Inhibition of Microtubule Affinity Regulating Kinase 4 by Metformin: Exploring the Neuroprotective Potential of Antidiabetic Drug through Spectroscopic and Computational Approaches
Abstract
:1. Introduction
2. Results and Discussion
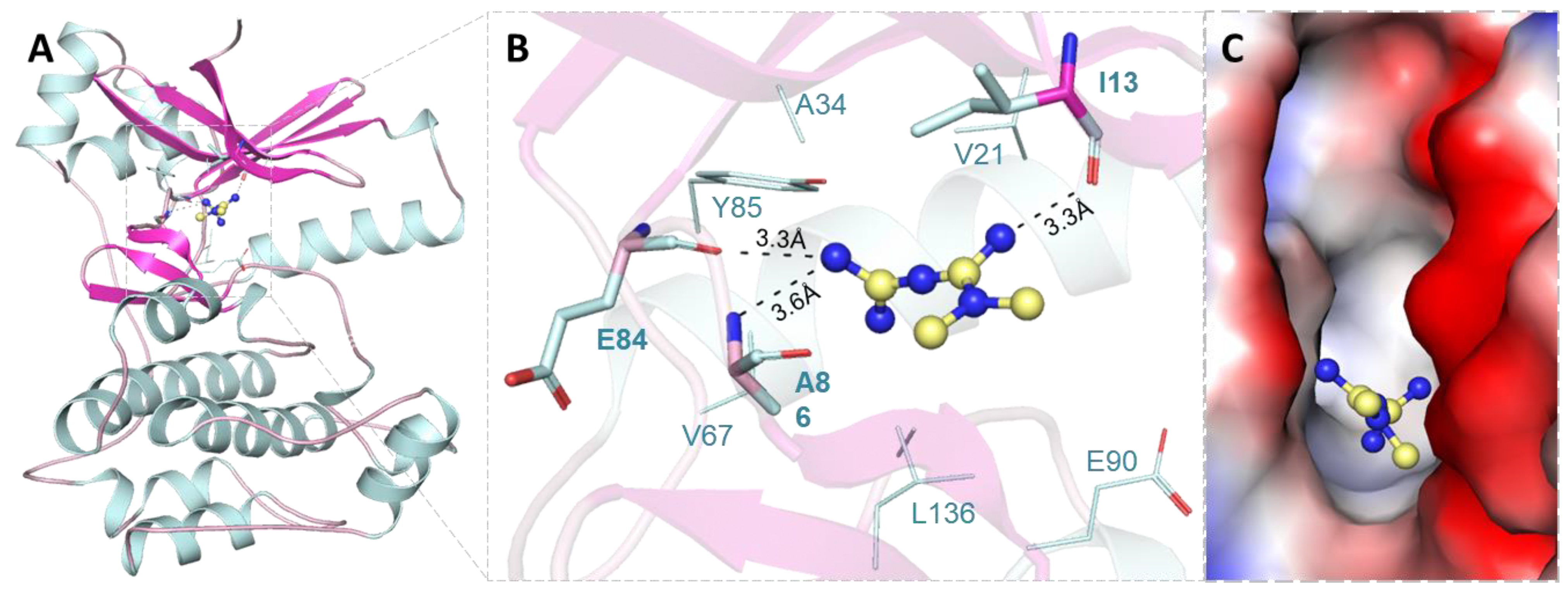
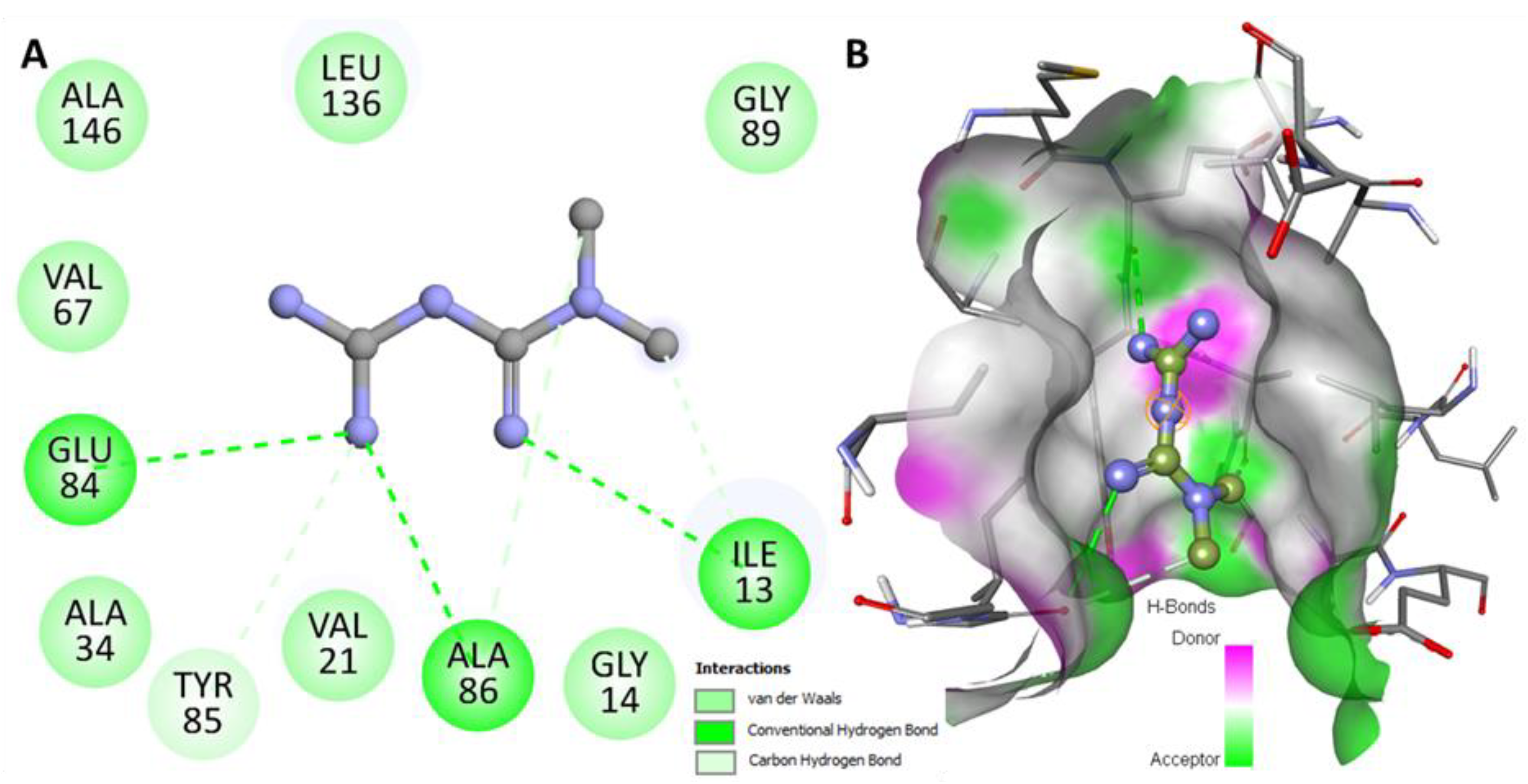
2.1. Molecular Docking Analysis
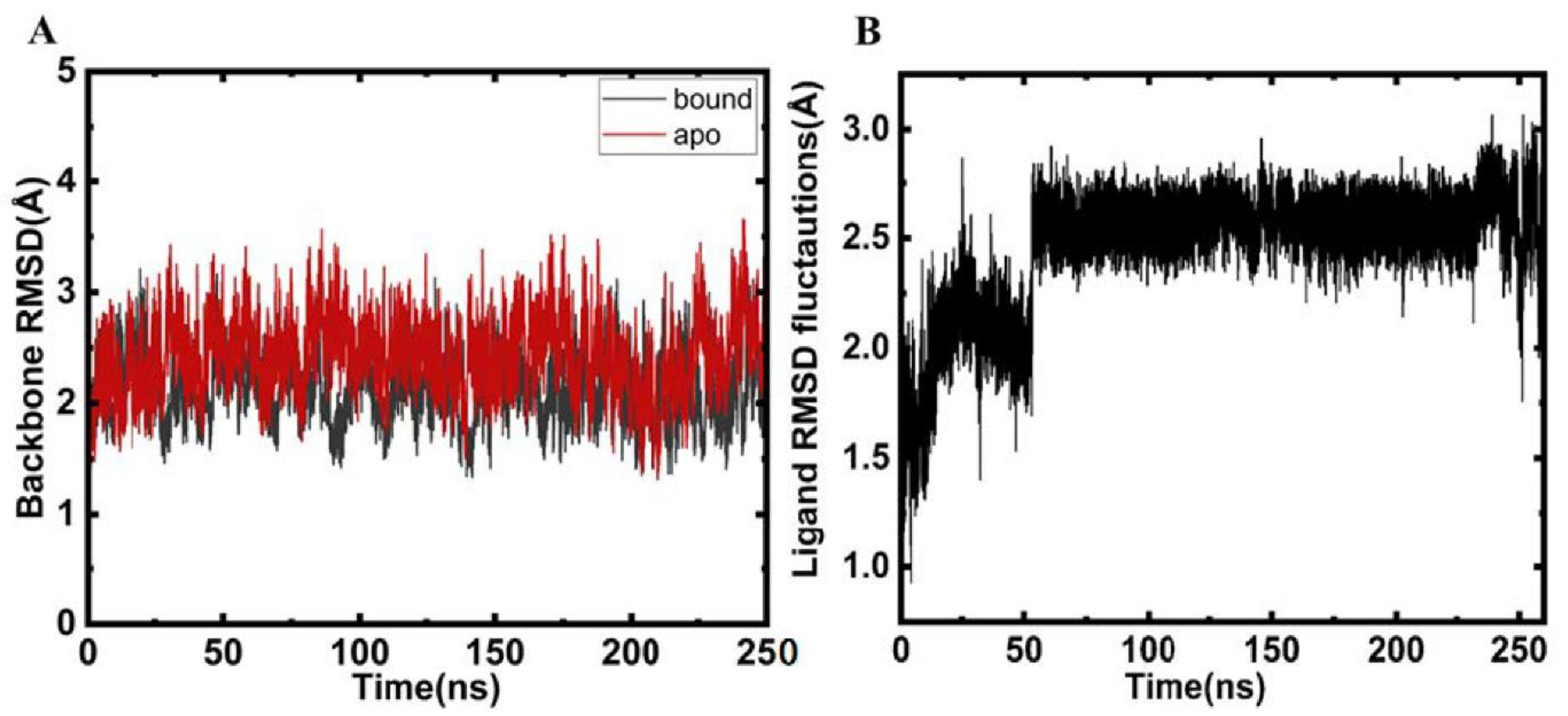
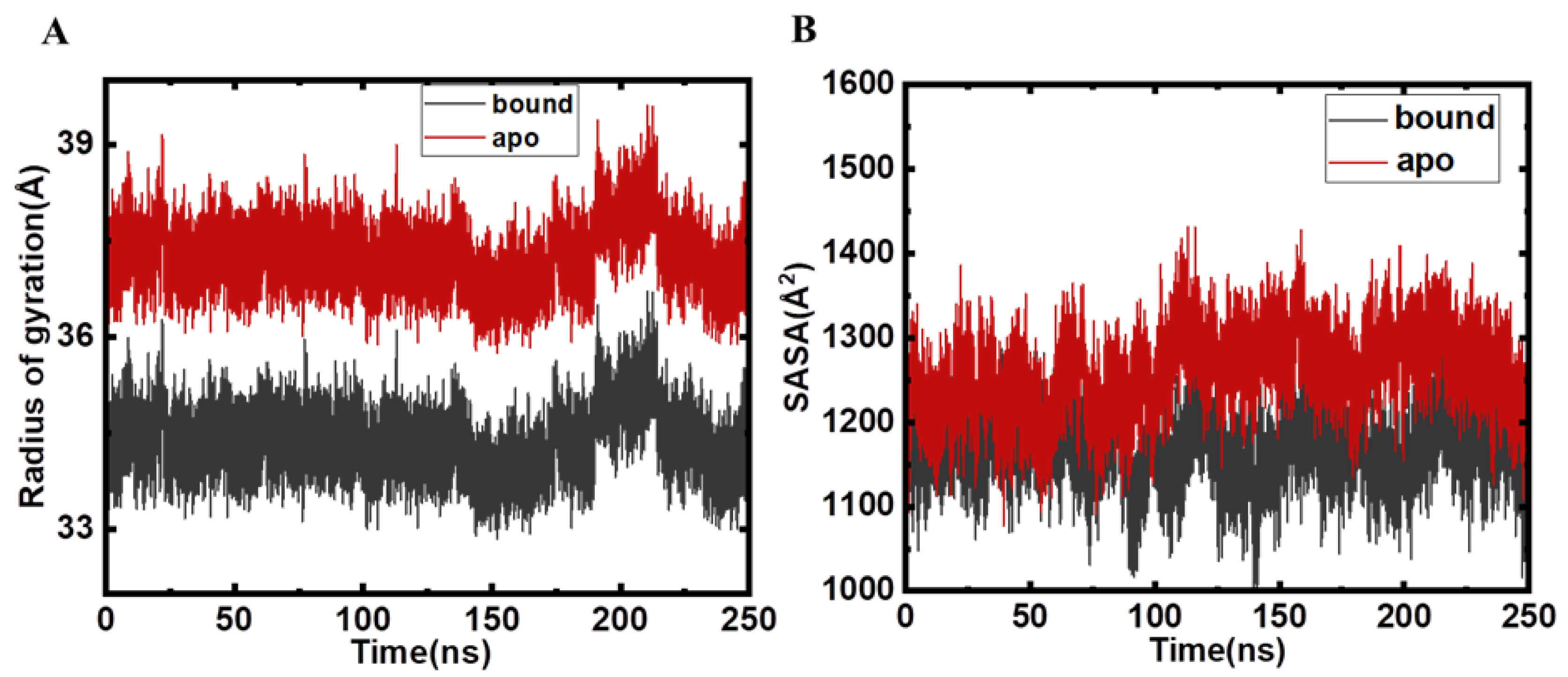
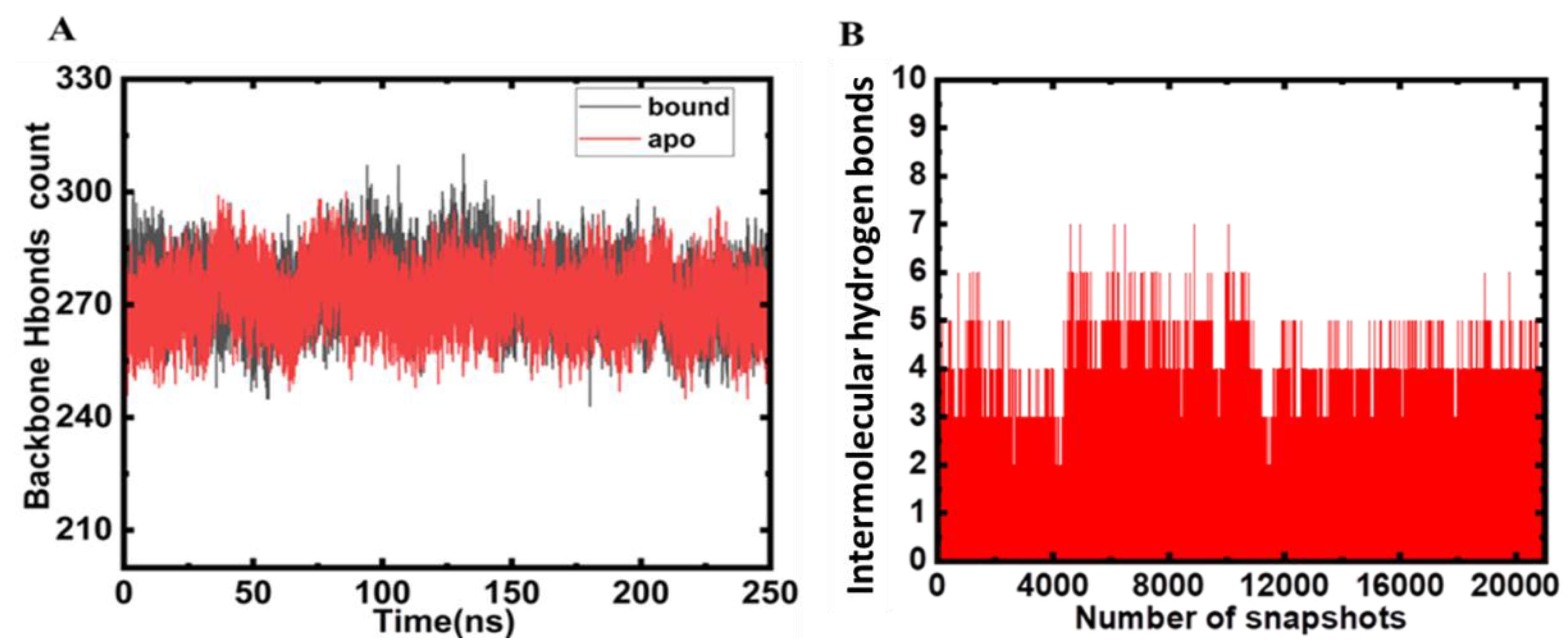
2.2. Structural Changes and Analyses Post MD
2.3. Hydrogen Bond Analysis
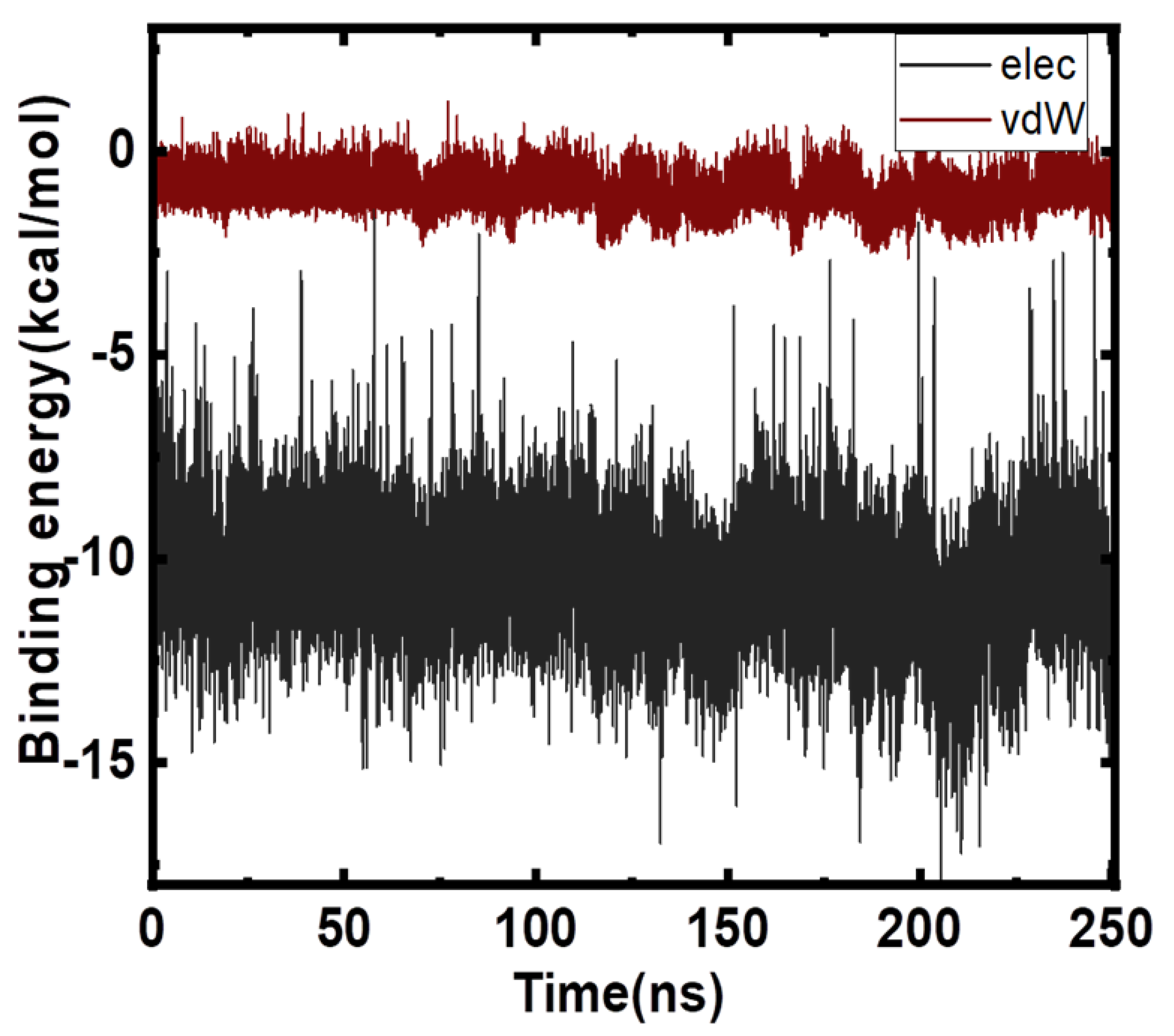
2.4. Free Energy Analyses
2.5. Fluorescence Based Binding
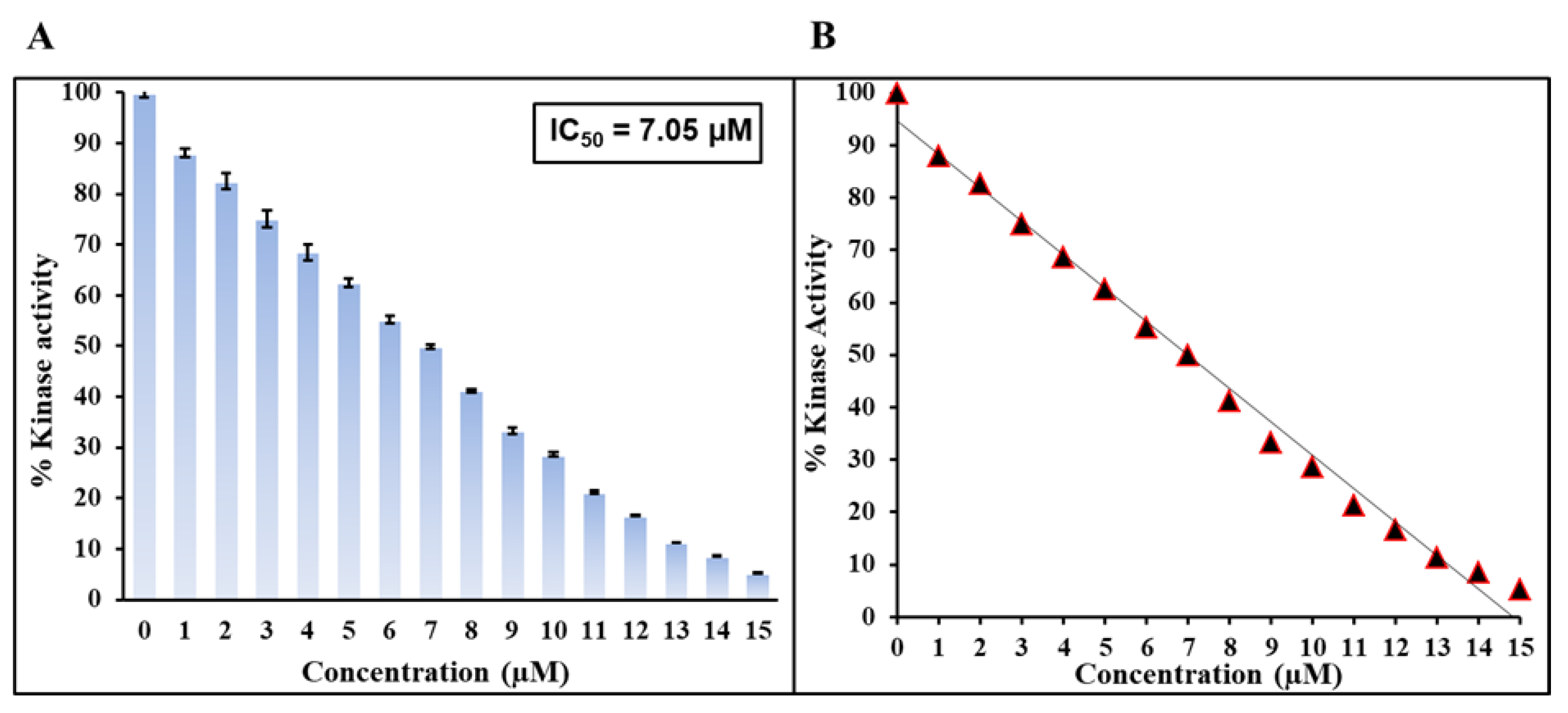
2.6. Kinase Assay
3. Materials and Methods
3.1. Materials
3.2. Molecular Docking
3.3. MD Simulations Setup of the Protein–Ligand Complex
3.4. System Preparation Prior to MD Setup
3.5. MD Simulation Details
3.6. Fluorescence Assay
3.7. Kinase Assay
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vanessa Fiorentino, T.; Prioletta, A.; Zuo, P.; Folli, F. Hyperglycemia-induced oxidative stress and its role in diabetes mellitus related cardiovascular diseases. Curr. Pharm. Des. 2013, 19, 5695–5703. [Google Scholar] [CrossRef] [PubMed]
- Soskolne, W.A.; Klinger, A. The relationship between periodontal diseases and diabetes: An overview. Ann. Periodontol. 2001, 6, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Rad, S.K.; Arya, A.; Karimian, H.; Madhavan, P.; Rizwan, F.; Koshy, S.; Prabhu, G. Mechanism involved in insulin resistance via accumulation of β-amyloid and neurofibrillary tangles: Link between type 2 diabetes and Alzheimer’s disease. Drug Des. Dev. Ther. 2018, 12, 3999. [Google Scholar]
- Kasznicki, J.; Sliwinska, A.; Drzewoski, J. Metformin in cancer prevention and therapy. Ann. Transl. Med. 2014, 2, 57. [Google Scholar]
- Association, A.D. Standards of medical care in diabetes—2014. Diabetes Care 2014, 37, S14–S80. [Google Scholar] [CrossRef]
- Ha, J.; Choi, D.-W.; Kim, K.J.; Cho, S.Y.; Kim, H.; Kim, K.Y.; Koh, Y.; Nam, C.M.; Kim, E. Association of metformin use with Alzheimer’s disease in patients with newly diagnosed type 2 diabetes: A population-based nested case–control study. Sci. Rep. 2021, 11, 24069. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhang, H.; Lu, Y. Molecular mechanisms of metformin for diabetes and cancer treatment. Front. Physiol. 2018, 9, 1039. [Google Scholar] [CrossRef]
- Zhou, G.; Myers, R.; Li, Y.; Chen, Y.; Shen, X.; Fenyk-Melody, J.; Wu, M.; Ventre, J.; Doebber, T.; Fujii, N. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 2001, 108, 1167–1174. [Google Scholar] [CrossRef]
- Patel, R.; Shah, G. Effect of metformin on clinical, metabolic and endocrine outcomes in women with polycystic ovary syndrome: A meta-analysis of randomized controlled trials. Curr. Med. Res. Opin. 2017, 33, 1545–1557. [Google Scholar] [CrossRef]
- Ferreira, C.; Sousa, M.; Rabaça, A.; Oliveira, P.F.; Alves, G.M.; Sá, R. Impact of metformin on male reproduction. Curr. Pharm. Des. 2015, 21, 3621–3633. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Chen, H.; Nie, F.; Wei, X.; Tao, Z.; Ma, J. The potential role of metformin in the treatment of Parkinson’s disease. J. Bio-X Res. 2020, 3, 27–35. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, S.; Fonseca, V.A.; Thethi, T.K.; Shi, L. Effect of metformin on neurodegenerative disease among elderly adult US veterans with type 2 diabetes mellitus. BMJ Open 2019, 9, e024954. [Google Scholar] [CrossRef] [PubMed]
- Evcimen, N.D.; King, G.L. The role of protein kinase C activation and the vascular complications of diabetes. Pharmacol. Res. 2007, 55, 498–510. [Google Scholar] [PubMed]
- Liang, W.-J.; Yang, H.-W.; Liu, H.-N.; Qian, W.; Chen, X.-L. HMGB1 upregulates NF-kB by inhibiting IKB-α and associates with diabetic retinopathy. Life Sci. 2020, 241, 117146. [Google Scholar] [CrossRef]
- Misra, P.; Chakrabarti, R. The role of AMP kinase in diabetes. Indian J. Med. Res. 2007, 125, 389. [Google Scholar] [PubMed]
- Sun, C.; Tian, L.; Nie, J.; Zhang, H.; Han, X.; Shi, Y. Inactivation of MARK4, an AMP-activated protein kinase (AMPK)-related kinase, leads to insulin hypersensitivity and resistance to diet-induced obesity. J. Biol. Chem. 2012, 287, 38305–38315. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Brooks III, C.L.; Frank, A.T. A rapid solvent accessible surface area estimator for coarse grained molecular simulations. J. Comput. Chem. 2017, 38, 1270–1274. [Google Scholar] [CrossRef]
- Hubbard, R.E.; Haider, M.K. Hydrogen Bonds in Proteins: Role and Strength. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, Ltd: Chichester, UK, 2010. [Google Scholar]
- Shamsi, A.; Shahwan, M.; Khan, M.S.; Alhumaydhi, F.A.; Alsagaby, S.A.; Al Abdulmonem, W.; Abdullaev, B.; Yadav, D.K. Mechanistic Insight into Binding of Huperzine A with Human Serum Albumin: Computational and Spectroscopic Approaches. Molecules 2022, 27, 797. [Google Scholar] [CrossRef]
- Shamsi, A.; Anwar, S.; Shahbaaz, M.; Mohammad, T.; Alajmi, M.F.; Hussain, A.; Hassan, I.; Ahmad, F.; Islam, A. Evaluation of Binding of Rosmarinic Acid with Human Transferrin and Its Impact on the Protein Structure: Targeting Polyphenolic Acid-Induced Protection of Neurodegenerative Disorders. Oxid. Med. Cell. Longev. 2020, 2020, 1245875. [Google Scholar] [CrossRef]
- Shamsi, A.; Ahmed, A.; Khan, M.S.; Al Shahwan, M.; Husain, F.M.; Bano, B. Understanding the binding between Rosmarinic acid and serum albumin: In vitro and in silico insight. J. Mol. Liquids 2020, 311, 113348. [Google Scholar] [CrossRef]
- Anwar, S.; Shamsi, A.; Shahbaaz, M.; Queen, A.; Khan, P.; Hasan, G.M.; Islam, A.; Alajmi, M.F.; Hussain, A.; Ahmad, F. Rosmarinic acid exhibits anticancer effects via MARK4 inhibition. Sci. Rep. 2020, 10, 10300. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Dasgupta, D.; Alhumaydhi, F.A.; Khan, M.S.; Alsagaby, S.A.; Al Ab-dulmonem, W.; Hassan, M.I.; Yadav, D.K. Inhibition of MARK4 by serotonin is an attractive therapeutic approach to combat Alzheimer’s disease and neuroinflammation. RSC Med. Chem. 2022, 13, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Swinney, D.C. Molecular mechanism of action (MMoA) in drug discovery. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2011; Volume 46, pp. 301–317. [Google Scholar]
- Anwar, S.; Shamsi, A.; Kar, R.K.; Queen, A.; Islam, A.; Ahmad, F.; Hassan, M.I. Structural and biochemical investigation of MARK4 inhibitory potential of cholic acid: Towards therapeutic implications in neurodegenerative diseases. Int. J. Biol. Macromol. 2020, 161, 596–604. [Google Scholar] [CrossRef]
- Anwar, S.; Khan, S.; Shamsi, A.; Anjum, F.; Shafie, A.; Islam, A.; Ahmad, F.; Hassan, M.I. Structure-based investigation of MARK4 inhibitory potential of Naringenin for therapeutic management of cancer and neurodegenerative diseases. J. Cell. Biochem. 2021, 122, 1445–1459. [Google Scholar] [CrossRef]
- Sack, J.S.; Gao, M.; Kiefer, S.E.; Myers, J.E., Jr.; Newitt, J.A.; Wu, S.; Yan, C. Crystal structure of microtubule affinity-regulating kinase 4 catalytic domain in complex with a pyrazolopyrimidine inhibitor. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, T.; Mathur, Y.; Hassan, M.I. InstaDock: A single-click graphical user interface for molecular docking-based virtual high-throughput screening. Brief. Bioinform. 2021, 22, bbaa279. [Google Scholar] [CrossRef]
- Anwar, S.; DasGupta, D.; Shafie, A.; Alhumaydhi, F.A.; Alsagaby, S.A.; Shahwan, M.; Anjum, F.; Al Abdulmonem, W.; Sharaf, S.E.; Hassan, M.I. Implications of Tempol in Pyruvate Dehydrogenase Kinase 3 Targeted Anticancer therapeutics: Computational, Spectroscopic and Calorimetric Studies. J. Mol. Liq. 2022, 350, 118581. [Google Scholar] [CrossRef]
- Anwar, S.; DasGupta, D.; Azum, N.; Alfaifi, S.Y.; Asiri, A.M.; Alhumaydhi, F.A.; Alsagaby, S.A.; Sharaf, S.E.; Shahwan, M.; Hassan, M.I. Inhibition of PDK3 by artemisinin, a repurposed antimalarial drug in cancer therapy. J. Mol. Liq. 2022, 355, 118928. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Kubista, M.; Sjöback, R.; Eriksson, S.; Albinsson, B. Experimental correction for the inner-filter effect in fluorescence spectra. Analyst 1994, 119, 417–419. [Google Scholar] [CrossRef]
- Shamsi, A.; Anwar, S.; Mohammad, T.; Alajmi, M.F.; Hussain, A.; Rehman, M.; Hasan, G.M.; Islam, A.; Hassan, M. MARK4 inhibited by AChE inhibitors, donepezil and Rivastigmine tartrate: Insights into Alzheimer’s disease therapy. Biomolecules 2020, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Waseem, R.; Anwar, S.; Khan, S.; Shamsi, A.; Hassan, M.; Anjum, F.; Shafie, A.; Islam, A.; Yadav, D.K. MAP/Microtubule Affinity Regulating Kinase 4 Inhibitory Potential of Irisin: A New Therapeutic Strategy to Combat Cancer and Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 10986. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Mohammad, T.; Shamsi, A.; Queen, A.; Parveen, S.; Luqman, S.; Hasan, G.M.; Alamry, K.A.; Azum, N.; Asiri, A.M. Discovery of Hordenine as a potential inhibitor of pyruvate dehydrogenase kinase 3: Implication in lung Cancer therapy. Biomedicines 2020, 8, 119. [Google Scholar] [CrossRef]

| Energy Factors | Average | Standard Deviation | Std Error of Mean |
|---|---|---|---|
| VDWAALS | −28.8312 | 5.9959 | 0.1971 |
| EEL | 34.7349 | 33.1069 | 1.0885 |
| EGB | −14.5293 | 32.5593 | 1.0705 |
| ESURF | −3.5261 | 0.6543 | 0.0215 |
| ΔGgas | 5.9037 | 31.4944 | 1.0355 |
| ΔGsolv | −18.0555 | 32.7427 | 1.0766 |
| ΔG (total) | −12.1517 | 4.7443 | 0.1560 |
| Mol Weight | H Bond Donors | H Bond Acceptors | No of Single Rotatable Bonds | No of Aromatic Rings | LogS | Flexibility Index | Lipinski Drug Like Test |
|---|---|---|---|---|---|---|---|
| 130.1 | 1 | 0 | 1 | 0 | 1.2 | 1.4 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, G.M.; DasGupta, D.; Alam, M.Z.; Baeesa, S.S.; Alghamdi, B.S.; Anwar, F.; Alqurashi, T.M.A.; Sharaf, S.E.; Al Abdulmonem, W.; Alyousef, M.A.; et al. Inhibition of Microtubule Affinity Regulating Kinase 4 by Metformin: Exploring the Neuroprotective Potential of Antidiabetic Drug through Spectroscopic and Computational Approaches. Molecules 2022, 27, 4652. https://doi.org/10.3390/molecules27144652
Ashraf GM, DasGupta D, Alam MZ, Baeesa SS, Alghamdi BS, Anwar F, Alqurashi TMA, Sharaf SE, Al Abdulmonem W, Alyousef MA, et al. Inhibition of Microtubule Affinity Regulating Kinase 4 by Metformin: Exploring the Neuroprotective Potential of Antidiabetic Drug through Spectroscopic and Computational Approaches. Molecules. 2022; 27(14):4652. https://doi.org/10.3390/molecules27144652
Chicago/Turabian StyleAshraf, Ghulam Md., Debarati DasGupta, Mohammad Zubair Alam, Saleh S. Baeesa, Badrah S. Alghamdi, Firoz Anwar, Thamer M. A. Alqurashi, Sharaf E. Sharaf, Waleed Al Abdulmonem, Mohammed A. Alyousef, and et al. 2022. "Inhibition of Microtubule Affinity Regulating Kinase 4 by Metformin: Exploring the Neuroprotective Potential of Antidiabetic Drug through Spectroscopic and Computational Approaches" Molecules 27, no. 14: 4652. https://doi.org/10.3390/molecules27144652
APA StyleAshraf, G. M., DasGupta, D., Alam, M. Z., Baeesa, S. S., Alghamdi, B. S., Anwar, F., Alqurashi, T. M. A., Sharaf, S. E., Al Abdulmonem, W., Alyousef, M. A., Alhumaydhi, F. A., & Shamsi, A. (2022). Inhibition of Microtubule Affinity Regulating Kinase 4 by Metformin: Exploring the Neuroprotective Potential of Antidiabetic Drug through Spectroscopic and Computational Approaches. Molecules, 27(14), 4652. https://doi.org/10.3390/molecules27144652









