Effects of Natural Rheum tanguticum on the Cell Wall Integrity of Resistant Phytopathogenic Pectobacterium carotovorum subsp. Carotovorum
Abstract
:1. Introduction
2. Results and Discussion
2.1. UPLC-ESI-MS Analysis of RTMB Active Ingredients
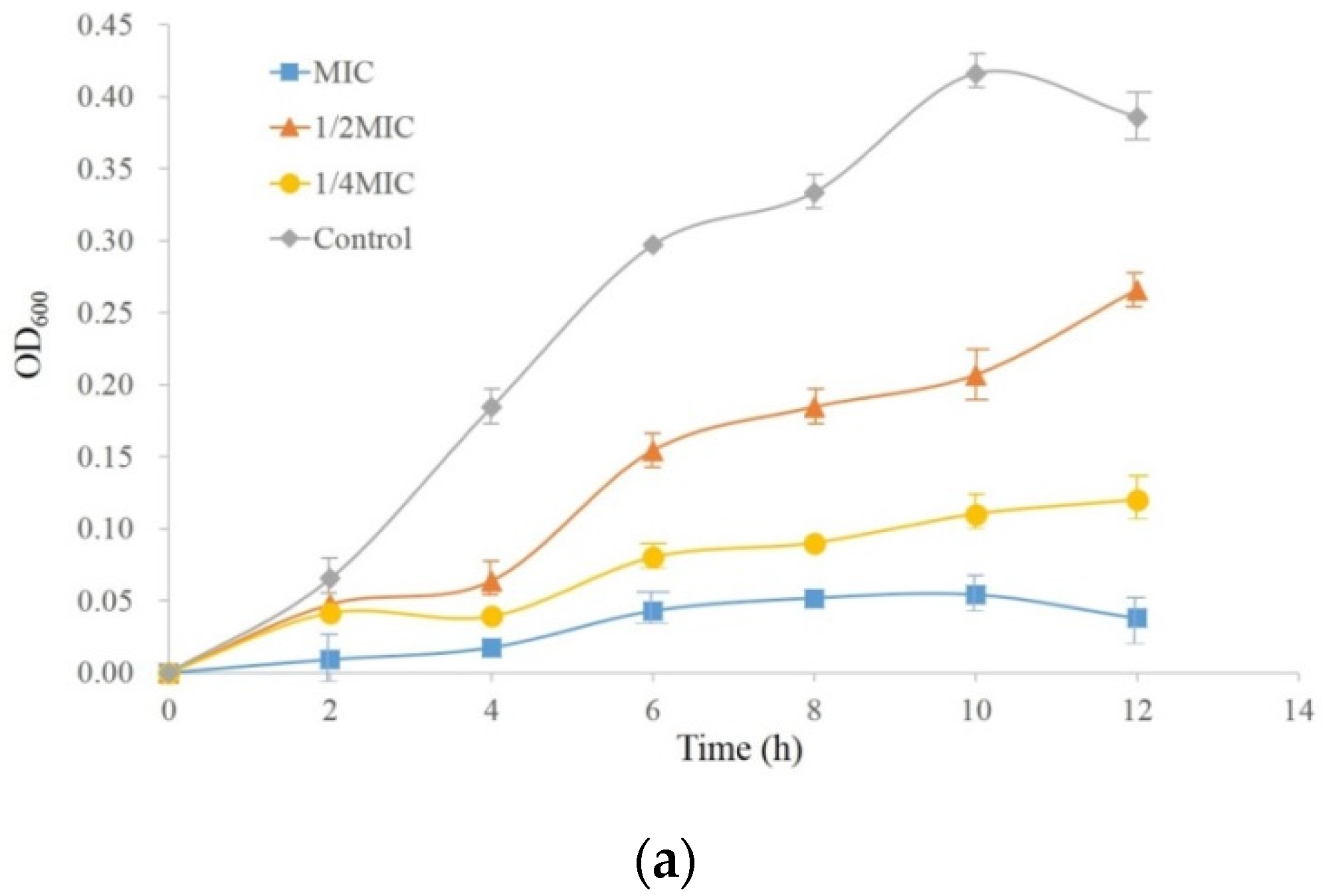
2.2. The MIC and the Growth Curves of PccS1
2.3. Fourier Transform Infrared Spectrophotometer (FTIR) Analysis
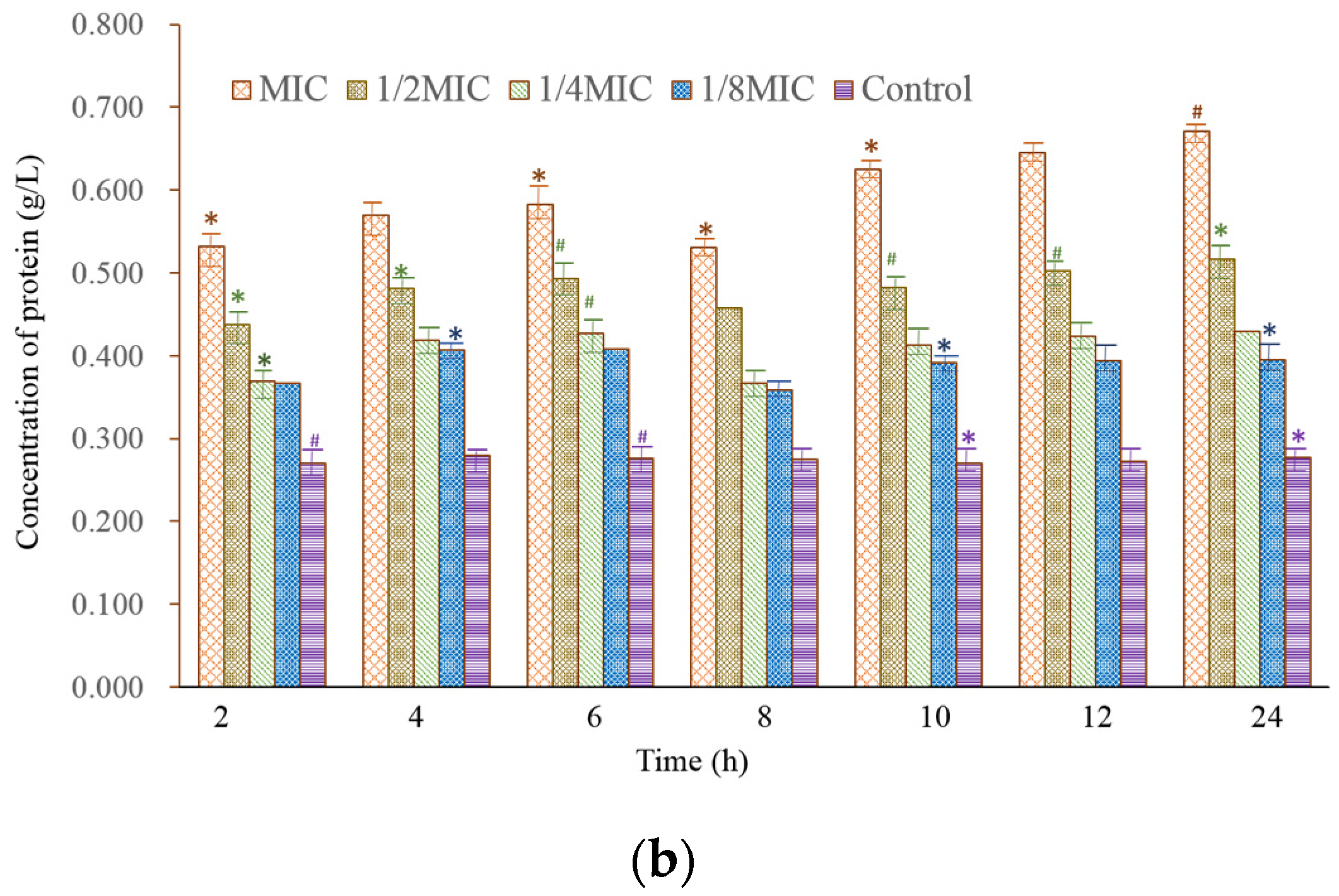
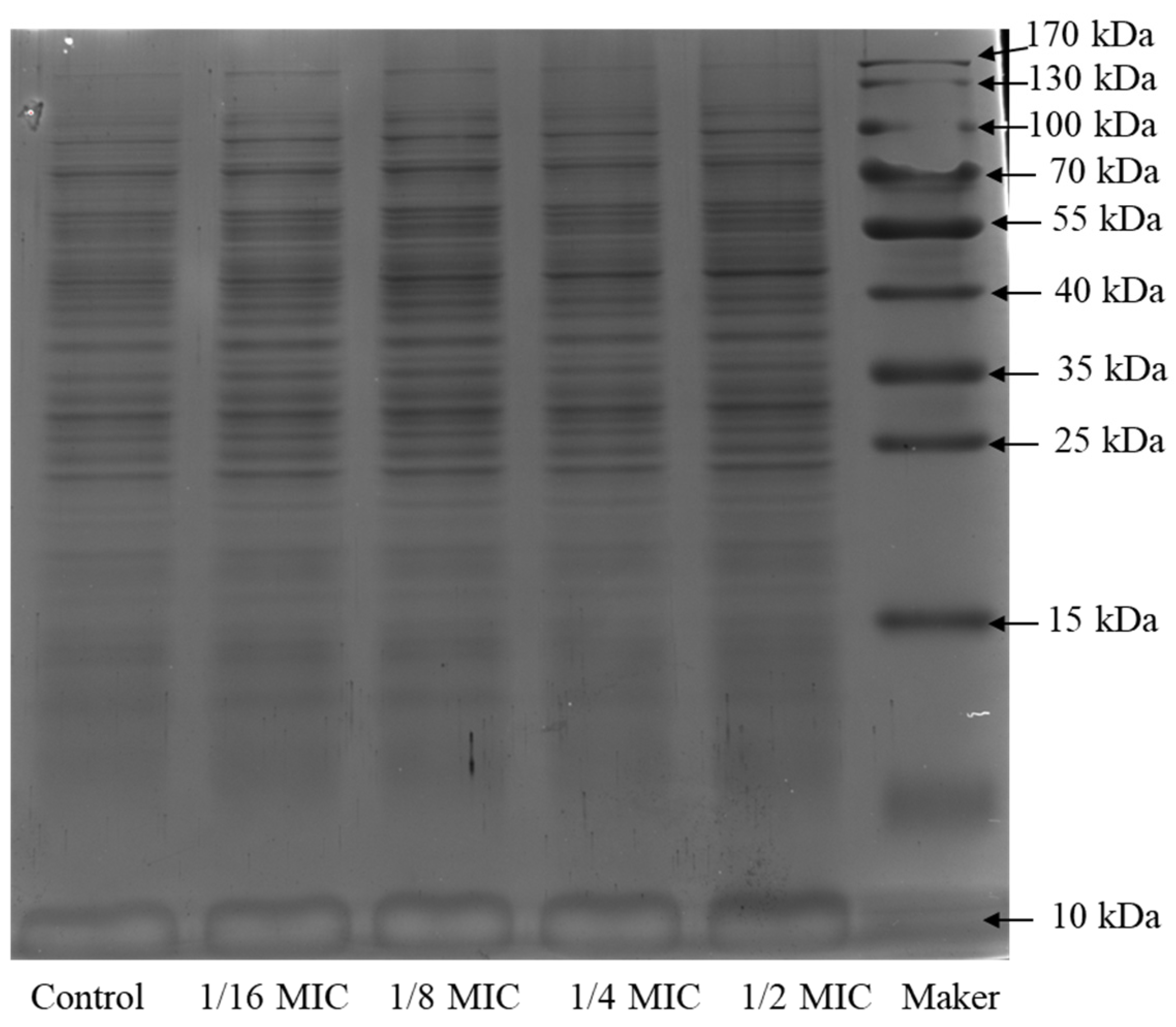
2.4. SDS-PAGE Electrophoresis of the Bacterial Protein
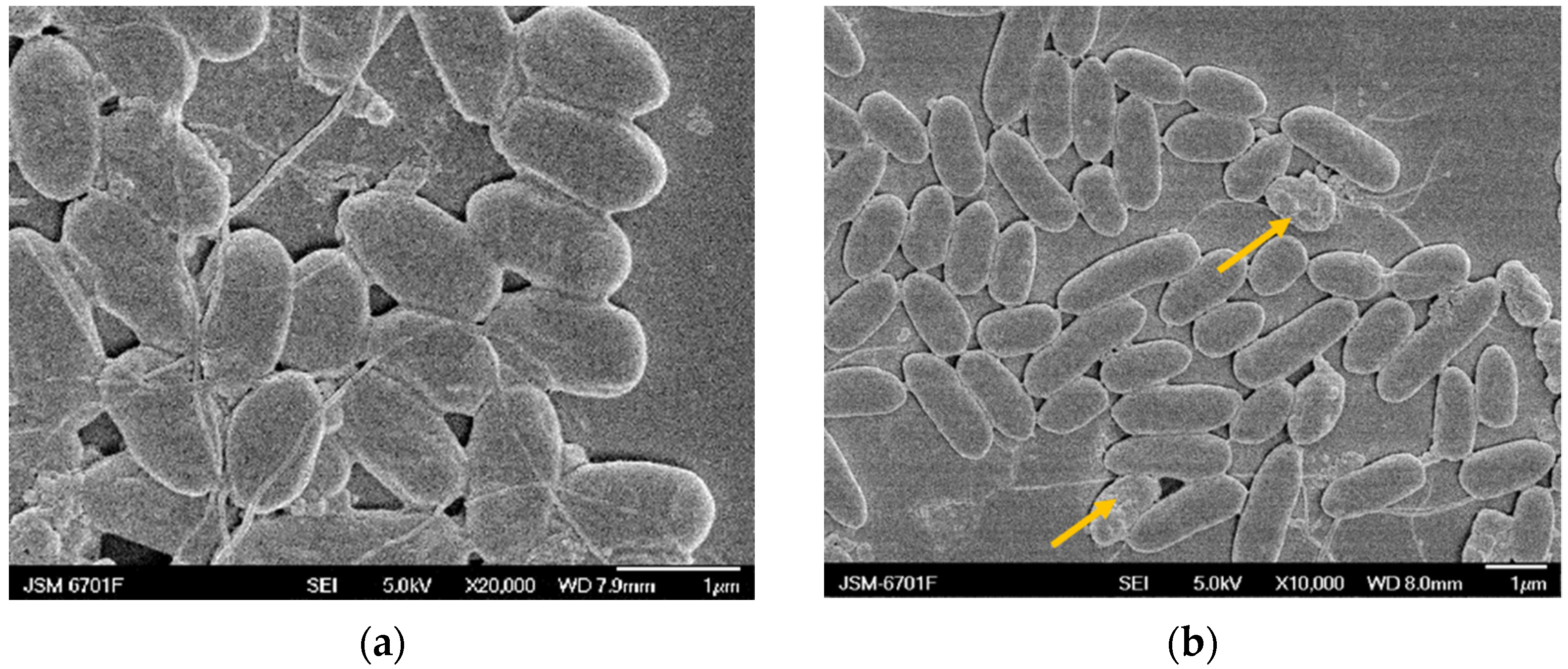
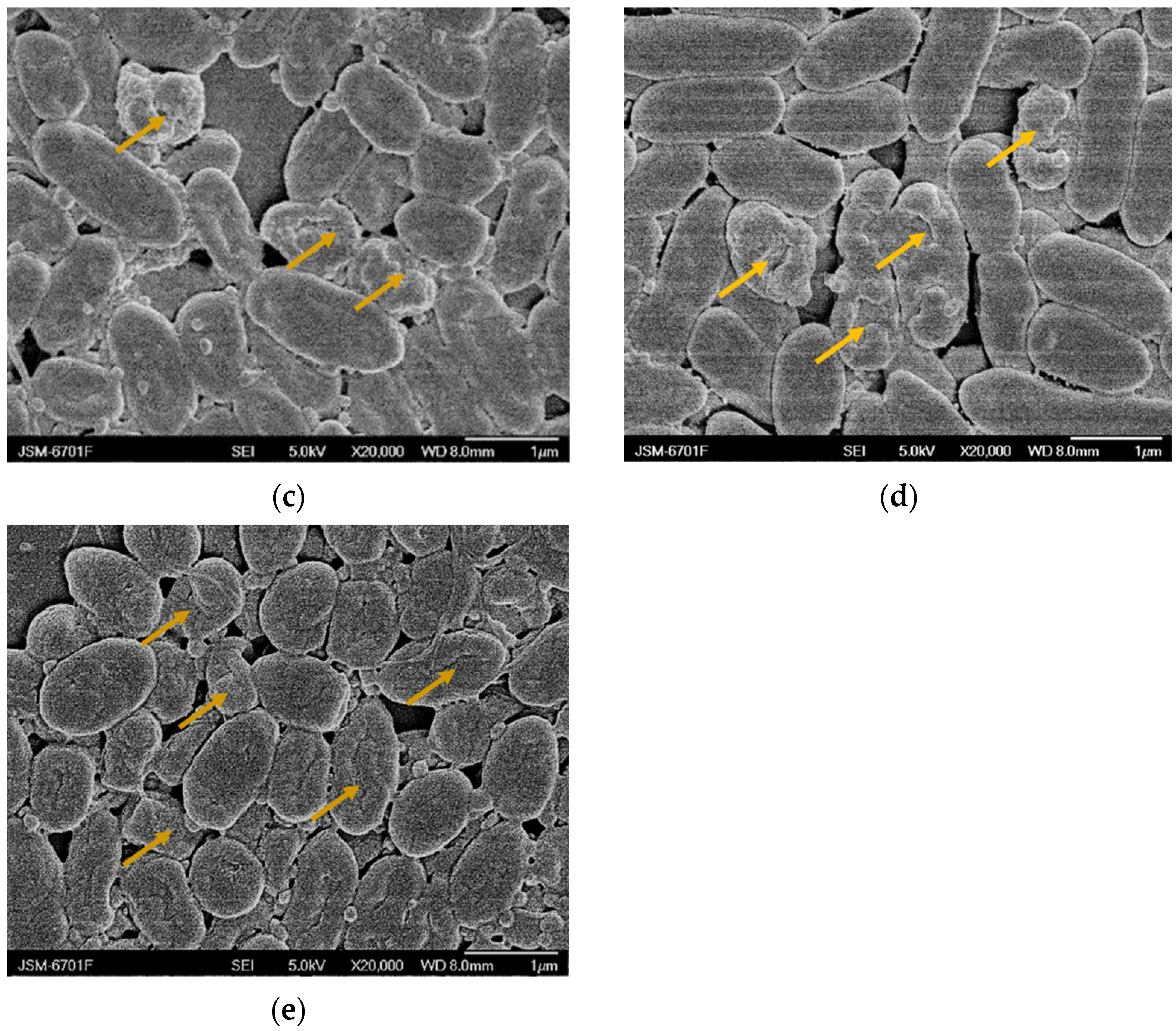
2.5. Scanning Electron Microscopy (SEM) Analysis
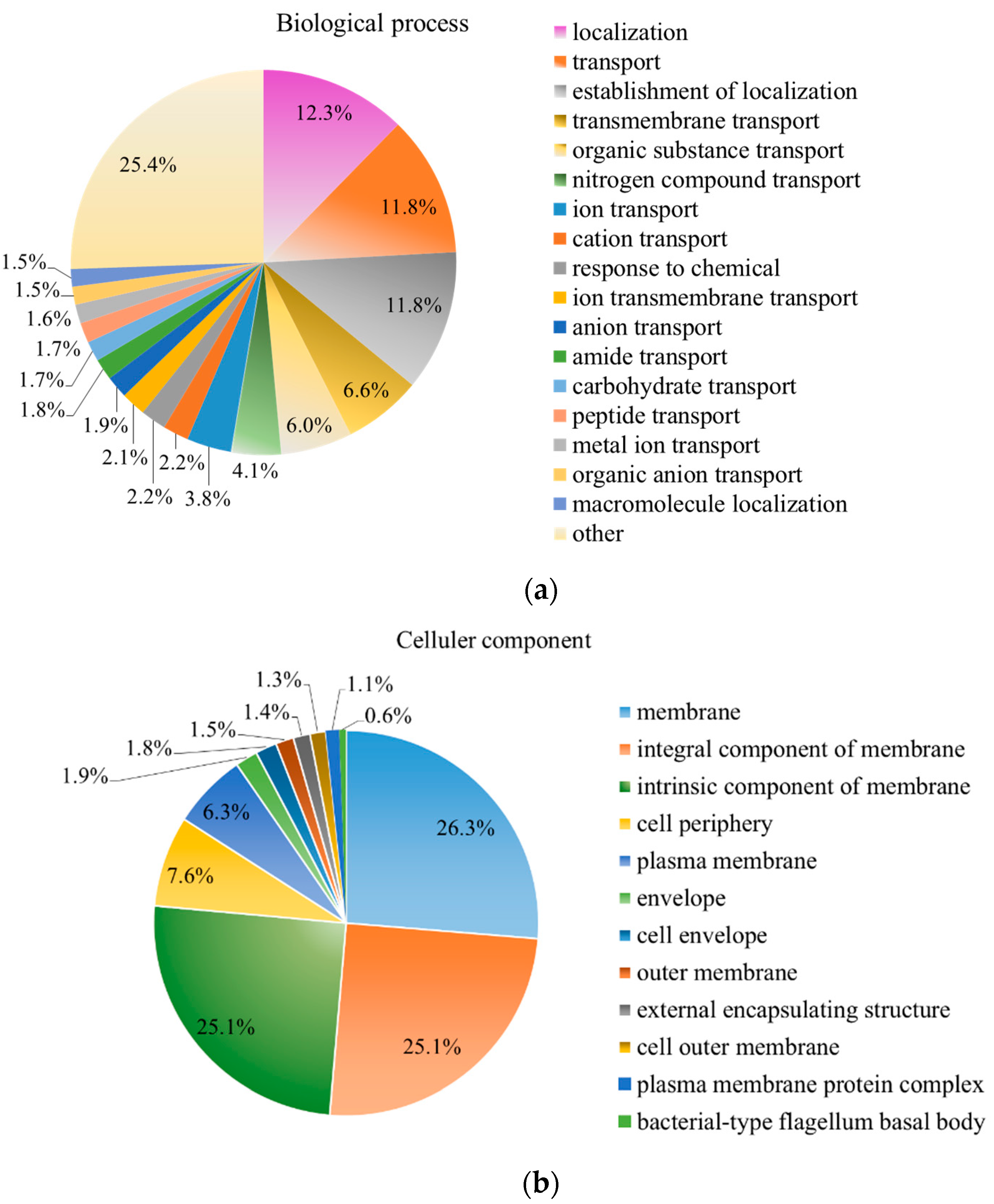
2.6. Proteomic Analysis
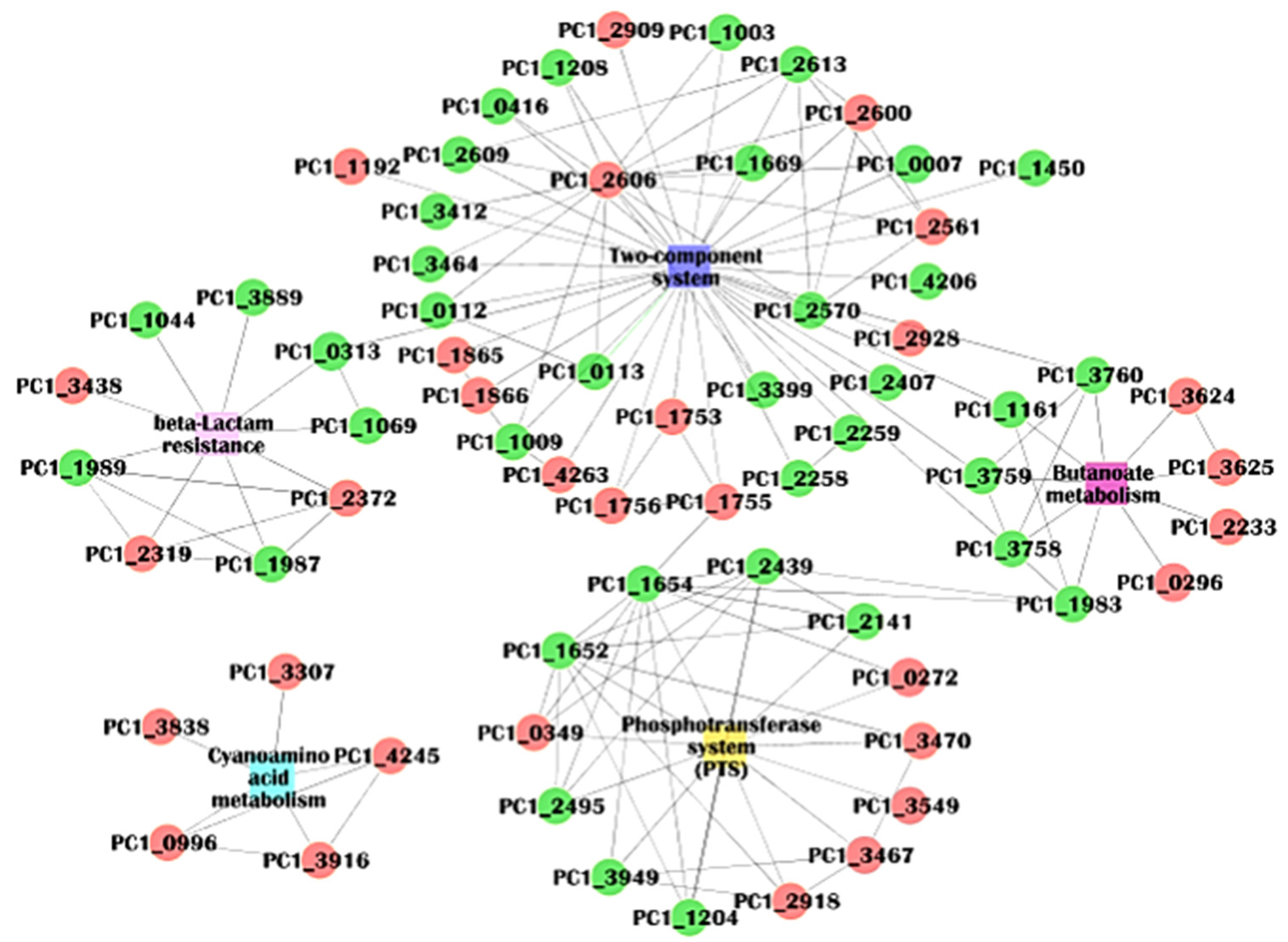
2.7. Protein–Protein Interactions (PPI) Analysis
3. Materials and Methods
3.1. Chemical Reagents, Bacterium and Growth Condition
3.2. Preparation of the Extract of the Natural Medicine
3.3. Ultra-Performance Liquid Chromatography-Electrospray-Mass Spectrometry (UPLC-ESI-MS) Analysis
3.4. The Minimum Inhibitory Concentration (MIC)
3.5. Measurement of Antibacterial Curves
3.6. Measurement of Extracellular Protein Content
3.7. SDS-PAGE Analysis of Bacterial Intracellular Proteins
3.8. Fourier Transform Infrared Spectrophotometer (FTIR) Analysis
3.9. Scanning Electron Microscopy (SEM)
3.10. Protein Digestion and LC-MS/MS Analysis
3.11. Bioinformatics Analysis
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Mdel, P.; Marquez-Villavicencio, B.; Weber, R.A.; Witherell, D.K.; Willis, A.O.; Charkowski, A.O. The 3-hydroxy-2-butanone pathway is required for Pectobacterium carotovorum pathogenesis. PLoS ONE 2011, 6, e22974. [Google Scholar]
- Ji, Z.; Ji, C.; Liu, B.; Zou, L.; Chen, G.; Yang, B. Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 2016, 7, 13435. [Google Scholar] [CrossRef]
- Lipsky, A.; Cohen, A.; Ion, A.; Yedidia, I. Genetic transformation of Ornithogalum via particle bombardment and generation of Pectobacterium carotovorum-resistant plants. Plant Sci. 2014, 228, 150–158. [Google Scholar] [CrossRef]
- El-Zaemey, S.; Heyworth, J.; Fritschi, L. Noticing pesticide spray drift from agricultural pesticide application areas and breast cancer: A case-control study. Aust. N. Z. J. Public Health 2013, 37, 547–555. [Google Scholar] [CrossRef]
- Verger, P.; Agamy, N.; Anshasi, M.; Al-Yousfi, A.B. Occurrence of pesticide residues in fruits and vegetables for the Eastern Mediterranean Region and potential impact on public health. Food Control 2021, 119, 107457. [Google Scholar]
- Kim, K.H.; Kabir, E.; Jahan, S.A. Exposure to pesticides and the associated human health effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef]
- Rangasamy, K.; Athiappan, M.; Devarajan, N.; Samykannu, G.; Parray, J.A.; Aruljothi, K.N.; Shameem, N.; Alqarawi, A.A.; Hashem, A.; Abd Allah, E.F. Pesticide degrading natural multidrug resistance bacterial flora. Microb. Pathog. 2018, 114, 304–310. [Google Scholar] [CrossRef]
- Bortoli-German, I.; Brun, E.; Py, B.; Chippaux, M.; Barras, F. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol. 1994, 11, 545–553. [Google Scholar] [CrossRef]
- Toth, I.K.; Bell, K.S.; Holeva, M.C.; Birch, P.R.J. Soft rot erwiniae: From genes to genomes. Mol. Plant Pathol. 2003, 4, 17–30. [Google Scholar] [CrossRef]
- Wang, C.; Pu, T.; Lou, W.; Wang, Y.; Gao, Z.; Hu, B.; Fan, J. Hfq, a RNA Chaperone, Contributes to Virulence by Regulating Plant Cell Wall-Degrading Enzyme Production, Type VI Secretion System Expression, Bacterial Competition, and Suppressing Host Defense Response in Pectobacterium carotovorum. Mol. Plant Microbe Interact. 2018, 31, 1166–1178. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Z.; Du, S.; Ma, L.; Liao, Y.; Wang, Y.; Toth, I.; Fan, J. Characterization of Pectobacterium carotovorum proteins differentially expressed during infection of Zantedeschia elliotiana in vivo and in vitro which are essential for virulence. Mol. Plant Pathol. 2018, 19, 35–48. [Google Scholar] [CrossRef]
- Al-Huqail, A.A.; Behiry, S.I.; Salem, M.Z.M.; Ali, H.M.; Siddiqui, M.H.; Salem, A.Z.M. Antifungal, antibacterial, and antioxidant activities of Acacia Saligna (Labill.) H. L. Wendl. flower extract: HPLC analysis of phenolic and flavonoid compounds. Molecules 2019, 24, 700. [Google Scholar] [CrossRef]
- El-Hefny, M.; Ashmawy, N.A.; Salem, M.Z.M.; Salem, A.Z.M. Antibacterial activities of the phytochemicals-characterized extracts of Callistemon viminalis, Eucalyptus camaldulensis and Conyza dioscoridis against the growth of some phytopathogenic bacteria. Microb. Pathog. 2017, 113, 348–356. [Google Scholar] [CrossRef]
- Ashmawy, N.A.; Salem, M.Z.M.; El-Hefny, M.; Abd El-Kareem, M.S.M.; El-Shanhorey, N.A.; Mohamed, A.A.; Salem, A.Z.M. Antibacterial activity of the bioactive compounds identified in three woody plants against some pathogenic bacteria. Microb. Pathog. 2018, 121, 331–340. [Google Scholar] [CrossRef]
- Jin, W.; Ge, R.L.; Wei, Q.J.; Bao, T.Y.; Shi, H.M.; Tu, P.F. Development of high-performance liquid chromatographic fingerprint for the quality control of Rheum tanguticum Maxim. ex Balf. J. Chromatogr. A. 2006, 1132, 320–324. [Google Scholar] [CrossRef]
- Chen, D.; Wang, L. Mechanisms of therapeutic effects of rhubarb on gut origin sepsis. Chin. J. Traumatol. 2009, 12, 365–369. [Google Scholar]
- Espinosa, A.; Paz-Y-Miño-C, G.; Santos, Y.; Ma, H.; Nadeau, M.; Seeram, N.P.; Rowley, D.C. Anti-amebic effects of Chinese rhubarb (Rheum palmatum) leaves’ extract, the anthraquinone rhein and related compounds. Heliyon 2020, 6, e03693. [Google Scholar] [CrossRef]
- Xiang, H.; Zuo, J.; Guo, F.; Dong, D. What we already know about rhubarb: A comprehensive review. Chin. Med. 2020, 15, 88. [Google Scholar] [CrossRef]
- Dou, Z.; Dai, Y.; Zhou, Y.; Wang, S. Quality evaluation of rhubarb based on qualitative analysis of the HPLC fingerprint and UFLC-Q-TOF-MS/MS combined with quantitative analysis of eight anthraquinone glycosides by QAMS. Biomed. Chromatogr. 2021, 35, e5074. [Google Scholar] [CrossRef]
- Kashiwada, Y.; Nonaka, G.; Nishioka, I. Studies on rhubarb (RheiRhizoma). XV. Simultaneous determination of phenolic constituents by high-performance liquid chromatography. Chem. Pharm. Bull. 1989, 37, 999–1004. [Google Scholar] [CrossRef]
- Jin, W.; Wang, Y.F.; Ge, R.L.; Shi, H.M.; Jia, C.Q.; Tu, P.F. Simultaneous analysis of multiple bioactive constituents in Rheum tanguticum Maxim. ex Balf. by high-performance liquid chromatography coupled to tandem mass spectrometry. Rapid Commun. Mass Sp. 2007, 21, 2351–2360. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Wu, C.; Lin, T.; Sheu, S. Determination of 19 rhubarb constituents by high-performance liquid chromatography-ultraviolet-mass spectrometry. J. Sep. Sci. 2006, 29, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.L.; Guo, T.; Xu, X.D.; Yang, J.S. Rapid identification and simultaneous analysis of multiple constituents from Rheum tanguticum Maxim. ex Balf. by UPLC/Q-TOF-MS. Nat. Prod. Res. 2017, 31, 1529–1535. [Google Scholar] [CrossRef]
- Komatsu, K.; Nagayama, Y.; Tanaka, K.; Ling, Y.; Basnet, P.; Meselhy, M. Development of a high performance liquid chromatographic method for systemtic quantitative analysis of chemical constituents in rhubarb. Chem. Pharm. Bull. 2006, 54, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhang, B.; Liu, Y.; Zhang, H.; Ding, G.T.; Qi, Y.J. Analysis of potential antibacterial components of Rheum tanguticum Maxim. ex Balf. based on spectral-activity relationship. Acta. Agric. Univ. Jiangxiensis 2022, 44, 593–604. [Google Scholar]
- Friedman, M.; Xu, A.; Lee, R.; Nguyen, D.N.; Phan, T.A.; Hamada, S.M.; Panchel, R.; Tam, C.C.; Kim, J.H.; Cheng, L.W.; et al. The inhibitory activity of anthraquinones against pathogenic protozoa, bacteria, and fungi and the relationship to structure. Molecules 2020, 25, 3101. [Google Scholar] [CrossRef]
- Yang, K.; Jin, M.J.; Quan, Z.S.; Piao, H.R. Design and synthesis of novel anti-proliferative emodin derivatives and studies on their cell cycle arrest, apoptosis pathway and migration. Molecules 2019, 24, 884. [Google Scholar] [CrossRef]
- Kumar, G.D.; Siva, B.; Vadlamudi, S.; Bathula, S.R.; Dutta, H.; Suresh Babu, K. Design, synthesis, and biological evaluation of pyrazole-linked aloe emodin derivatives as potential anticancer agents. RSC Med. Chem. 2021, 12, 791–796. [Google Scholar] [CrossRef]
- Park, C.G.; Kim, J.J.; Kim, H.K. Lipase-mediated synthesis of ricinoleic acid vanillyl ester and evaluation of antioxidant and antibacterial activity. Enzyme Microb. Tech. 2020, 133, 109454. [Google Scholar] [CrossRef]
- Kwiatkowski, P.; Pruss, A.; Wojciuk, B.; Dołęgowska, B.; Wajs-Bonikowska, A.; Sienkiewicz, M.; Mężyńska, M.; Łopusiewicz, Ł. The influence of essential oil compounds on antibacterial activity of mupirocin-susceptible and induced low-level mupirocin-resistant MRSA Strains. Molecules 2019, 24, 3105. [Google Scholar] [CrossRef]
- Yang, D.; Castro, D.J.; el-Sayed, I.H.; el-Sayed, M.A.; Saxton, R.E.; Zhang, N.Y. A Fourier transform infrared spectroscopic comparison of cultured human fibroblast and fibrosarcoma cells: A new method for detection of malignancies. J. Clin. Laser Med. Sur. 1995, 13, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Hackshaw, K.V.; Miller, J.S.; Aykas, D.P.; Rodriguez-Saona, L. Vibrational Spectroscopy for Identification of Metabolites in Biologic Samples. Molecules 2020, 25, 4725. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, X.; Li, J.; Zhang, Q.; Li, X.; An, Q.; Ye, X.; Zhao, Z.; Cai, L.; Han, Y.; et al. Antibacterial mechanism of the synergistic combination between streptomycin and alcohol extracts from the Chimonanthus salicifolius S. Y. Hu. Leaves. J. Ethnopharmacol. 2020, 250, 112467. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Kang, S.; Yin, Z.; Jia, R.; Song, X.; Li, L.; Li, Z.; Zou, Y.; Liang, X.; Li, L.; et al. Antibacterial activity and mechanism of berberine against Streptococcus agalactiae. Int. J. Clin. Exp. Pathol. 2015, 8, 5217–5223. [Google Scholar] [PubMed]
- Xu, C.; Li, J.; Yang, L.; Shi, F.; Yang, L.; Ye, M. Antibacterial activity and a membrane damage mechanism of Lachnum YM30 melanin against Vibrio parahaemolyticus and Staphylococcus aureus. Food Control 2017, 73, 1445–1451. [Google Scholar] [CrossRef]
- Coenye, T. Social interactions in the Burkholderia cepacia complex: Biofilms and quorum sensing. Future Microbiol. 2010, 5, 1087–1099. [Google Scholar] [CrossRef]
- Xu, M.; Xue, H.; Li, X.; Zhao, Y.; Lin, L.; Yang, L.; Zheng, G. Chemical composition, antibacterial properties and mechanism of Smilax china L. polyphenols. Appl. Microbiol. Biotechnol. 2019, 103, 9013–9022. [Google Scholar] [CrossRef]
- Poole, K. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 2004, 61, 2200–2223. [Google Scholar] [CrossRef]
- Li, L.; Ge, H.; Gu, D.; Meng, H.; Li, Y.; Jia, M.; Zheng, C.; Zhou, X. The role of two-component regulatory system in β-lactam antibiotics resistance. Microbiol. Res. 2018, 215, 126–129. [Google Scholar]
- Baucheron, S.; Mouline, C.; Praud, K.; Chaslus-Dancla, E.; Cloeckaert, A. TolC but not AcrB is essential for multidrug-resistant Salmonella enterica serotype Typhimurium colonization of chicks. J. Antimicrob. Chemother. 2005, 55, 707–712. [Google Scholar] [CrossRef]
- Dörr, T.; Lam, H.; Alvarez, L.; Cava, F.; Davis, B.M.; Waldor, M.K. A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in vibrio cholera. PLoS Genet. 2014, 10, e1004433. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ying, J.; Yang, G.; Li, A.; Wang, J.; Lu, J.; Wang, J.; Xu, T.; Yi, H.; Li, K.; et al. Structure-Function Analysis of the Transmembrane Protein AmpG from Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0168060. [Google Scholar] [CrossRef] [PubMed]
- Esquivel, R.N.; Pohlschroder, M. A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility. Mol. Microbiol. 2014, 93, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Q.; Ma, L.; Zhao, C.D.; Yan, J.Y.; Che, S.; Zhou, Z.W.; Wang, H.; Yang, L.K.; Hu, B.S. Transcriptome of pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol. Plant Pathol. 2020, 21, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, C.Y. Generally stressed out bacteria: Environmental stress response mechanisms in gram-positive bacteria. Integr. Comp. Biol. 2020, 60, 126–133. [Google Scholar] [CrossRef]
- Hirakawa, H.; Kurushima, J.; Hashimoto, Y.; Tomita, H. Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics 2020, 9, 635. [Google Scholar] [CrossRef]
- Kim, H.S.; Thammarat, P.; Lommel, S.A.; Hogan, C.S.; Charkowski, A.O. Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant-microbe interactions. Mol. Plant Microbe Interact. 2011, 24, 773–786. [Google Scholar] [CrossRef]
- Schoeters, F.; Van Dijck, P. Protein-Protein Interactions in Candida albicans. Front Microbiol. 2019, 10, 1792. [Google Scholar] [CrossRef]
- Koeth, L.M.; DiFranco-Fisher, J.M.; McCurdy, S. A reference broth microdilution method for dalbavancin in vitro susceptibility testing of bacteria that grow aerobically. J. Vis. Exp. 2015, 103, 53028. [Google Scholar] [CrossRef]
- Reddy, G.M.; Garcia, J.R.; Reddy, V.H.; de Andrade, A.M.; Camilo, A., Jr.; Pontes Ribeiro, R.A.; de Lazaro, S.R. Synthesis, antimicrobial activity and advances in structure-activity relationships (SARs) of novel tri-substituted thiazole derivatives. Eur. J. Med. Chem. 2016, 23, 508–513. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Luber, C.A.; Cox, J.; Lauterbach, H.; Fancke, B.; Selbach, M.; Tschopp, J.; Akira, S.; Wiegand, M.; Hochrein, H.; O’Keeffe, M.; et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32, 279–289. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P. The perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/ Swiss-Prot, the manually annotated section of the UniProt knowledge base: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, 109–114. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar]




| No. | RT (min) | Formula | Measured Mass [M-H]− (m/z) | Theoretical Mass (m/z) | MS/MS | Identification | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 0.66 | C13H16O10 | 331.0638 | 332.0638 | 125.0208,169.0153 | Gallic acid-3-O-glucosidea | [19] |
| 2 | 1.96 | C13H16O10 | 331.0640 | 332.0671 | 125.0617, 169.0345 | Gallic acid-4-O-glucosidea | [19]. |
| 3 | 7.05 | C15H14O6 | 289.0718 | 290.0718 | 271.0652, 179.0033, 161.0523 | catechin | [19] |
| 4 | 7.50 | C30H26O12 | 577.1264 | 578.1352 | 425.0423, 289.0742, 125.0241 | Procyanidin B | [19,20] |
| 5 | 17.03 | C23H26O11 | 477.1453 | 478.1402 | 313.0147, 169.0135 | Lindleyin | [19,21] |
| 6 | 18.09 | C23H26O11 | 477.1453 | 478.1402 | 313.0147, 169.0135 | Isolindleyin | [19,21] |
| 7 | 18.91 | C44H34O20 | 881.1753 | 882.1571 | 729.1460, 559.1257, 541.0923 477.1452, 289.0909 | procyanidin B-2-3,3′-di-O-gllate | [19,24] |
| 8 | 22.44 | C19H22O19 | 393.1192 | 394.4113 | 231.0256 | 6-Hydroxymusizin-8-O-b-d-glucopyranoside | [21,24] |
| 9 | 25.25 | C21H18O11 | 445.0702 | 446.0776 | 283.0281, 239.0978, 211.0514 | Rhein-8-O-glucopyranoside | [19] |
| 10 | 27.72 | C27H26O12 | 541.1352 | 542.4321 | 313.0562, 227.0296, 169.0123 | Resveratrol-4′-O-(2″-galloyl)-b-d-glucopyranoside | [20,23] |
| 11 | 28.75 | C27H26O12 | 541.1353 | 542.1352 | 313.0562, 227.0296, 169.0122 | Resveratrol-4′-O-(6″-galloyl)-b-d-glucopyranoside | [20,23] |
| 12 | 32.86 | C20H24O9 | 407.1348 | 408.3990 | 313.0256, 245.0823, 230.0572 | Torachrysone 8-O-glucoside | [21,22] |
| 13 | 33.79 | C21H20O10 | 431.0928 | 432.0984 | 269.0261, 240.9125, 211.0392 169.0077, 125.0299 | Aloe-emodin-8-O-β-d-glucopyranoside | [19,23] |
| 14 | 35.75 | C21H20O10 | 431.0931 | 432.0984 | 269.0261, 240.0439, 225.0518 | Emodin-1-O-β-d-glucoside | [19] |
| 15 | 42.68 | C21H20O10 | 431.0934 | 432.3780 | 269.0261, 239.0077, 211.0158 | Aloe-emodin-3-(hydroxymethyl)-O-β-d-glucopyranoside | [19] |
| 16 | 44.03 | C21H20O10 | 431.0934 | 432.3780 | 269.0261, 253.0253, 225.0565 169.0077 | Emodin-8-O-β- d-glucoside | [19,23] |
| 17 | 44.95 | C32H32O13 | 623.1873 | 624.1770 | 459.0034, 307.3884, 235.0591 169.0167, 125.0231 | 4-(4-Hydroxyphenyl)-2-butanone-4′-O-β-d-(6″-O-galloyl-2″-O-cinnamoyl)-glucopyranoside | [19] |
| 18 | 45.55 | C32H32O13 | 623.1763 | 624.1770 | 459.0032, 235.0581, 169.0167 | 4-(4-Hydroxyphenyl)-2-butanone-4 -O-β-d-(2″-O-galloyl-6″-O-p-coumaroyl)-glucopyranoside | [19] |
| 19 | 46.82 | C22H22O10 | 445.0703 | 446.1140 | 283.0281, 240.0381, 225.0498 212.0453 | Rhein-8-O-β-d-glucopyranosid | [19,23] |
| 20 | 47.81 | C30H30O15 | 629.1561 | 630.2315 | 465.1231, 313.2586, 169.1358 | 4-(4′-Hydroxyphenyl)-2-butanone-4′-O-b-d-(2″-O-galloyl-6″-O-galloyl)glucopyranoside | [21] |
| 21 | 48.27 | C32H32O12 | 607.1840 | 608.1821 | 443.1002, 295.0456, 169.0345 | 4-(4′-Hydroxyphenyl)-2-butanone-4′-O-b-d-(2″-O-cinnamoyl-6″-O-galloyl)glucopyranoside | [19,21] |
| 22 | 50.78 | C16H12O5 | 283.0274 | 284.0612 | 240.0426 | 6-methyl-aloe emodin | [19] |
| 23 | 53.14 | C15H10O5 | 269.0427 | 270.0445 | 241.0371, 225.0597, 182.0562 | emodin | [19] |
| 24 | 54.17 | C15H10O4 | 253.0505 | 254.0506 | 225.0534, 181.3284 | Chrysophanol | [19] |
| 25 | 55.49 | C16H12O5 | 283.0608 | 284.0612 | 239.1295, 211.0185 | Physcion | [19,23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Y.; Wang, M.; Zhang, B.; Liu, Y.; Fan, J.; Wang, Z.; Song, L.; Mohamed Abdul, P.; Zhang, H. Effects of Natural Rheum tanguticum on the Cell Wall Integrity of Resistant Phytopathogenic Pectobacterium carotovorum subsp. Carotovorum. Molecules 2022, 27, 5291. https://doi.org/10.3390/molecules27165291
Qi Y, Wang M, Zhang B, Liu Y, Fan J, Wang Z, Song L, Mohamed Abdul P, Zhang H. Effects of Natural Rheum tanguticum on the Cell Wall Integrity of Resistant Phytopathogenic Pectobacterium carotovorum subsp. Carotovorum. Molecules. 2022; 27(16):5291. https://doi.org/10.3390/molecules27165291
Chicago/Turabian StyleQi, Yanjiao, Mingyang Wang, Bo Zhang, Yue Liu, Jiaqin Fan, Zifan Wang, Li Song, Peer Mohamed Abdul, and Hong Zhang. 2022. "Effects of Natural Rheum tanguticum on the Cell Wall Integrity of Resistant Phytopathogenic Pectobacterium carotovorum subsp. Carotovorum" Molecules 27, no. 16: 5291. https://doi.org/10.3390/molecules27165291
APA StyleQi, Y., Wang, M., Zhang, B., Liu, Y., Fan, J., Wang, Z., Song, L., Mohamed Abdul, P., & Zhang, H. (2022). Effects of Natural Rheum tanguticum on the Cell Wall Integrity of Resistant Phytopathogenic Pectobacterium carotovorum subsp. Carotovorum. Molecules, 27(16), 5291. https://doi.org/10.3390/molecules27165291







