Peptide-Mediated Targeted Delivery of Aloe-Emodin as Anticancer Drug
Abstract
1. Introduction
2. Results and Discussion
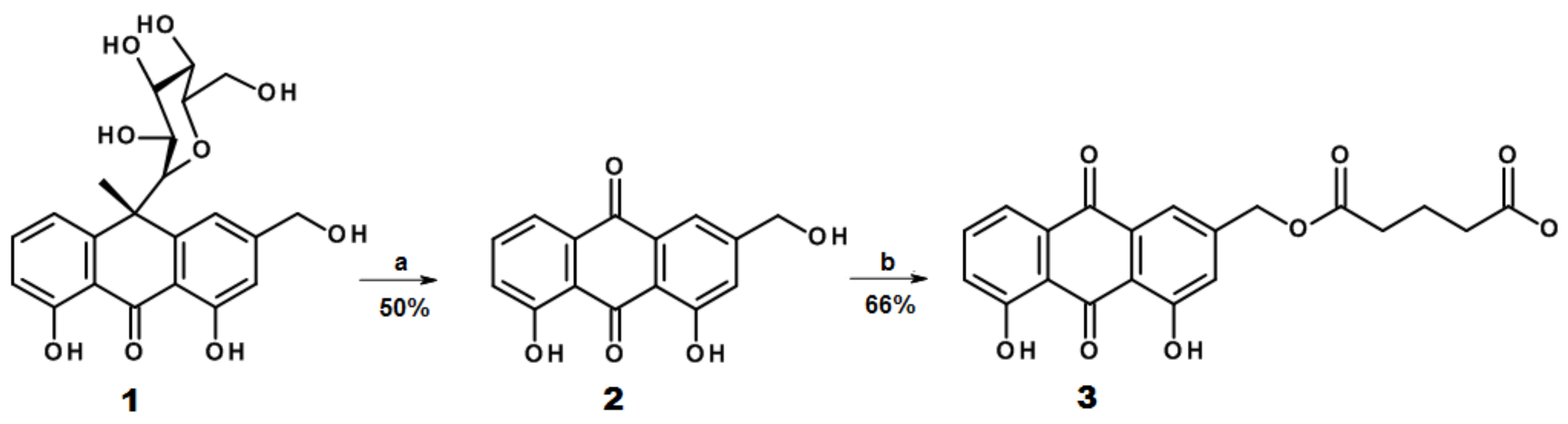
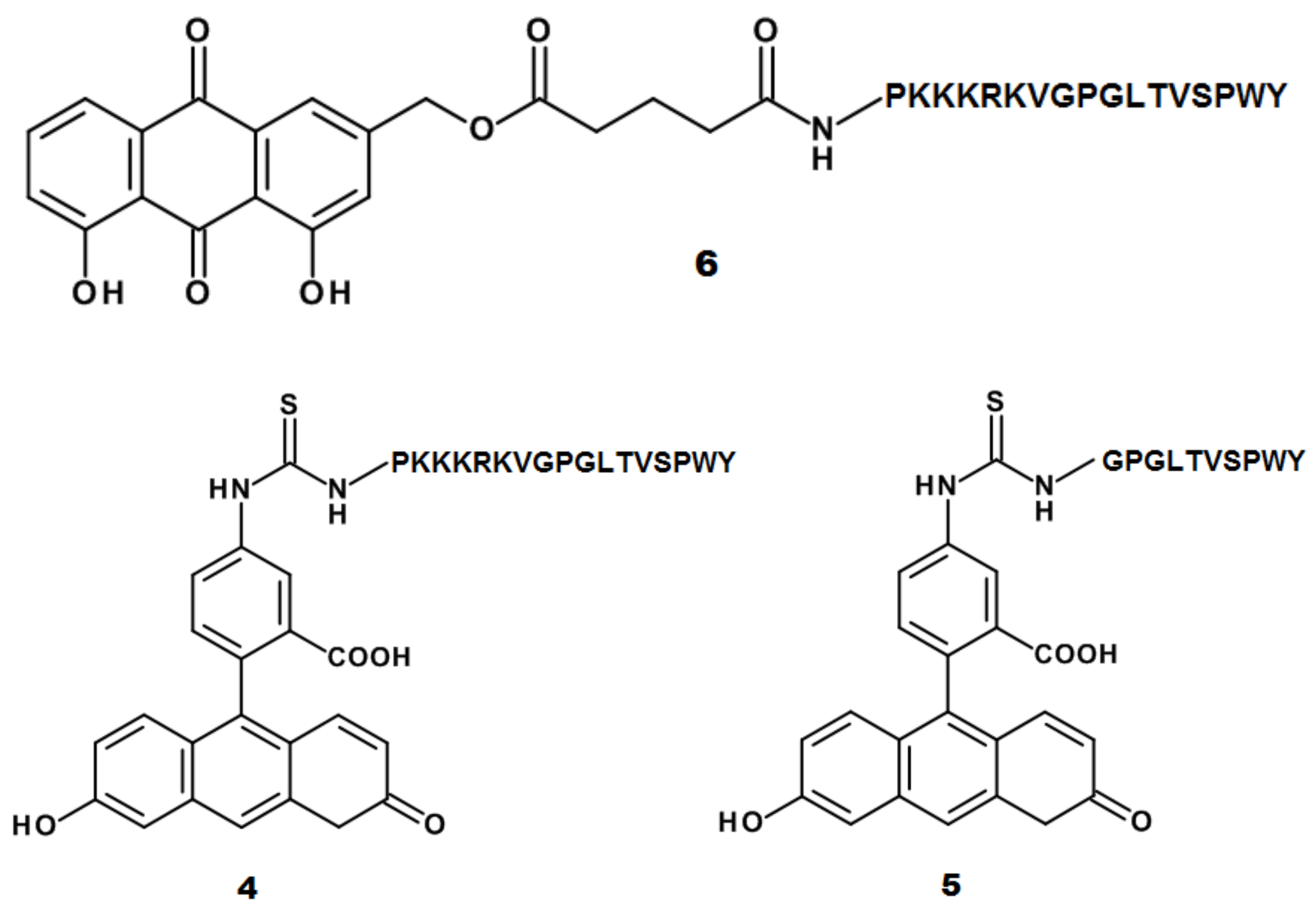
2.1. Design and Synthesis of AE Derivatives and Conjugates
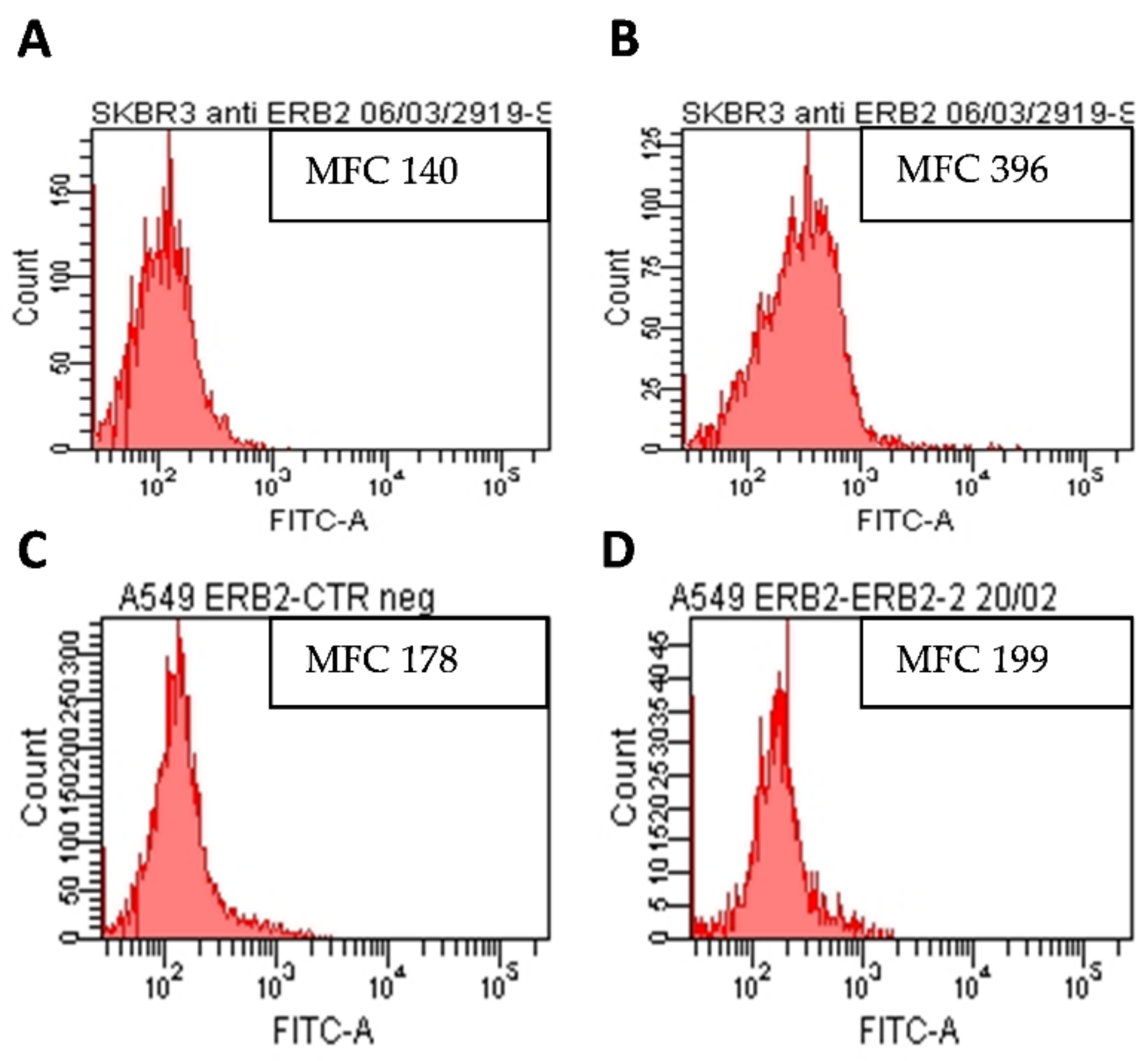
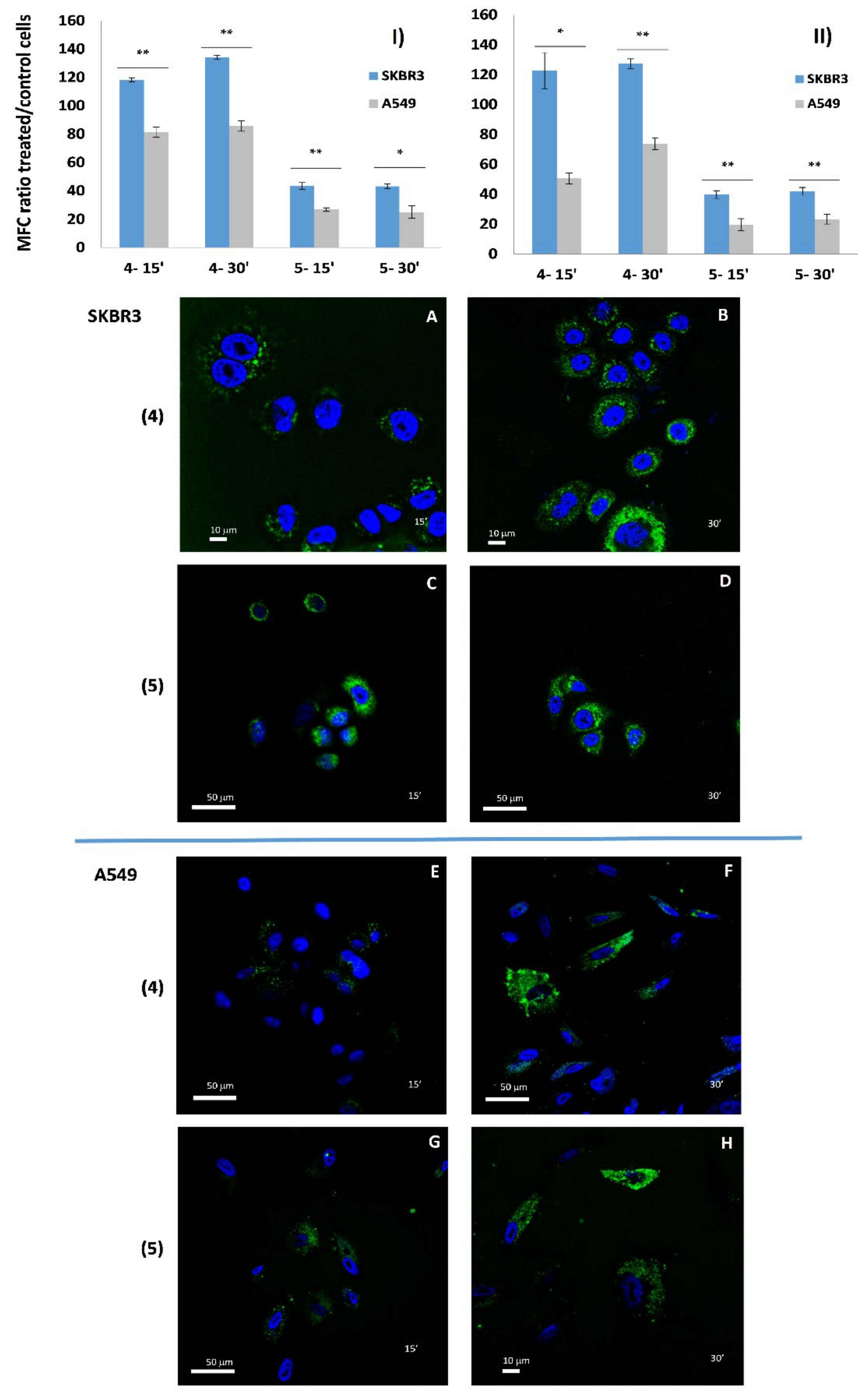
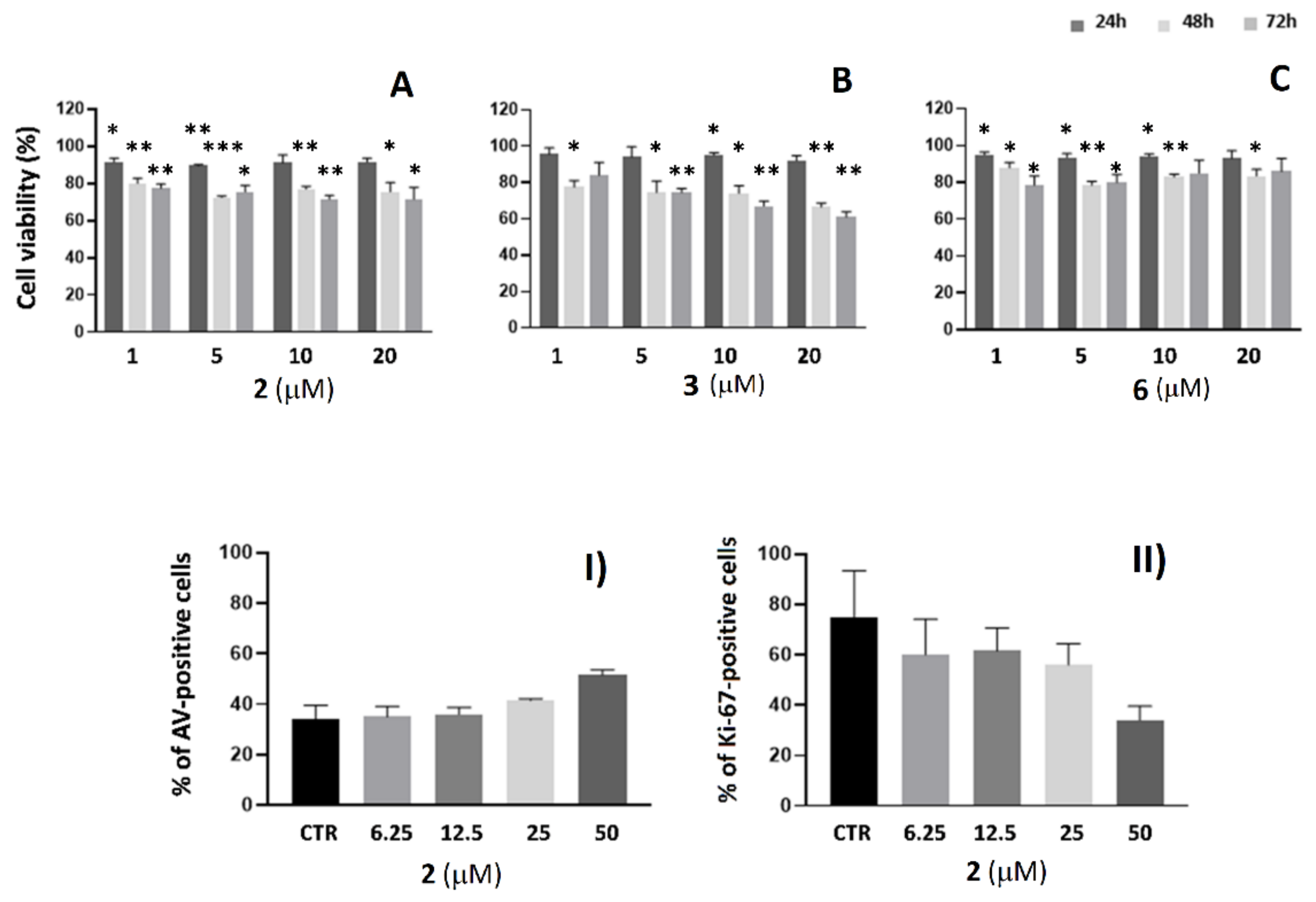
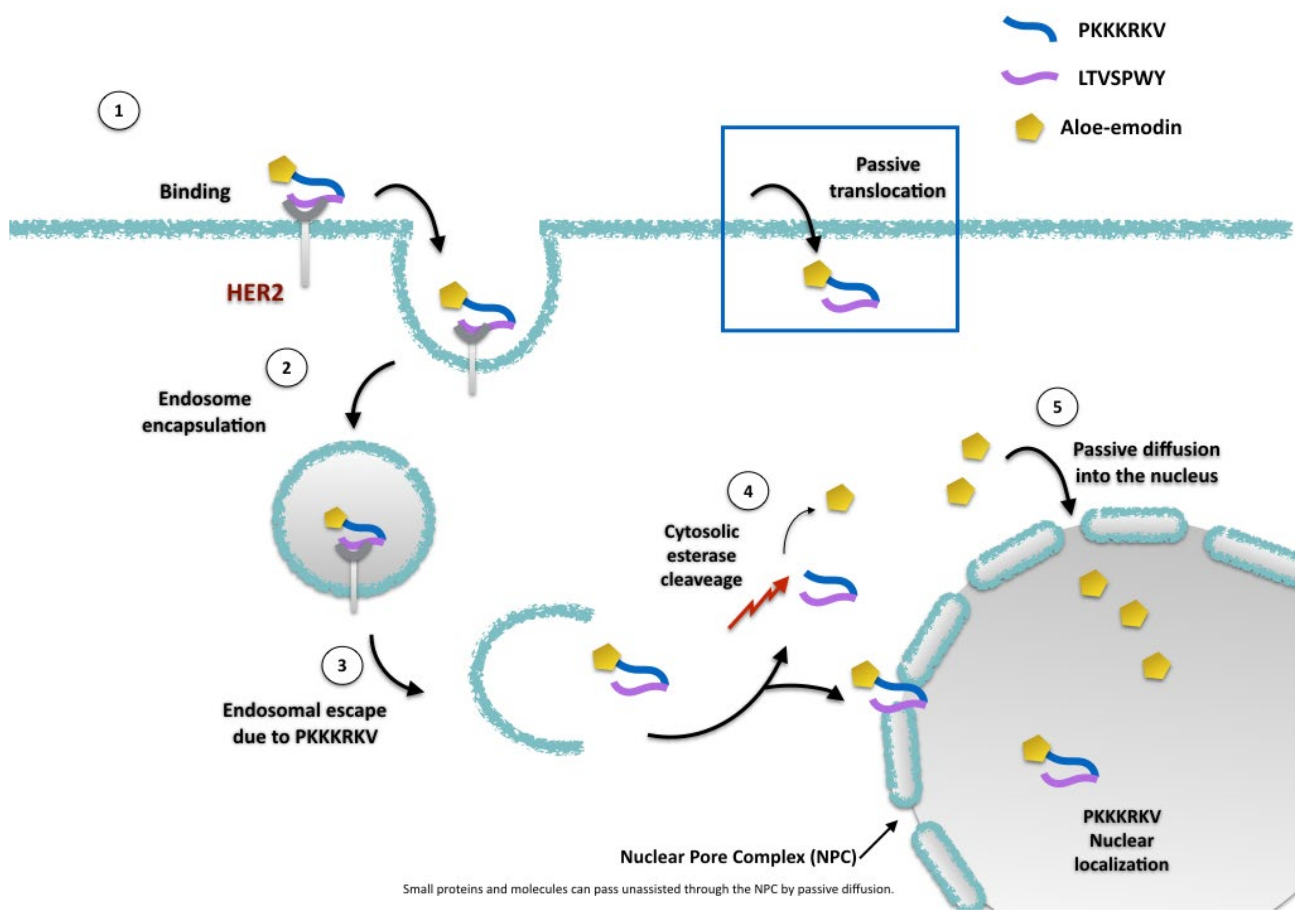
2.2. AE and AE Conjugates Internalization Experiments
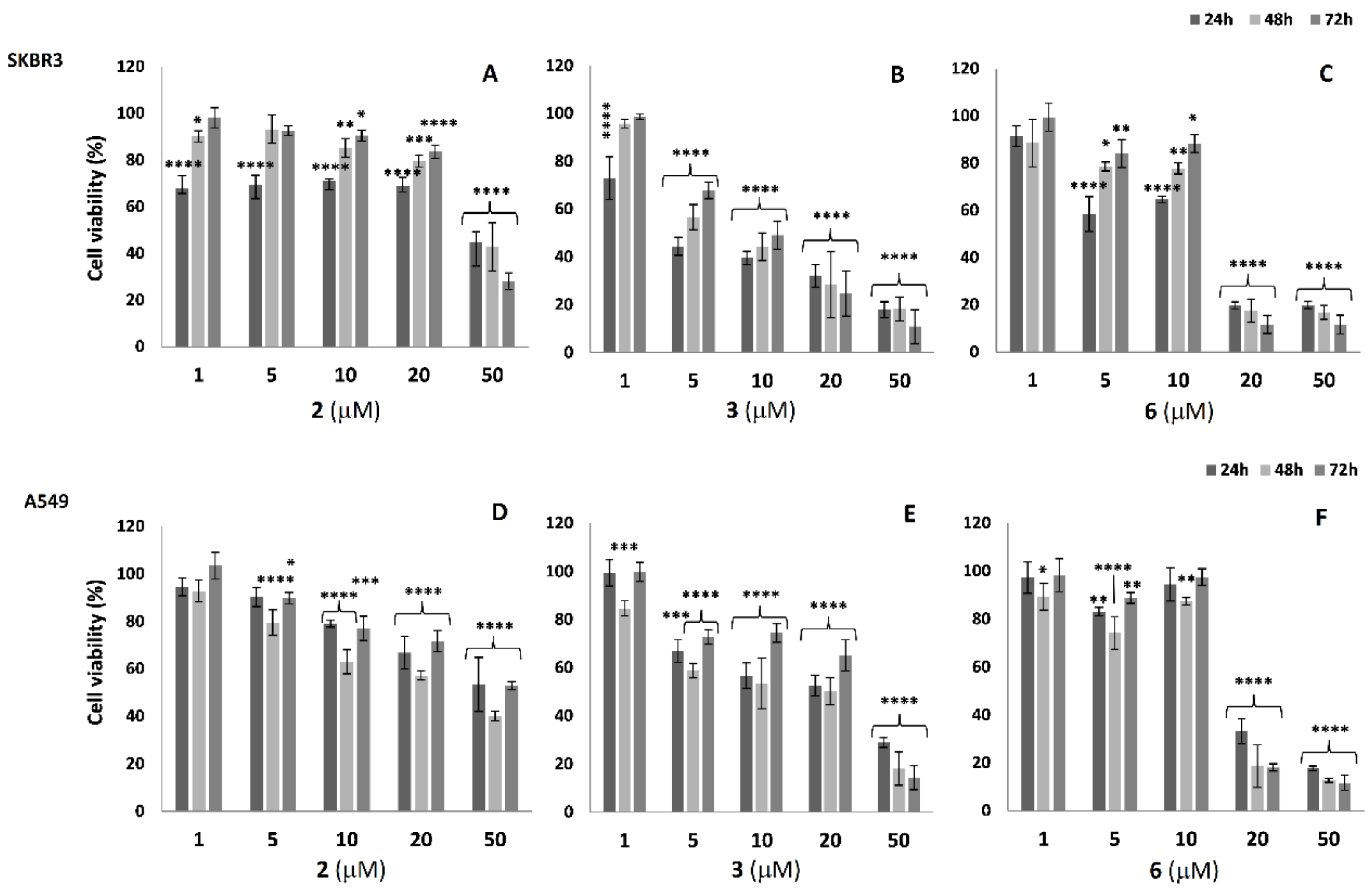
2.3. Cell Viability Analysis in SKBR3 and A549 Cells
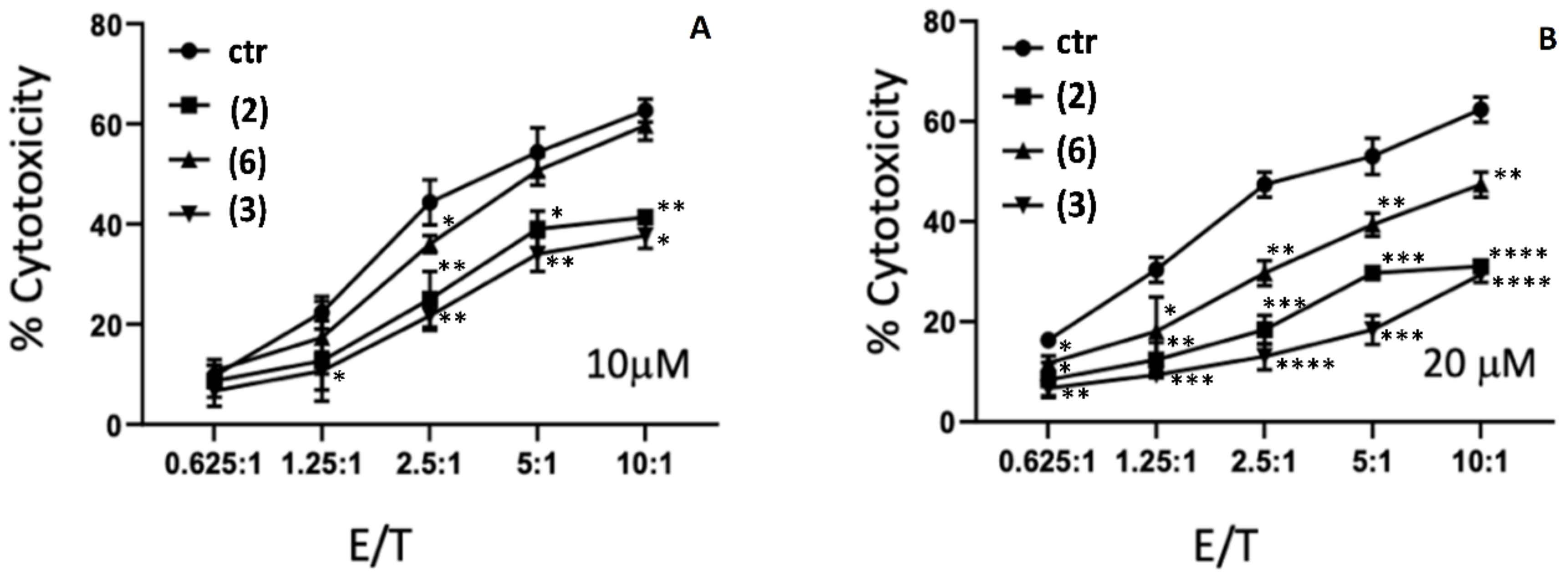
2.4. Cytotoxicity Studies of AE and AE Derivatives on NK92 and K562 Cells
3. Materials and Methods
3.1. Aloe-Emodin (AE) Derivatives Synthesis and Characterization
Analytical Methods and Characterization of the Products
3.2. Peptides and Conjugates Synthesis and Characterization
Analytical Methods and Characterization of the Products
3.3. Cell Lines
3.4. Flow Cytometry
3.4.1. Analysis of HER2 Expression on SKBR3 and A549 Cell Lines
3.4.2. Cellular Uptake of AE Derivatives in SKBR3 and A549 Cell Lines
3.4.3. Analysis of Apoptosis and Cell Proliferation in NK-92 Cells
3.5. Cell Viability Assay
3.6. Cytotoxicity Assay
3.7. Growth Inhibition Assay
3.8. Confocal Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ullah, M.F. Breast Cancer: Current Perspectives on the Disease Status. Adv. Exp. Med. Biol. 2019, 1152, 51–64. [Google Scholar] [CrossRef]
- Yap, K.M.; Sekar, M.; Fuloria, S.; Wu, Y.S.; Gan, S.H.; Rani Rani, N.I.M.; Subramaniyan, V.; Kokare, C.; Lum, P.T.; Begum, M.Y.; et al. Drug Delivery of Natural Products Through Nanocarriers for Effective Breast Cancer Therapy: A Comprehensive Review of Literature. Int. J. Nanomed. 2021, 16, 7891–7941. [Google Scholar] [CrossRef] [PubMed]
- Solís-Cruz, G.Y.; Pérez-López, L.A.; Alvarez-Roman, R.; Rivas-Galindo, V.M.; Silva-Mares, D.A.; Ibarra-Rivera, T.R. Nanocarriers as Administration Systems of Natural Products. Curr. Top. Med. Chem. 2021, 21, 2365–2373. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.; Brown, S. Antibody–drug conjugates as novel anti-cancer chemotherapeutics. Biosci. Rep. 2015, 35, e00225. [Google Scholar] [CrossRef]
- Wang, Y.; Cheetham, A.G.; Angacian, G.; Su, H.; Xie, L.; Cui, H. Peptide–drug conjugates as effective prodrug strategies for targeted delivery. Adv. Drug Deliv. Rev. 2017, 110–111, 112–126. [Google Scholar] [CrossRef]
- Iqbal, N.; Iqbal, N. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar] [CrossRef]
- Nieto, C.; Centa, A.; Rodríguez-Rodríguez, J.A.; Pandiella, A.; del Valle, E.M.M. Paclitaxel-Trastuzumab Mixed Nanovehicle to Target HER2-Overexpressing Tumors. Nanomaterials 2019, 9, 948. [Google Scholar] [CrossRef]
- Berillo, D.; Kozhahmetova, M.; Lebedeva, L. Overview of the Biological Activity of Anthraquinons and Flavanoids of the Plant Rumex Species. Molecules 2022, 27, 1204. [Google Scholar] [CrossRef]
- Dong, X.; Zeng, Y.; Liu, Y.; You, L.; Yin, X.; Fu, J.; Ni, J. Aloe-emodin: A review of its pharmacology, toxicity, and pharmacokinetics. Phytother. Res. 2020, 34, 270–281. [Google Scholar] [CrossRef]
- Sanders, B.; Ray, A.M.; Goldberg, S.; Clark, T.; McDaniel, H.R.; Atlas, S.E.; Farooqi, A.; Konefal, J.; Lages, L.C.; Lopez, J.; et al. Anti-cancer effects of aloe-emodin: A systematic review. J. Clin. Transl. Res. 2017, 3, 283–296. [Google Scholar]
- Özenver, N.; Saeed, M.; Demirezer, L.; Efferth, T. Aloe-emodin as drug candidate for cancer therapy. Oncotarget 2018, 9, 17770–17796. [Google Scholar] [CrossRef]
- Tabolacci, C.; Oliverio, S.; Lentini, A.; Rossi, S.; Galbiati, A.; Montesano, C.; Mattioli, P.; Provenzano, B.; Facchiano, F.; Beninati, S. Aloe-emodin as antiproliferative and differentiating agent on human U937 monoblastic leukemia cells. Life Sci. 2011, 89, 812–820. [Google Scholar] [CrossRef]
- Tabolacci, C.; Rossi, S.; Lentini, A.; Provenzano, B.; Turcano, L.; Facchiano, F.; Beninati, S. Aloin enhances cisplatin antineoplastic activity in B16-F10 melanoma cells by transglutaminase-induced differentiation. Amino Acids 2011, 44, 293–300. [Google Scholar] [CrossRef]
- Chihara, T.; Shimpo, K.; Beppu, H.; Yamamoto, N.; Kaneko, T.; Wakamatsu, K.; Sonoda, S. Effects of Aloe-emodin and Emodin on Proliferation of the MKN45 Human Gastric Cancer Cell Line. Asian Pac. J. Cancer Prev. 2015, 16, 3887–3891. [Google Scholar] [CrossRef]
- Suboj, P.; Babykutty, S.; Valiyaparambil Gopi, D.R.; Nair, R.S.; Srinivas, P.; Gopala, S. Aloe emodin inhibits colon cancer cell migration/angiogenesis by downregulating MMP-2/9, RhoB and VEGF via reduced DNA binding activity of NF-κB. Eur. J. Pharm. Sci. 2012, 45, 581–591. [Google Scholar] [CrossRef]
- Chung, J.-G.; Chen, Y.-Y.; Chiang, S.-Y.; Lin, J.-G.; Ma, Y.-S.; Liao, C.-L.; Weng, S.-W.; Lai, T.-Y.; Chung, S.C. Emodin, aloe-emodin and rhein inhibit migration and invasion in human tongue cancer SCC-4 cells through the inhibition of gene expression of matrix metalloproteinase-9. Int. J. Oncol. 2010, 36, 1113–1120. [Google Scholar] [CrossRef]
- Zou, G.; Zhang, X.; Wang, L.; Li, X.; Xie, T.; Zhao, J.; Yan, J.; Wang, L.; Ye, H.; Jiao, S.; et al. Herb-sourced emodin inhibits angiogenesis of breast cancer by targeting VEGFA transcription. Theranostics 2020, 10, 6839–6853. [Google Scholar] [CrossRef]
- Ma, J.-W.; Hung, C.-M.; Lin, Y.-C.; Ho, C.-T.; Kao, J.-Y.; Way, T.-D. Aloe-emodin inhibits HER-2 expression through the downregulation of Y-box binding protein-1 in HER-2-overexpressing human breast cancer cells. Oncotarget 2016, 7, 58915–58930, Erratum in Oncotarget 2019, 10, 5727–5729. [Google Scholar] [CrossRef][Green Version]
- Panigrahi, G.K.; Yadav, A.; Srivastava, A.; Tripathi, A.; Raisuddin, S.; Das, M. Mechanism of Rhein-Induced Apoptosis in Rat Primary Hepatocytes: Beneficial Effect of Cyclosporine A. Chem. Res. Toxicol. 2015, 28, 1133–1143. [Google Scholar] [CrossRef]
- Dong, X.; Fu, J.; Yin, X.; Qu, C.; Yang, C.; He, H.; Ni, J. Induction of Apoptosis in HepaRG Cell Line by Aloe-Emodin through Generation of Reactive Oxygen Species and the Mitochondrial Pathway. Cell. Physiol. Biochem. 2017, 42, 685–696. [Google Scholar] [CrossRef]
- Vanderhoeven, F.; Redondo, A.L.; Martinez, A.L.; Vargas-Roig, L.M.; Sanchez, A.M.; Flamini, M.I. Synergistic antitumor activity by combining trastuzumab with retinoic acid in HER2 positive human breast cancer cells. Oncotarget 2018, 9, 26527–26542. [Google Scholar] [CrossRef][Green Version]
- Ren, W.; Liu, Y.; Wan, S.; Fei, C.; Wang, W.; Chen, Y.; Zhang, Z.; Wang, T.; Wang, J.; Zhou, L.; et al. BMP9 Inhibits Proliferation and Metastasis of HER2-Positive SK-BR-3 Breast Cancer Cells through ERK1/2 and PI3K/AKT Pathways. PLoS ONE 2014, 9, e96816. [Google Scholar] [CrossRef]
- Shadidi, M.; Sioud, M. Identification of novel carrier peptides for the specific delivery of therapeutics into cancer cells. FASEB J. 2002, 17, 256–258. [Google Scholar] [CrossRef]
- Wang, X.-F.; Birringer, M.; Dong, L.-F.; Veprek, P.; Low, P.; Swettenham, E.; Stantic, M.; Yuan, L.-H.; Zobalova, R.; Wu, K.; et al. A Peptide Conjugate of Vitamin E Succinate Targets Breast Cancer Cells with High ErbB2 Expression. Cancer Res. 2007, 67, 3337–3344. [Google Scholar] [CrossRef][Green Version]
- Gong, C.; Pan, D.; Qiu, F.; Sun, P.; Zhang, Y.-H. Selective DNA Delivery to Tumor Cells Using an Oligoarginine-LTVSPWY Peptide. PLoS ONE 2014, 9, e110632. [Google Scholar] [CrossRef]
- Jie, L.Y.; Cai, L.L.; Wang, L.J.; Ying, X.Y.; Yu, R.S.; Zhang, M.M.; Du, Y.Z. Actively-targeted LTVSPWY peptide-modified magnetic nanoparticles for tumor imaging. Int J. Nanomed. 2012, 7, 3981–3989. [Google Scholar]
- Leary, J.F. Design of sophisticated shaped, multilayered, and multifunctional nanoparticles for combined in-vivo imaging and advanced drug delivery. Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications XVI, 108910R. In Proceedings of SPIE, San Francisco, CA, USA, 5 March 2019. [Google Scholar] [CrossRef]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Şeker Karatoprak, G.; Akkol, E.K.; Yücel, Ç.; Acıkara, B.; Sobarzo-Sánchez, E. Advances in Understanding the Role of Aloe Emodin and Targeted Drug Delivery Systems in Cancer. Oxidative Med. Cell. Longev. 2022, 2022, 7928200. [Google Scholar] [CrossRef]
- Pecere, T.; Sarinella, F.; Salata, C.; Gatto, B.; Bet, A.; Vecchia, F.D.; Diaspro, A.; Carli, M.; Palumbo, M.; Palù, G. Involvement of p53 in specific anti-neuroectodermal tumor activity of aloe-emodin. Int. J. Cancer 2003, 106, 836–847. [Google Scholar] [CrossRef]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef]
- Nakase, I.; Kobayashi, S.; Futaki, S. Endosome-disruptive peptides for improving cytosolic delivery of bioactive macromolecules. Biopolymers 2010, 94, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Vidal, P.; Chaloin, L.; Heitz, F.; Divita, G. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 1997, 25, 2730–2736. [Google Scholar] [CrossRef] [PubMed]
- Midoux, P.; Kichler, A.; Boutin, V.; Maurizot, A.J.-C.; Monsigny, M. Membrane Permeabilization and Efficient Gene Transfer by a Peptide Containing Several Histidines. Bioconjugate Chem. 1998, 9, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Open source drug discovery consortium; Raghava, G.P.S. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef]
- Szöke, B.; Horváth, J.; Halmos, G.; Rékási, Z.; Groot, K.; Nagy, A.; Schally, A.V. LH-RH analogue carrying a cytotoxic radical is internalized by rat pituitary cells in vitro. Peptides 1994, 15, 359–366. [Google Scholar] [CrossRef]
- Feng, B.; Zhao, X.; Zhao, W.; Jiang, H.; Ren, Z.; Chen, Y.; Yuan, Y.; Du, Z. Ethyl 2-Succinate-Anthraquinone Attenuates Inflammatory Response and Oxidative Stress via Regulating NLRP3 Signaling Pathway. Front. Pharmacol. 2021, 12, 719822. [Google Scholar] [CrossRef]
- Wei, X.; Henke, V.G.; Strübing, C.; Brown, E.B.; Clapham, D.E. Real-Time Imaging of Nuclear Permeation by EGFP in Single Intact Cells. Biophys. J. 2003, 84, 1317–1327. [Google Scholar] [CrossRef][Green Version]
- Wang, H.-Y.; Chen, J.-X.; Sun, Y.-X.; Deng, J.-Z.; Li, C.; Zhang, X.-Z.; Zhuo, R.-X. Construction of cell penetrating peptide vectors with N-terminal stearylated nuclear localization signal for targeted delivery of DNA into the cell nuclei. J. Control. Release 2011, 155, 26–33. [Google Scholar] [CrossRef]
- Lin, S.-Z.; Zhang, W.; Li, H.; Bu, H.; Chen, H.; Tong, H.; Liu, D.; Guo, H. Emodin inhibits the differentiation and maturation of dendritic cells and increases the production of regulatory T cells. Int. J. Mol. Med. 2011, 29, 159–164. [Google Scholar] [CrossRef][Green Version]
- Yu, C.-S.; Yu, F.-S.; Chan, J.K.-S.; Li, T.-M.; Lin, S.-S.; Chen, S.-C.; Hsia, T.-C.; Chang, Y.-H.; Chung, J.-G. Aloe-emodin affects the levels of cytokines and functions of leukocytes from Sprague-Dawley rats. In Vivo 2006, 20, 505–509. [Google Scholar]
- Ruscetti, M.; Leibold, J.; Bott, M.J.; Fennell, M.; Kulick, A.; Salgado, N.R.; Chen, C.-C.; Ho, Y.-J.; Sanchez-Rivera, F.J.; Feucht, J.; et al. NK cell–mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 2018, 362, 1416–1422. [Google Scholar] [CrossRef]
- Cassidy, S.A.; Cheent, K.S.; Khakoo, S. Effects of Peptide on NK Cell-Mediated MHC I Recognition. Front. Immunol. 2014, 5, 133. [Google Scholar] [CrossRef]
- Kim, H.; Byun, J.-E.; Yoon, S.R.; Koohy, H.; Jung, H.; Choi, I. SARS-CoV-2 peptides bind to NKG2D and increase NK cell activity. Cell. Immunol. 2021, 371, 104454. [Google Scholar] [CrossRef]
- Koller, A.; Bianchini, R.; Schlager, S.; Münz, C.; Kofler, B.; Wiesmayr, S. The neuropeptide galanin modulates natural killer cell function. Neuropeptides 2017, 64, 109–115. [Google Scholar] [CrossRef]
- Jang, S.-H.; Oh, M.-S.; Baek, H.-I.; Ha, K.-C.; Lee, J.-Y.; Jang, Y.-S. Silk peptide treatment potentiates natural killer cell activity in vitro and induces natural killer cell maturation and activation in mouse splenocytes. Pharm. Biol. 2019, 57, 369–379. [Google Scholar] [CrossRef]
- Rychener, M.; Steiger, W. Purification, glucose-6-phosphate dehydrogenase inhibition, and HPLC analysis of four 1,8-dihydroxyanthrones. Pharm. Acta Helv. 1989, 64, 8–15. [Google Scholar]
- Iqbal, M.; Iqbal, R.; Bilal, M.; Akram, M.S.; Baloch, I.B.; Baloch, M.K. Antimicrobial Activities of Monoesters of Succinic Acid. Asian J. Chem. 2014, 26, 8025–8028. [Google Scholar] [CrossRef]
- Gong, J.H.; Maki, G.; Klingemann, H.G. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia 1994, 8, 652–658. [Google Scholar]

| Name | Sequence | Function |
|---|---|---|
| LTV | 1 LTVSPWY | HER2 recognition |
| CPP | PKKKRKV | CPP and Nuclear targeting |
| CPP-LTV | PKKKRKVGPGLTVSPWY | full functional sequence |
| 2gLTV | GPGLTVSPWY | linker + HER2 recognition. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stringaro, A.; Serra, S.; Gori, A.; Calcabrini, A.; Colone, M.; Dupuis, M.L.; Spadaro, F.; Cecchetti, S.; Vitali, A. Peptide-Mediated Targeted Delivery of Aloe-Emodin as Anticancer Drug. Molecules 2022, 27, 4615. https://doi.org/10.3390/molecules27144615
Stringaro A, Serra S, Gori A, Calcabrini A, Colone M, Dupuis ML, Spadaro F, Cecchetti S, Vitali A. Peptide-Mediated Targeted Delivery of Aloe-Emodin as Anticancer Drug. Molecules. 2022; 27(14):4615. https://doi.org/10.3390/molecules27144615
Chicago/Turabian StyleStringaro, Annarita, Stefano Serra, Alessandro Gori, Annarica Calcabrini, Marisa Colone, Maria Luisa Dupuis, Francesca Spadaro, Serena Cecchetti, and Alberto Vitali. 2022. "Peptide-Mediated Targeted Delivery of Aloe-Emodin as Anticancer Drug" Molecules 27, no. 14: 4615. https://doi.org/10.3390/molecules27144615
APA StyleStringaro, A., Serra, S., Gori, A., Calcabrini, A., Colone, M., Dupuis, M. L., Spadaro, F., Cecchetti, S., & Vitali, A. (2022). Peptide-Mediated Targeted Delivery of Aloe-Emodin as Anticancer Drug. Molecules, 27(14), 4615. https://doi.org/10.3390/molecules27144615










