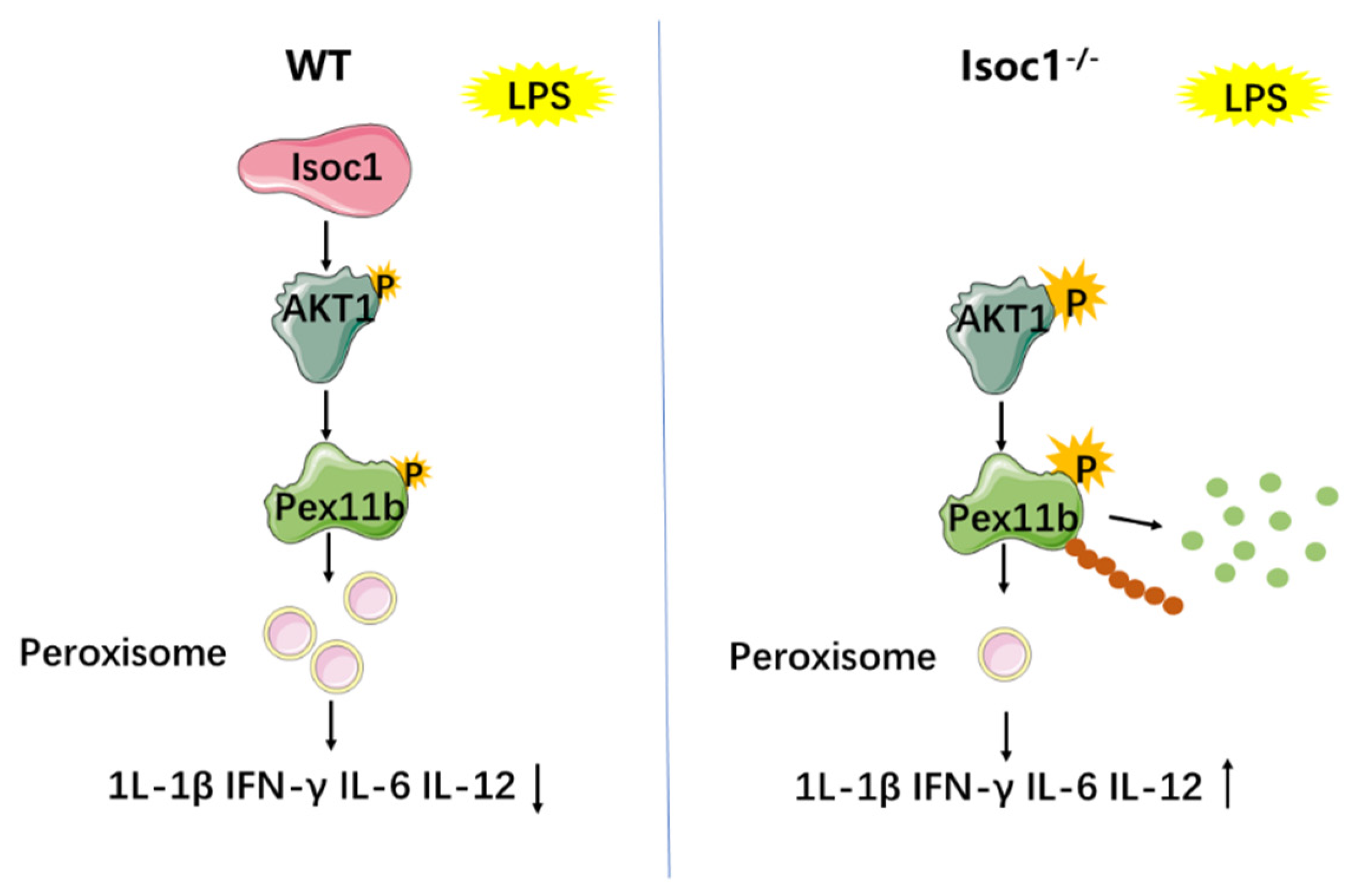
:1. Background
2. Results
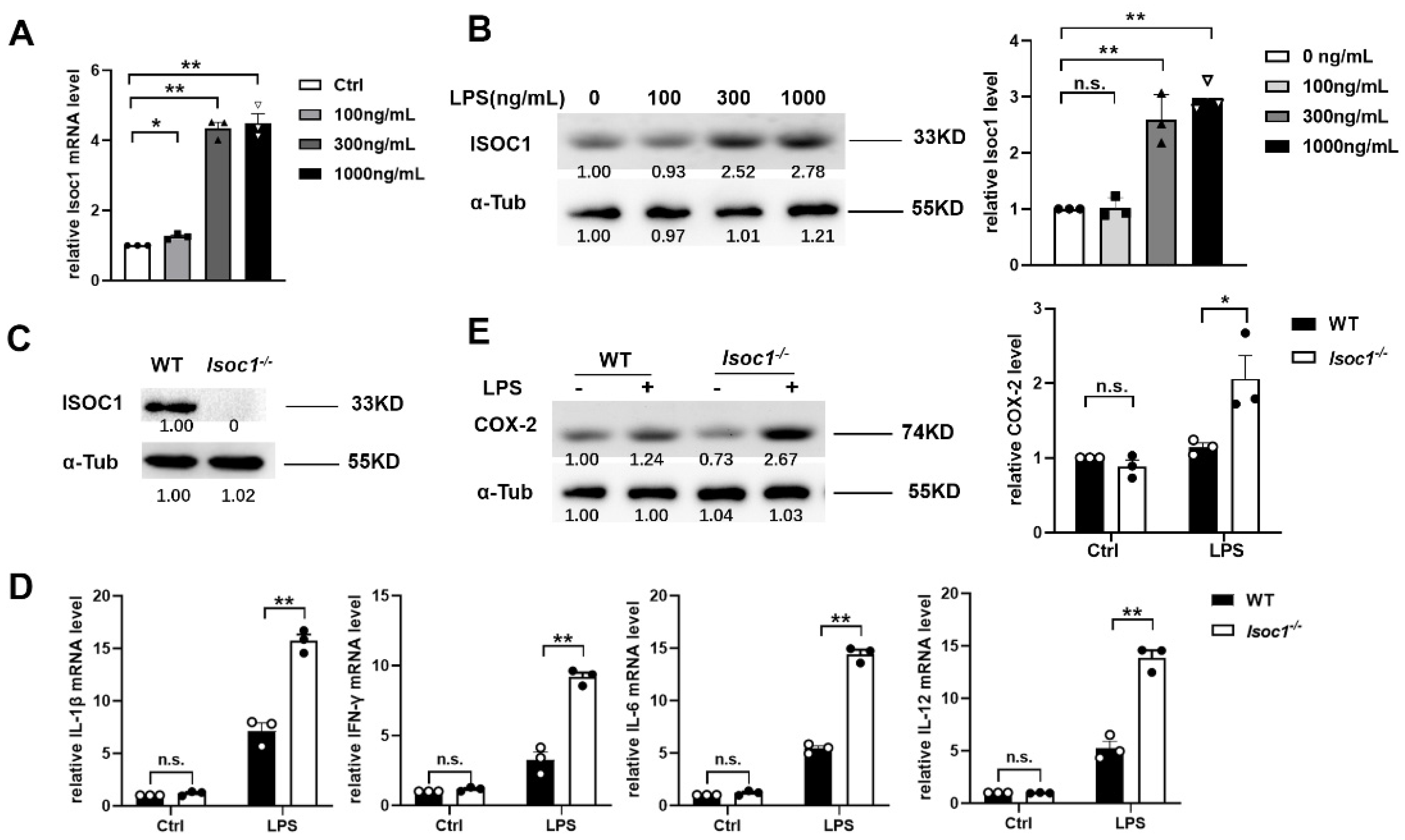
2.1. ISOC1 Expression Is Increased under LPS Stimulation and ISOC1 Regulates the Expression of Inflammatory Cytokines
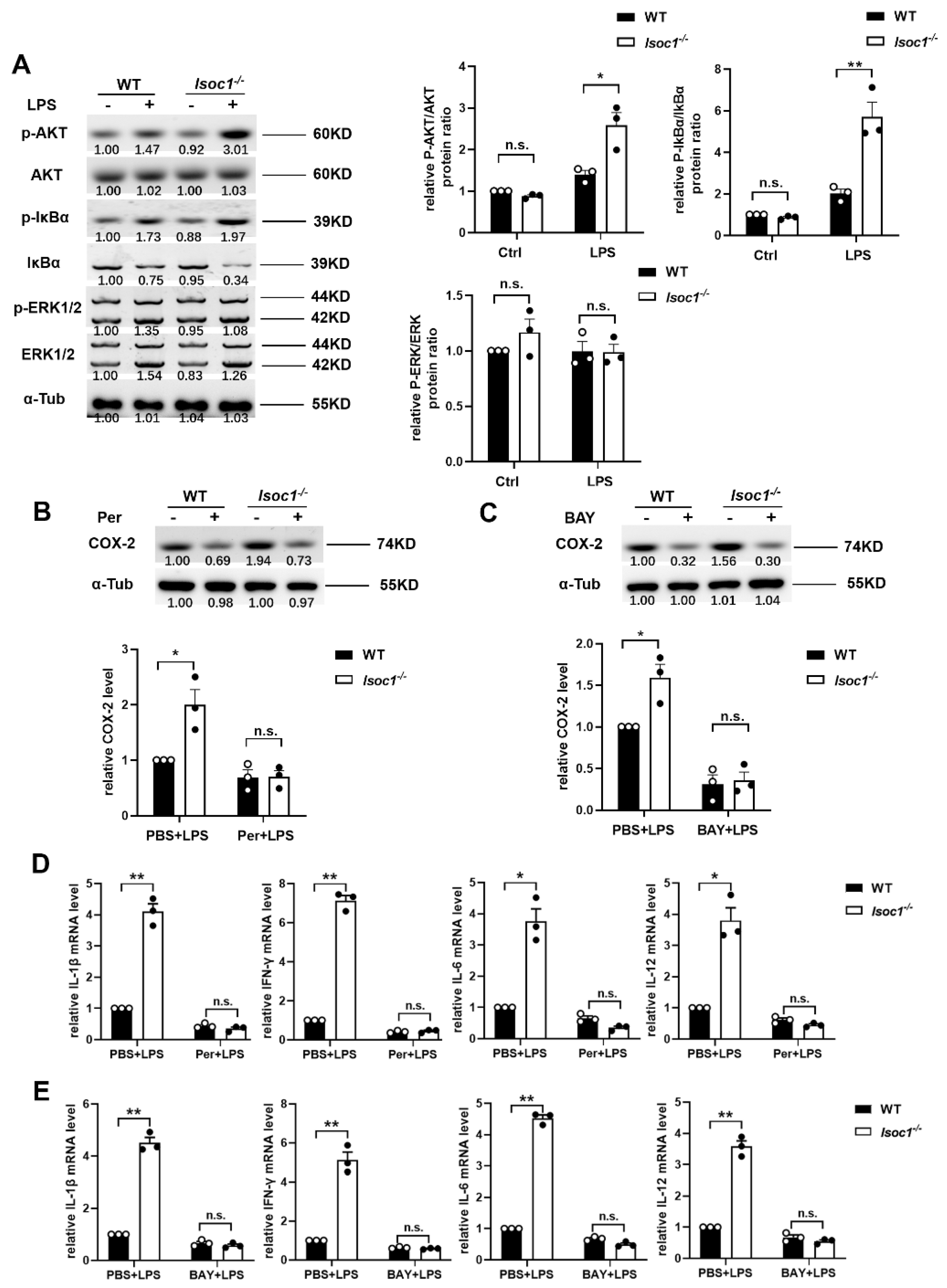
2.2. ISOC1 Mediates Macrophage Inflammatory Response via AKT1 and NF-κB Pathway
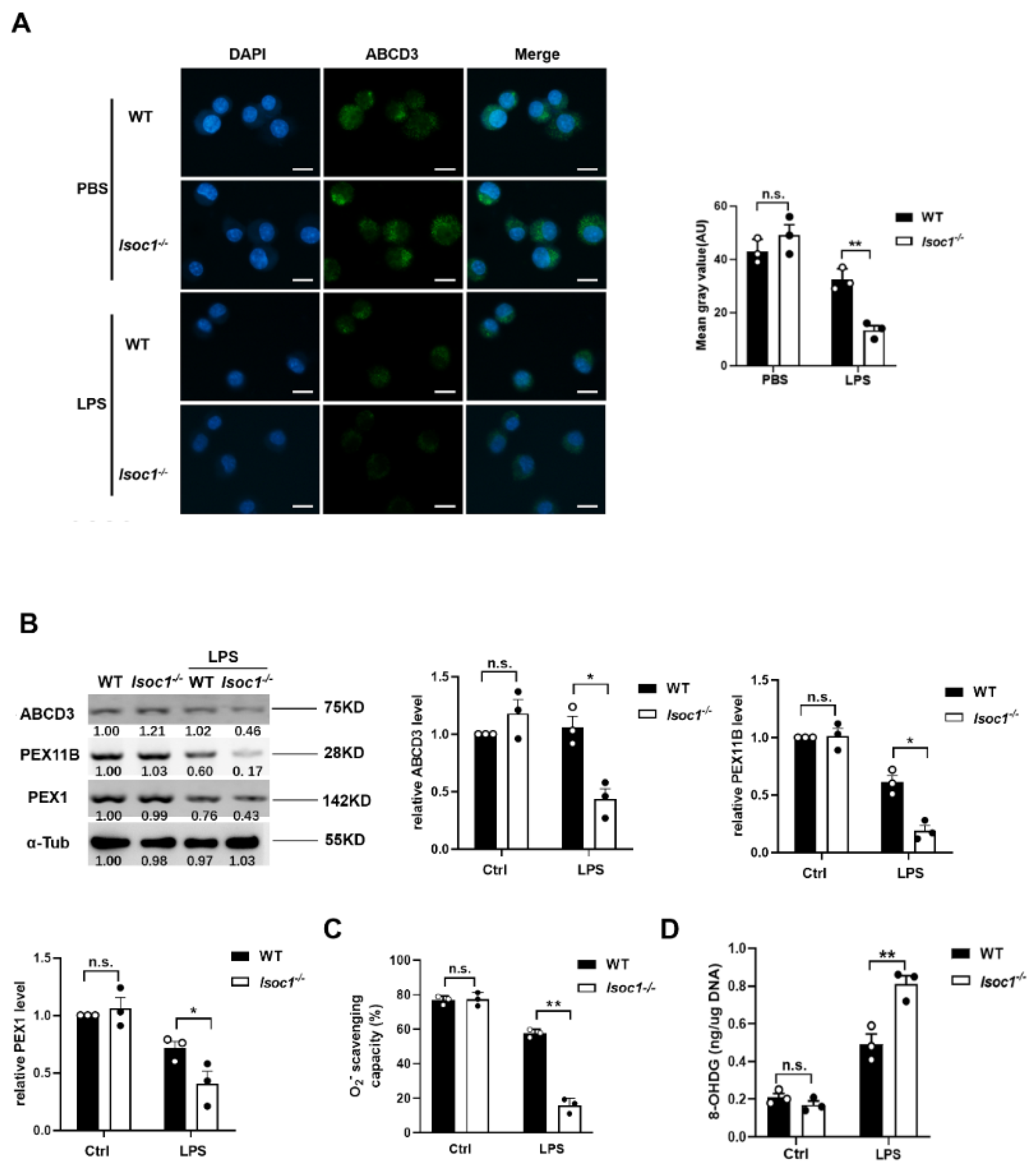
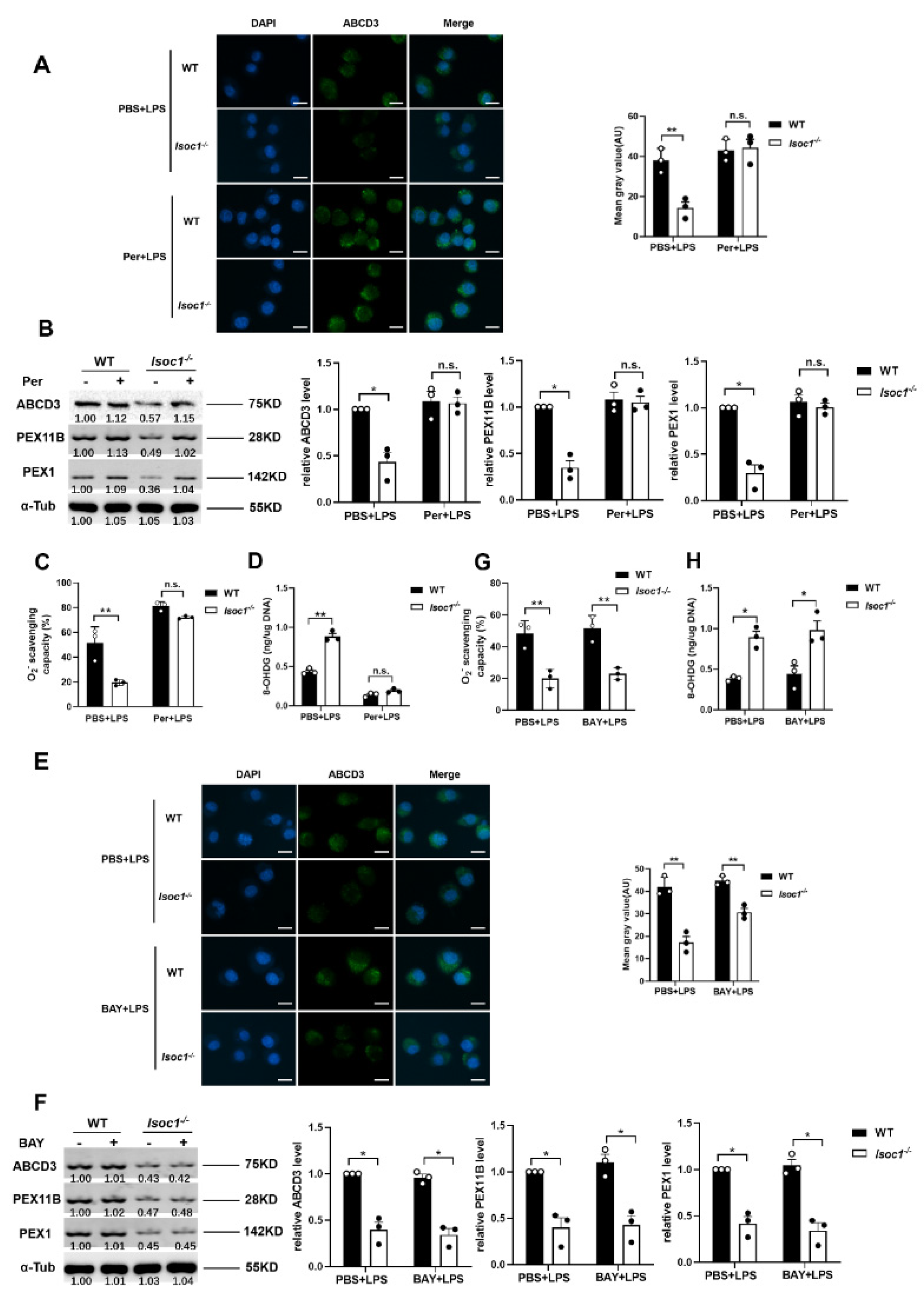
2.3. The Response to LPS Stimulation Reduces Peroxisomal Production in Isoc1-/- Macrophages
2.4. AKT1 Regulates Peroxisome Biogenesis
2.5. AKT1 Promotes the Stability of PEX11B through Phosphorylation and Ubiquitination
3. Methods
3.1. Cell Culture and Transfection
3.2. Plasmids Construction
3.3. Real-Time Quantitative PCR
3.4. Immunoblot and Immunoprecipitation
3.5. Immunofluorescence Assay
3.6. O2-Scavenging Capacity
3.7. Measurement and Quantification of Oxidative DNA Damage
3.8. Cell Proliferation Assay
3.9. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Zuo, A.; Zhang, J.; Wen, W.; Jiang, W.; Chen, H.; Liang, D.; Sun, J.; Wang, M. HIF-1alpha/JMJD1A signaling regulates inflammation and oxidative stress following hyperglycemia and hypoxia-induced vascular cell injury. Cell Mol. Biol. Lett. 2021, 26, 40. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.S. Caspase-11 non-canonical inflammasome: A critical sensor of intracellular lipopolysaccharide in macrophage-mediated inflammatory responses. Immunology 2017, 152, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, F.B.; Cunha, P.; Jensen, K.; Glass, E.J.; Foucras, G.; Robert-Granie, C.; Rupp, R.; Rainard, P. Differential response of bovine mammary epithelial cells to Staphylococcus aureus or Escherichia coli agonists of the innate immune system. Vet. Res. 2013, 44, 40. [Google Scholar] [CrossRef]
- Komazin, G.; Maybin, M.; Woodard, R.W.; Scior, T.; Schwudke, D.; Schombel, U.; Gisch, N.; Mamat, U.; Meredith, T.C. Substrate structure-activity relationship reveals a limited lipopolysaccharide chemotype range for intestinal alkaline phosphatase. J. Biol. Chem. 2019, 294, 19405–19423. [Google Scholar] [CrossRef]
- Stifano, G.; Affandi, A.J.; Mathes, A.L.; Rice, L.M.; Nakerakanti, S.; Nazari, B.; Lee, J.; Christmann, R.B.; Lafyatis, R. Chronic Toll-like receptor 4 stimulation in skin induces inflammation, macrophage activation, transforming growth factor beta signature gene expression, and fibrosis. Arthritis Res. Ther. 2014, 16, R136. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Takeuchi, S.; Kubota, K.; Kobayashi, Y.; Kozakai, S.; Ukai, I.; Shichiku, A.; Okubo, M.; Numasaki, M.; Kanemitsu, Y.; et al. Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent Toll-like receptor 4 internalization and LPS-induced TBK1-IKK-IRF3 axis activation. J. Biol. Chem. 2018, 293, 10186–10201. [Google Scholar] [CrossRef]
- Gronemeyer, T.; Wiese, S.; Ofman, R.; Bunse, C.; Pawlas, M.; Hayen, H.; Eisenacher, M.; Stephan, C.; Meyer, H.E.; Waterham, H.R.; et al. The proteome of human liver peroxisomes: Identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS ONE 2013, 8, e57395. [Google Scholar] [CrossRef]
- Pedersen, C.C.; Refsgaard, J.C.; Ostergaard, O.; Jensen, L.J.; Heegaard, N.H.; Borregaard, N.; Cowland, J.B. Impact of microRNA-130a on the neutrophil proteome. BMC Immunol. 2015, 16, 70. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Zhao, Y.; Tang, M.; Luo, Z.; Wang, X. Knockdown of ISOC1 suppresses cell proliferation in pancreatic cancer in vitro. Oncol. Lett. 2019, 17, 4263–4270. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhao, L.; Wang, F.; Bai, H.; Li, J.; Li, M.; Hu, X.; Cao, J.; Wang, G. Knockdown of ISOC1 inhibits the proliferation and migration and induces the apoptosis of colon cancer cells through the AKT/GSK-3β pathway. Carcinogenesis 2020, 41, 1123–1133. [Google Scholar] [CrossRef]
- Zhao, J.Q.; Li, X.N.; Fu, L.P.; Zhang, N.; Cai, J.H. ISOC1 promotes the proliferation of gastric cancer cells by positively regulating CDK19. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11602–11609. [Google Scholar] [PubMed]
- Shi, J.; Yang, F.; Zhou, N.; Jiang, Y.; Zhao, Y.; Zhu, J.; Prelaj, A.; Malhotra, J.; Normanno, N.; Danese, E.; et al. Isochorismatase domain-containing protein 1 (ISOC1) participates in DNA damage repair and inflammation-related pathways to promote lung cancer development. Transl. Lung Cancer Res. 2021, 10, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Jia, A.; Wang, Y.; Wang, Y.; Li, Y.; Yang, Q.; Cao, Y.; He, Y.; Dong, L.; Dong, Y.; Hou, Y.; et al. The kinase AKT1 potentiates the suppressive functions of myeloid-derived suppressor cells in inflammation and cancer. Cell Mol. Immunol. 2021, 18, 1074–1076. [Google Scholar] [CrossRef]
- Yong, H.; Wu, G.; Chen, J.; Liu, X.; Bai, Y.; Tang, N.; Liu, L.; Wei, J. lncRNA MALAT1 Accelerates Skeletal Muscle Cell Apoptosis and Inflammatory Response in Sepsis by Decreasing BRCA1 Expression by Recruiting EZH2. Mol. Ther. Nucleic Acids 2020, 19, 97–108. [Google Scholar] [CrossRef]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bulow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef]
- Smith, J.J.; Aitchison, J.D. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 2013, 14, 803–817. [Google Scholar] [CrossRef]
- Yang, J.; Cheng, M.; Gu, B.; Wang, J.; Yan, S.; Xu, D. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 2020, 11, 833. [Google Scholar] [CrossRef]
- Jin, L.; Chen, C.; Li, Y.; Yuan, F.; Gong, R.; Wu, J.; Zhang, H.; Kang, B.; Yuan, G.; Zeng, H.; et al. A Biodegradable Mg-Based Alloy Inhibited the Inflammatory Response of THP-1 Cell-Derived Macrophages Through the TRPM7-PI3K-AKT1 Signaling Axis. Front. Immunol. 2019, 10, 2798. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Lv, P.; Huang, J.; Ke, J.; Yan, J. GYY4137 exhibits anti-atherosclerosis effect in apolipoprotein E (−/−) mice via PI3K/Akt and TLR4 signalling. Clin. Exp. Pharmacol. Physiol. 2020, 47, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Dunster, K.; Lai, F.P.L.; Sentry, J.W. Limkain b1, a novel human autoantigen localized to a subset of ABCD3 and PXF marked peroxisomes. Clin. Exp. Immunol. 2005, 140, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Ha, Z.; Di Pietro, E.; Nichol, J.N.; Bolt, A.M.; Goncalves, C.; Dupéré-Richer, D.; Pettersson, F.; Mann, K.K.; Braverman, N.E.; et al. Peroxisomes protect lymphoma cells from HDAC inhibitor-mediated apoptosis. Cell Death Differ. 2017, 24, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.P.; Xu, Z.; Hou, S.; Limonta, D.; Kumar, A.; Power, C.; Hobman, T.C. Interplay between Zika Virus and Peroxisomes during Infection. Cells 2019, 8, 725. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Jiang, Y.; Zhou, H.; Li, A.; Wei, Y.; Bao, Z.; Wang, D.; Zhao, J.; Chen, X.; et al. Tegaserod Maleate Inhibits Esophageal Squamous Cell Carcinoma Proliferation by Suppressing the Peroxisome Pathway. Front. Oncol. 2021, 11, 683241. [Google Scholar] [CrossRef]
- Xu, Z.; Asahchop, E.L.; Branton, W.G.; Gelman, B.B.; Power, C.; Hobman, T.C. MicroRNAs upregulated during HIV infection target peroxisome biogenesis factors: Implications for virus biology, disease mechanisms and neuropathology. PLoS Pathog. 2017, 13, e1006360. [Google Scholar] [CrossRef]
- Esmaeili, M.; Ghaedi, K.; Shoaraye Nejati, A.; Nematollahi, M.; Shiralyian, H.; Nasr-Esfahani, M.H. Pioglitazone significantly prevented decreased rate of neural differentiation of mouse embryonic stem cells which was reduced by Pex11β knock-down. Neuroscience 2016, 312, 35–47. [Google Scholar] [CrossRef]
- Ahlemeyer, B.; Gottwald, M.; Baumgart-Vogt, E. Deletion of a single allele of the Pex11β gene is sufficient to cause oxidative stress, delayed differentiation and neuronal death in mouse brain. Dis. Model Mech. 2012, 5, 125–140. [Google Scholar] [CrossRef]
- Asare, A.; Levorse, J.; Fuchs, E. Coupling organelle inheritance with mitosis to balance growth and differentiation. Science 2017, 355, eaah4701. [Google Scholar] [CrossRef]
- Angermuller, S.; Islinger, M.; Volkl, A. Peroxisomes and reactive oxygen species, a lasting challenge. Histochem. Cell Biol. 2009, 131, 459–463. [Google Scholar] [CrossRef]
- Hideshima, T.; Catley, L.; Yasui, H.; Ishitsuka, K.; Raje, N.; Mitsiades, C.; Podar, K.; Munshi, N.C.; Chauhan, D.; Richardson, P.G.; et al. Perifosine, an oral bioactive novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood 2006, 107, 4053–4062. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, D.; Hideshima, T.; Rodig, S.; Santo, L.; Pozzi, S.; Vallet, S.; Ikeda, H.; Perrone, G.; Gorgun, G.; Patel, K.; et al. Dual inhibition of akt/mammalian target of rapamycin pathway by nanoparticle albumin-bound-rapamycin and perifosine induces antitumor activity in multiple myeloma. Mol. Cancer Ther. 2010, 9, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Suzuki, T.; Okamura, T.; Kurita, A.; Nohara, G.; Ishii, S.; Kado, S.; Takagi, A.; Tsugane, M.; Shishido, Y. Perifosine, a Bioavailable Alkylphospholipid Akt Inhibitor, Exhibits Antitumor Activity in Murine Models of Cancer Brain Metastasis Through Favorable Tumor Exposure. Front. Oncol. 2021, 11, 754365. [Google Scholar] [CrossRef]
- Jo, H.; Mondal, S.; Tan, D.; Nagata, E.; Takizawa, S.; Sharma, A.K.; Hou, Q.; Shanmugasundaram, K.; Prasad, A.; Tung, J.K.; et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl. Acad. Sci. USA 2012, 109, 10581–10586. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Li, X.; Yin, M.; Wang, H.; Qin, L.; Li, H.; Liu, W.; Zhao, Z.; Zhao, Q.; Chen, H.; et al. Selective autophagy receptor SQSTM1/ p62 inhibits Seneca Valley virus replication by targeting viral VP1 and VP3. Autophagy 2021, 17, 3763–3775. [Google Scholar] [CrossRef]
- Hung, Y.L.; Fang, S.H.; Wang, S.C.; Cheng, W.C.; Liu, P.L.; Su, C.C.; Chen, C.S.; Huang, M.Y.; Hua, K.F.; Shen, K.H.; et al. Corylin protects LPS-induced sepsis and attenuates LPS-induced inflammatory response. Sci. Rep. 2017, 7, 46299. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Ahn, S.H.; Deshmukh, H.; Johnson, N.; Cowell, L.G.; Rude, T.H.; Scott, W.K.; Nelson, C.L.; Zaas, A.K.; Marchuk, D.A.; Keum, S.; et al. Two genes on A/J chromosome 18 are associated with susceptibility to Staphylococcus aureus infection by combined microarray and QTL analyses. PLoS Pathog. 2010, 6, e1001088. [Google Scholar] [CrossRef]
- Delille, H.K.; Agricola, B.; Guimaraes, S.C.; Borta, H.; Luers, G.H.; Fransen, M.; Schrader, M. Pex11pbeta-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J. Cell Sci. 2010, 123, 2750–2762. [Google Scholar] [CrossRef]
- Ganguli, G.; Pattanaik, K.P.; Jagadeb, M.; Sonawane, A. Mycobacterium tuberculosis Rv3034c regulates mTORC1 and PPAR-gamma dependant pexophagy mechanism to control redox levels in macrophages. Cell Microbiol. 2020, 22, e13214. [Google Scholar] [CrossRef]
- Mao, S.; Luo, X.; Li, Y.; He, C.; Huang, F.; Su, C. Role of PI3K/AKT/mTOR Pathway Associated Oxidative Stress and Cardiac Dysfunction in Takotsubo Syndrome. Curr. Neurovasc. Res. 2020, 17, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lu, Y.C.; Wang, J.; Wang, L.; Yu, H.; Li, Y.F.; Kong, A.; Chan, J.; Lee, S. Type 2 Diabetes Promotes Cell Centrosome Amplification via AKT-ROS-Dependent Signalling of ROCK1 and 14–3-3σ. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 47, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S.; Trzyna, W.C.; McClintock, D.S.; Schumacker, P.T. Role of Oxidants in NF-κB Activation and TNF-α Gene Transcription Induced by Hypoxia and Endotoxin. J. Immunol. 2000, 165, 1013. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Yao, A.; Liu, F.; Chen, K.; Tang, L.; Liu, L.; Zhang, K.; Yu, C.; Bian, G.; Guo, H.; Zheng, J.; et al. Programmed death 1 deficiency induces the polarization of macrophages/microglia to the M1 phenotype after spinal cord injury in mice. Neurotherapeutics 2014, 11, 636–650. [Google Scholar] [CrossRef]
- Ebberink, M.S.; Koster, J.; Visser, G.; Spronsen, F.; Stolte-Dijkstra, I.; Smit, G.P.; Fock, J.M.; Kemp, S.; Wanders, R.J.; Waterham, H.R. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11beta gene. J. Med. Genet. 2012, 49, 307–313. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Zhao, Q.; Fu, B.; Xiong, Y.; Zhang, S.; Xu, S.; Wu, H. ISOC1 Modulates Inflammatory Responses in Macrophages through the AKT1/PEX11B/Peroxisome Pathway. Molecules 2022, 27, 5896. https://doi.org/10.3390/molecules27185896
Lin X, Zhao Q, Fu B, Xiong Y, Zhang S, Xu S, Wu H. ISOC1 Modulates Inflammatory Responses in Macrophages through the AKT1/PEX11B/Peroxisome Pathway. Molecules. 2022; 27(18):5896. https://doi.org/10.3390/molecules27185896
Chicago/Turabian StyleLin, Xiaoyuan, Qingting Zhao, Beibei Fu, Yan Xiong, Shanfu Zhang, Shiyao Xu, and Haibo Wu. 2022. "ISOC1 Modulates Inflammatory Responses in Macrophages through the AKT1/PEX11B/Peroxisome Pathway" Molecules 27, no. 18: 5896. https://doi.org/10.3390/molecules27185896
APA StyleLin, X., Zhao, Q., Fu, B., Xiong, Y., Zhang, S., Xu, S., & Wu, H. (2022). ISOC1 Modulates Inflammatory Responses in Macrophages through the AKT1/PEX11B/Peroxisome Pathway. Molecules, 27(18), 5896. https://doi.org/10.3390/molecules27185896





