Transcriptome Analyses Revealed the Key Metabolic Genes and Transcription Factors Involved in Terpenoid Biosynthesis in Sacred Lotus
Abstract
1. Introduction
2. Results
2.1. Identification of Terpenoid Components in Various Plant Parts
2.2. RNA-Seq and Transcriptomic Assembly
2.3. Gene Annotation and Functional Classification
2.4. Identification of Differential Expression Genes (DEGs)
2.5. Functional Classification of DEGs
2.6. DEGs Involved in Terpenoid biosynthesis
2.7. Co-Expression Network of TPS and WRKY Transcription Factors
2.8. qRT-PCR Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Plants Materials
4.2. LC-MS/MS Analysis
4.3. Total RNA Extraction and Transcriptome Sequencing
4.4. RNA-Seq Analysis
4.5. Validation of DEGs by qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ferrer-Gallego, P.P.; Boisset, F.; Jarvis, C. Typification of the sacred lotus nelumbo nucifera (nelumbonaceae). Taxon 2015, 64, 156–159. [Google Scholar] [CrossRef]
- Lin, Z.; Damaris, R.N.; Shi, T.; Li, J.; Yang, P. Transcriptomic analysis identifies the key genes involved in stamen petaloid in lotus (Nelumbo nucifera). BMC Genom. 2018, 19, 554. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, T.; Huang, L.; He, D.l.; Nyong, A.T.M.; Yang, P. MaSystematic transcriptomic analysis provides insights into lotus (Nelumbo nucifera) seed development. Plant Growth Regul. 2018, 86, 339–350. [Google Scholar] [CrossRef]
- Huang, B.; Ban, X.; He, J.; Jing, T.; Tian, J.; Wang, Y. Hepatoprotective and antioxidant activity of ethanolic extracts of edible lotus (Nelumbo nucifera Gaertn.) leaves. Food Chem. 2010, 120, 873–878. [Google Scholar] [CrossRef]
- Huang, B.; He, J.; Ban, X.; Zeng, H.; Yao, X.; Wang, Y. Antioxidant activity of bovine and porcine meat treated with extracts from edible lotus (Nelumbo nucifera) rhizome knot and leaf. Meat Sci. 2011, 87, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, B.H.; Fang, J.B.; Liu, Y.L.; Zhang, H.H.; Fang, L.C.; Guan, L.; Li, S.H. Analysis of flavonoids from lotus (Nelumbo nucifera) leaves using high performance liquid chromatography/photodiode array detector tandem electrospray ionization mass spectrometry and an extraction method optimized by orthogonal design. J. Chromatogr. A 2012, 1227, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhu, M.; Guo, M. Research advances in traditional and modern use of Nelumbo nucifera: Phytochemicals, health promoting activities and beyond. Crit. Rev. Food Sci. Nutr. 2019, 59, S189–S209. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Mukherjee, D.; Maji, A.K.; Rai, S.; Heinrich, M. The sacred lotus (Nelumbo nucifera)-phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009, 61, 407–422. [Google Scholar] [CrossRef]
- Neill, S.O.; Gould, K.S.; Kilmartin, P.A.; Mitchell, K.A.; Markham, K.R. Antioxidant capacities of green and cyanic leaves in the sun species, Quintinia serrata. Funct. Plant Biol. 2002, 29, 1437–1443. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef]
- Huang, L.; Li, J.; Ye, H.; Li, C.; Wang, H.; Liu, B.; Zhang, Y. Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in catharanthus roseus. Planta 2012, 236, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Zwawiak, J.; Pawelczyk, A.; Olender, D.; Zaprutko, L. Structure and activity of pentacyclic triterpenes codrugs. A review. Mini Rev. Med. Chem. 2021, 21, 1509–1526. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.S.; de Jesus, B.; da Silva Luiz, S.; Viana, C.B.; Adao, M.C.; Figueiredo, F.S.; Carvalho, T.; Silva, M.L.; Londero, V.S.; de Souza Figueiredo, C.T.; et al. Antiinflammatory activity of natural triterpenes—An overview from 2006 to 2021. Phytother. Res. 2022, 36, 1459–1506. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Lu, J.; Wang, J.; Gao, W.Y. Advances in biosynthesis of triterpenoid saponins in medicinal plants. Chin. J. Nat. Med. 2020, 18, 417–424. [Google Scholar] [CrossRef]
- Szakiel, A.; Pączkowski, C.; Henry, M. Influence of environment-al abiotic factors on the content of saponins in plants. Phyto-chem. Rev. 2010, 10, 471–491. [Google Scholar] [CrossRef]
- Bouvier, F.; Rahier, A.; Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 2005, 44, 357–429. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Pulido, P.; Perello, C.; Rodriguez-Concepcion, M. New insights into plant isoprenoid metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef]
- Gantet, P.; Memelink, J. Transcription factors: Tools to engineer the production of pharmacologically active plant metabolites. Trends Pharmacol. Sci. 2002, 23, 563–569. [Google Scholar] [CrossRef]
- He, X.Y.; Wang, H.; Yang, J.F.; Deng, K.; Wang, T. RNA sequencing on Amomum villosum Lour. induced by MeJA identifies the genes of WRKY and terpene synthases involved in terpene biosynthesis. Genome 2018, 61, 91–102. [Google Scholar] [CrossRef]
- Ali, M.; Li, P.; She, G.; Chen, D.; Wan, X.; Zhao, J. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 2017, 7, 16074. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Hussain, R.M.; Rehman, N.U.; She, G.; Li, P.; Wan, X.; Guo, L.; Zhao, J. De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in Blue Anise Sage (Salvia guaranitica L.). DNA Res. 2018, 25, 597–617. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, A.; Patel, P.A.; Sharma, P.; Thoutireddy, S.; Das, N. Lotus (Nelumbo nucifera Gaertn.) and Its Bioactive Phytocompounds: A Tribute to Cancer Prevention and Intervention. Cancers 2022, 14, 529. [Google Scholar] [CrossRef] [PubMed]
- Limwachiranon, J.; Huang, H.; Shi, Z.; Li, L.; Luo, Z. Lotus Flavonoids and Phenolic Acids: Health Promotion and Safe Consumption Dosages. Compr. Rev. Food Sci. Food Saf. 2018, 17, 458–471. [Google Scholar] [CrossRef]
- Menéndez-Perdomo, I.M.; Facchini, P.J. Benzylisoquinoline Alkaloids Biosynthesis in Sacred Lotus. Molecules 2018, 23, 2899. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Pinthong, D.; Hano, C. Flavonoids from Nelumbo nucifera Gaertn., a Medicinal Plant: Uses in Traditional Medicine, Phytochemistry and Pharmacological Activities. Medicines 2018, 5, 127. [Google Scholar] [CrossRef]
- Damasco, G.; Shivakumar, V.S.; Misciewicz, T.M.; Daly, D.C.; Fine, P. Leaf Transcriptome Assembly of Protium copal (Burseraceae) and Annotation of Terpene Biosynthetic Genes. Genes 2019, 10, 392. [Google Scholar] [CrossRef]
- George, J.K.; Shelvy, S.; Fayad, A.M.; Umadevi, P.; Angadi, U.B.; Iquebal, M.A.; Jaiswal, S.; Rai, A.; Kumar, D. De novo transcriptome sequencing assisted identification of terpene synthases from black pepper (Piper nigrum) berry. Physiol. Mol. Biol. Plants 2021, 27, 1153–1161. [Google Scholar] [CrossRef]
- Han, X.J.; Wang, Y.D.; Chen, Y.C.; Lin, L.Y.; Wu, Q.K. Transcriptome sequencing and expression analysis of terpenoid biosynthesis genes in Litsea cubeba. PLoS ONE 2013, 8, e76890. [Google Scholar] [CrossRef]
- Li, R.; Li, Z.; Leng, P.; Hu, Z.; Wu, J.; Dou, D. Transcriptome sequencing reveals terpene biosynthesis pathway genes accounting for volatile terpene of tree peony. Planta 2021, 254, 67. [Google Scholar] [CrossRef]
- Qiu, F.; Wang, X.; Zheng, Y.; Wang, H.; Liu, X.; Su, X. Full-Length Transcriptome Sequencing and Different Chemotype Expression Profile Analysis of Genes Related to Monoterpenoid Biosynthesis in Cinnamomum porrectum. Int. J. Mol. Sci. 2019, 20, 6230. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, B.; Zhang, X.; Zhang, G.; Yan, A.; Wang, H.; Wang, X.; Xu, H. Transcriptome profiles of three Muscat table grape cultivars to dissect the mechanism of terpene biosynthesis. Sci. Data 2019, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xie, C.; Huang, Y.; An, W.; Liu, S.; Huang, S.; Zheng, X. Metabolism and transcriptome profiling provides insight into the genes and transcription factors in volved in monoterpene biosynthesis of borneol chemotype of Cinnamomum camphora induced by mechanical damage. PeerJ 2021, 9, e11465. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng./Biotechnol. 2015, 148, 63–106. [Google Scholar] [CrossRef]
- Cordoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Friesen, J.A.; Rodwell, V.W. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004, 5, 248. [Google Scholar] [CrossRef]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef]
- Srivastava, Y.; Tripathi, S.; Mishra, B.; Sangwan, N.S. Cloning and homologous characterization of geranylgeranyl pyrophosphate synthase (GGPPS) from Withania somnifera revealed alterations in metabolic flux towards gibberellic acid biosynthesis. Planta 2022, 256, 4. [Google Scholar] [CrossRef]
- Rai, A.; Smita, S.S.; Singh, A.K.; Shanker, K.; Nagegowda, D.A. Heteromeric and homomeric geranyl diphosphate synthases from Catharanthus roseus and their role in monoterpene indole alkaloid biosynthesis. Mol. Plant 2013, 6, 1531–1549. [Google Scholar] [CrossRef]
- Wang, G.; Dixon, R.A. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 2009, 106, 9914–9919. [Google Scholar] [CrossRef]
- Zhu, X.F.; Suzuki, K.; Saito, T.; Okada, K.; Tanaka, K.; Nakagawa, T.; Matsuda, H.; Kawamukai, M. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol. Biol. 1997, 35, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Li, N.; Zhang, D.H.; Xu, J.W. Increased production of ganoderic acids by overexpression of homologous farnesyl diphosphate synthase and kinetic modeling of ganoderic acid production in Ganoderma lucidum. Microb. Cell Factories 2019, 18, 115. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.L.; Wong, W.S.; Jang, I.C.; Chua, N.H. Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 2017, 213, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, M.C.; Alfieri, M.; De Tommasi, N.; Moses, T.; Goossens, A.; Leone, A. Boosting the Synthesis of Pharmaceutically Active Abietane Diterpenes in S. sclarea Hairy Roots by Engineering the GGPPS and CPPS Genes. Front. Plant Sci. 2020, 11, 924. [Google Scholar] [CrossRef]
- Engprasert, S.; Taura, F.; Kawamukai, M.; Shoyama, Y. Molecular cloning and functional expression of geranylgeranyl pyrophosphate synthase from Coleus forskohlii Briq. BMC Plant Biol. 2004, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Liao, P.; Zhang, T.; Zhou, W.; Wang, J.; Xu, H.; Liu, Y.; Lin, Z. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioprocess Eng. 2010, 15, 236–245. [Google Scholar] [CrossRef]
- Rahimi, S.; Kim, Y.J.; Devi, B.S.R.; Khorolragchaa, A.; Sukweenadhi, J.; Yang, D.C. Isolation and characterization of Panax ginseng geranylgeranyl-diphosphate synthase genes responding to drought stress. Eur. J. Plant Pathol. 2015, 142, 747–758. [Google Scholar] [CrossRef]
- Takaya, A.; Zhang, Y.W.; Asawatreratanakul, K.; Wititsuwannakul, D.; Wititsuwannakul, R.; Takahashi, S.; Koyama, T. Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyl diphosphate synthase of Hevea brasiliensis. Biochim. Biophys. Acta 2003, 1625, 214–220. [Google Scholar] [CrossRef]
- Wang, Y.; Miao, Z.; Tang, K. Molecular cloning and functional expression analysis of a new gene encoding geranylgeranyl diphosphate synthase from hazel (Corylus avellana L. Gasaway). Mol. Biol. Rep. 2010, 37, 3439–3444. [Google Scholar] [CrossRef]
- Beck, G.; Coman, D.; Herren, E.; Ruiz-Sola, M.A.; Rodríguez-Concepción, M.; Gruissem, W.; Vranová, E. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 2013, 82, 393–416. [Google Scholar] [CrossRef]
- Okada, K.; Saito, T.; Nakagawa, T.; Kawamukai, M.; Kamiya, Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 2000, 122, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chappell, J. Biochemical and genomic characterization of terpene synthases in Magnolia grandiflora. Plant Physiol. 2008, 147, 1017–1033. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Liu, Y.J.; Wang, C.L.; Zeng, Q.Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef]
- Zang, B.; Li, H.; Li, W.; Deng, X.W.; Wang, X. Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 2011, 76, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef]
- Karunanithi, P.S.; Zerbe, P. Terpene Synthases as Metabolic Gatekeepers in the Evolution of Plant Terpenoid Chemical Diversity. Front. Plant Sci. 2019, 10, 1166. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 57, 2081–2093. [Google Scholar] [CrossRef]
- Wang, M.; Qiu, X.; Pan, X.; Li, C. Transcriptional Factor-Mediated Regulation of Active Component Biosynthesis in Medicinal Plants. Curr. Pharm. Biotechnol. 2021, 22, 848–866. [Google Scholar] [CrossRef]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 2013, 1829, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, J.; Xu, Z.; Li, J.; Wang, J. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in white root (Salvia miltiorrhiza). Ind. Crops Prod. 2021, 170, 113784. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
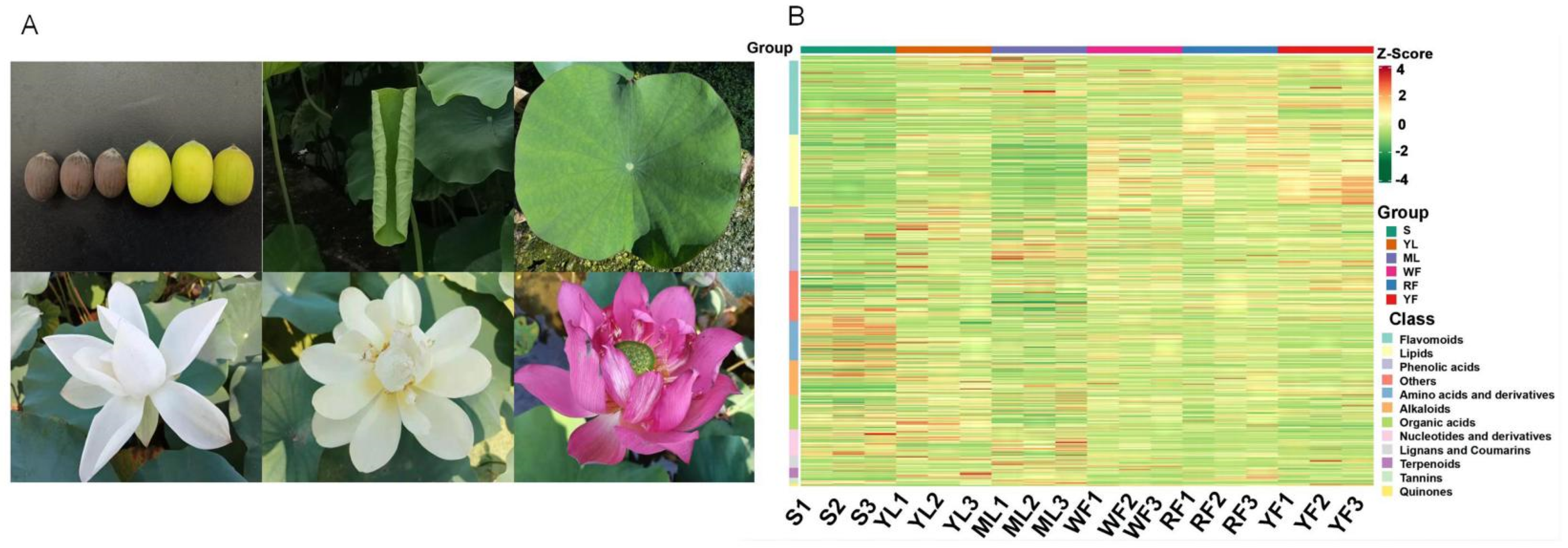

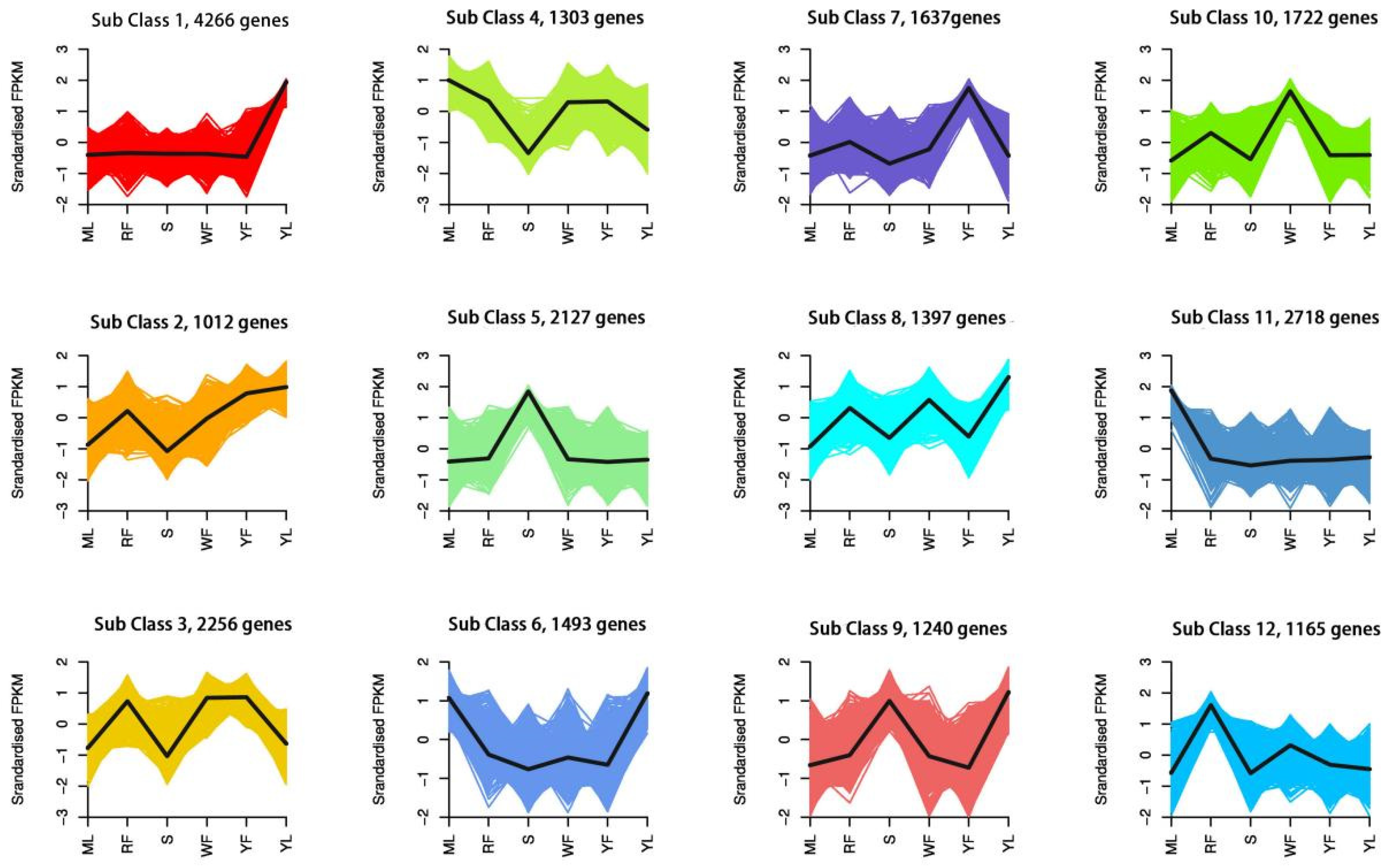
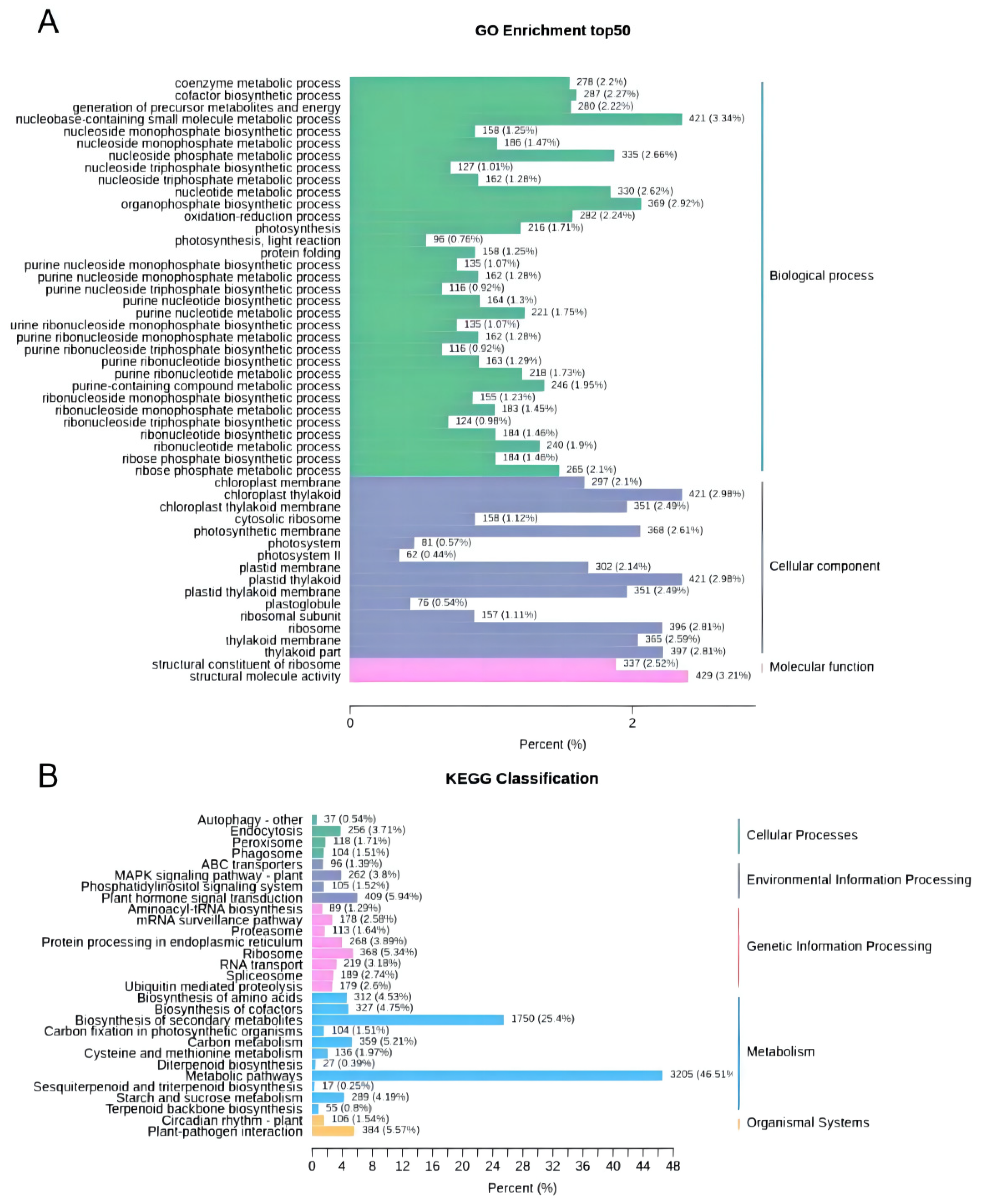
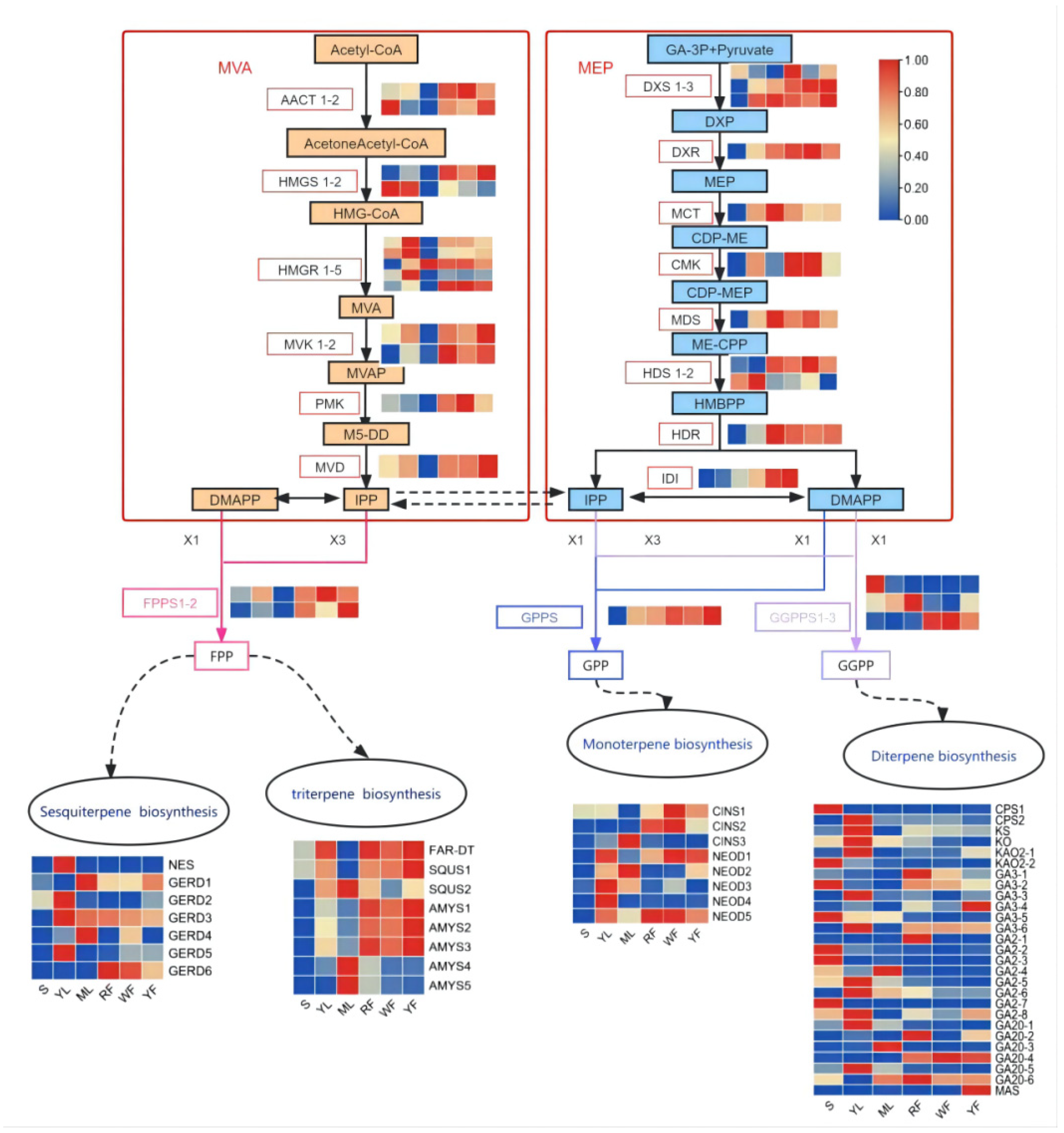
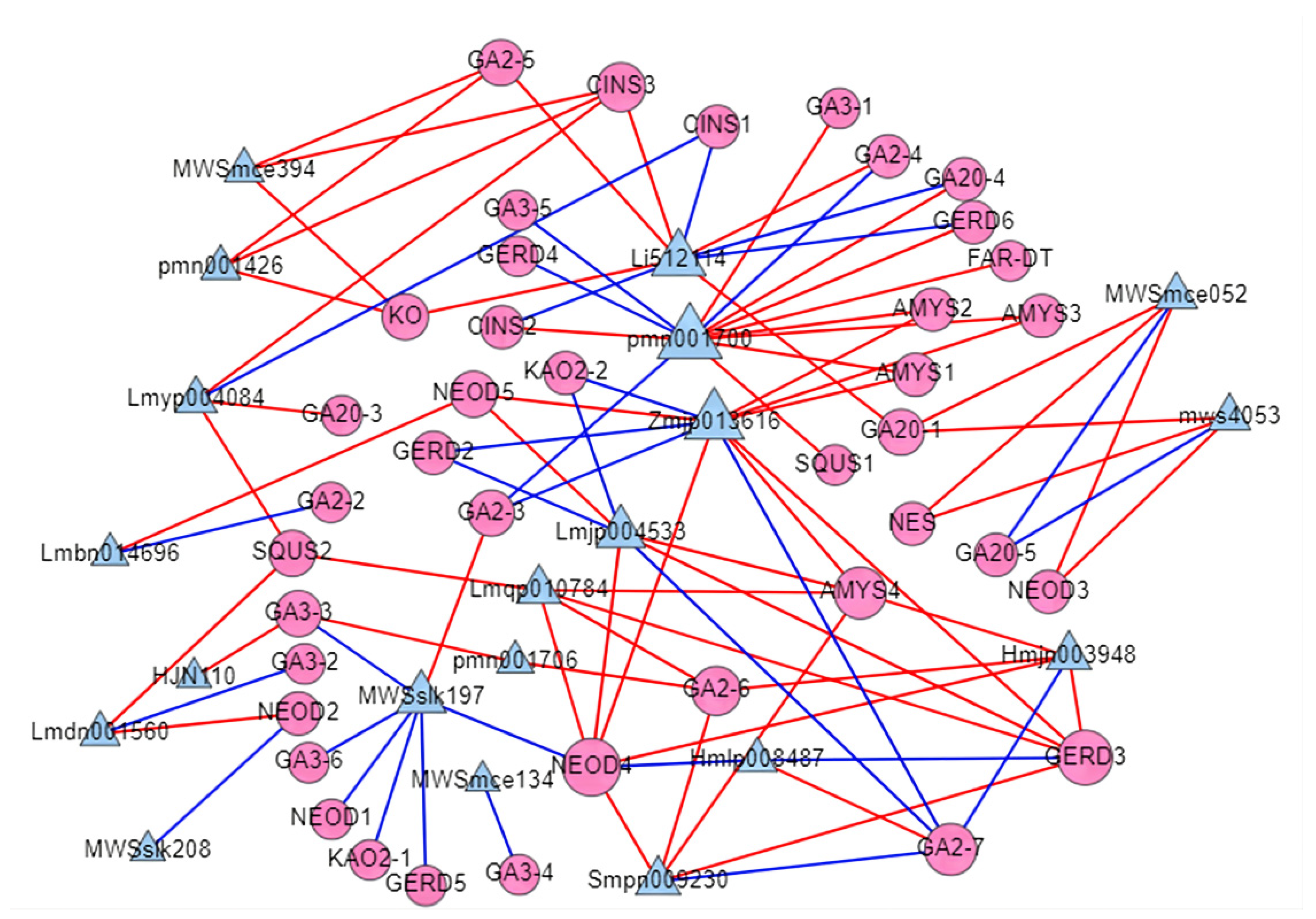
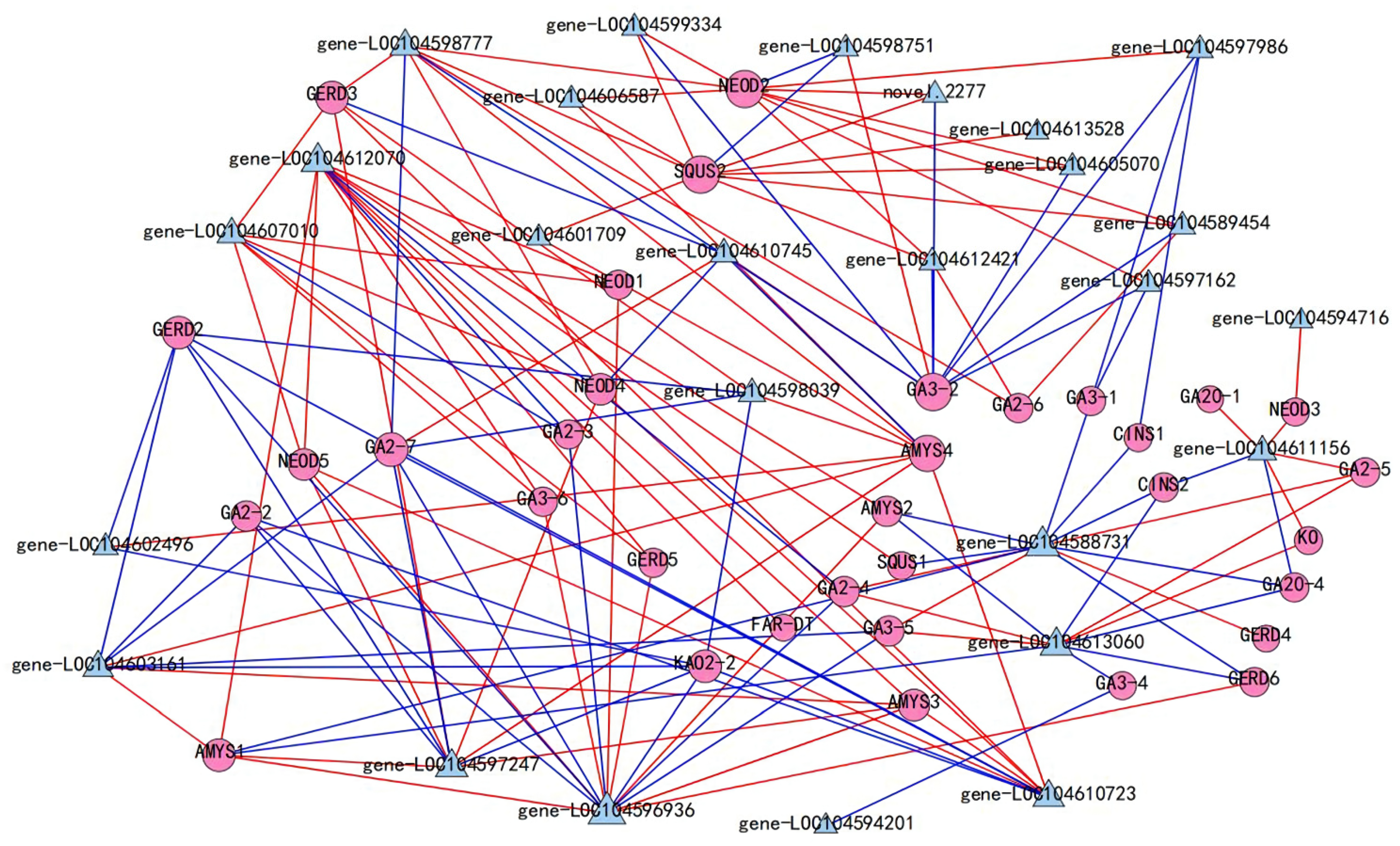

| Index | Compounds | Class II | Q1 (Da) | Q3 (Da) | Level | S | YL | ML | WF | RF | YF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lmyp004084 | Perillyl alcohol | Monoterpenoids | 1.53 × 102 | 1.07 × 102 | 3 | 3.77 × 104 | 7.46 × 104 | 2.75 × 105 | 1.42 × 104 | 1.28 × 105 | 3.54 × 104 |
| Hmlp008487 | Blumenol C | Monoterpenoids | 2.11 × 102 | 8.11 × 101 | 2 | 5.35 × 104 | 3.68 × 104 | 4.29 × 104 | 4.13 × 104 | 4.54 × 104 | 4.00 × 104 |
| MWSslk208 | Kaurenoic Acid | Ditepenoids | 3.01 × 102 | 3.01 × 102 | 2 | 1.39 × 105 | 3.99 × 104 | 3.45 × 104 | 1.92 × 105 | 1.25 × 105 | 2.12 × 104 |
| Lmbn014696 | Pimaric acid | Ditepenoids | 3.01 × 102 | 3.01 × 102 | 2 | 1.31 × 105 | 4.26 × 105 | 2.61 × 105 | 6.73 × 105 | 5.65 × 105 | 2.27 × 105 |
| Lmqp010784 | Progesterone | Ditepenoids | 3.15 × 102 | 1.09 × 102 | 2 | 4.26 × 104 | 1.78 × 106 | 1.10 × 106 | 1.65 × 105 | 2.05 × 106 | 2.96 × 105 |
| Lmdn001560 | 6-DeoxyCatalpol | Sesquiterpenoids | 3.45 × 102 | 1.65 × 102 | 2 | 7.79 × 104 | 2.12 × 105 | 3.96 × 105 | 9.90 × 104 | 1.53 × 105 | 2.15 × 105 |
| Hmmn003964 | 7-Deoxyloganic acid | Sesquiterpenoids | 3.59 × 102 | 1.97 × 102 | 3 | 1.17 × 105 | 2.09 × 104 | 4.41 × 104 | 6.32 × 104 | 4.45 × 104 | 7.78 × 103 |
| MWSslk197 | Secoxyloganin | Sesquiterpenoids | 4.05 × 102 | 1.65 × 102 | 2 | 1.31 × 105 | 9.96 × 103 | 5.50 × 104 | 3.76 × 104 | 2.46 × 104 | 2.01 × 104 |
| Lmjp004533 | Kisasagenol A Triacetate | Ditepenoids | 4.47 × 102 | 2.85 × 102 | 2 | 9.00 × 100 | 6.08 × 104 | 1.13 × 105 | 3.12 × 104 | 1.89 × 104 | 6.32 × 104 |
| MWSmce134 | Betulonic acid | Triterpene | 4.53 × 102 | 4.53 × 102 | 3 | 4.16 × 103 | 7.04 × 103 | 3.15 × 103 | 1.13 × 103 | 9.60 × 102 | 9.00 × 100 |
| pmn001700 | 24,30-Dihydroxy-12(13)-enolupinol | Triterpene | 4.55 × 102 | 4.55 × 102 | 1 | 1.84 × 104 | 5.13 × 104 | 1.33 × 104 | 3.60 × 105 | 4.79 × 105 | 3.42 × 105 |
| HJN110 | Betulinic acid | Triterpene | 4.55 × 102 | 4.55 × 102 | 2 | 1.30 × 105 | 4.16 × 105 | 1.77 × 105 | 5.79 × 104 | 1.64 × 105 | 1.84 × 105 |
| mws4053 | Ursolic acid | Triterpene | 4.55 × 102 | 4.55 × 102 | 3 | 5.60 × 105 | 4.24 × 106 | 1.08 × 106 | 5.73 × 105 | 2.41 × 105 | 4.01 × 105 |
| MWSmce052 | 3-Epiursolic acid | Triterpene | 4.55 × 102 | 4.55 × 102 | 3 | 5.39 × 105 | 4.05 × 106 | 1.14 × 106 | 5.63 × 105 | 2.45 × 105 | 3.94 × 105 |
| Zmjp013616 | 12,13-Dihydroursolic acid | Triterpene | 4.59 × 102 | 4.23 × 102 | 3 | 9.00 × 100 | 3.98 × 104 | 1.62 × 104 | 1.49 × 104 | 1.89 × 105 | 1.15 × 105 |
| pmn001706 | 2-Hydroxyoleanolic acid | Triterpene | 4.71 × 102 | 4.71 × 102 | 1 | 6.68 × 105 | 1.94 × 107 | 5.08 × 106 | 1.10 × 106 | 1.00 × 107 | 1.41 × 106 |
| Li512114 | Corosolic Acid Methyl Ester | Triterpene | 4.85 × 102 | 4.53 × 102 | 3 | 3.10 × 103 | 5.62 × 103 | 5.89 × 103 | 1.18 × 103 | 2.29 × 103 | 1.32 × 103 |
| MWSmce394 | Tormentic acid | Triterpene | 4.87 × 102 | 4.69 × 102 | 3 | 3.62 × 103 | 3.18 × 104 | 1.52 × 104 | 6.23 × 102 | 6.18 × 103 | 9.00 × 100 |
| Hmjn003948 | Madasiatic acid | Triterpene | 4.87 × 102 | 4.87 × 102 | 2 | 3.49 × 104 | 1.87 × 106 | 5.72 × 105 | 2.35 × 105 | 2.15 × 106 | 6.11 × 105 |
| pmn001426 | Euscaphic acid | Triterpene | 4.87 × 102 | 4.69 × 102 | 3 | 3.18 × 103 | 3.23 × 104 | 1.60 × 104 | 3.93 × 102 | 6.52 × 103 | 9.00 × 100 |
| Smpn009230 | 2α,3α,23-trihydroxyolean-12-en-28-oic acid | Triterpene | 4.87 × 102 | 4.87 × 102 | 2 | 3.34 × 104 | 1.88 × 106 | 5.09 × 105 | 2.27 × 105 | 2.01 × 106 | 5.99 × 105 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Du, F.; Yang, N.; Zhang, C.; Wang, Z.; Zheng, X.; Tang, J.; Yang, L.; Dong, C. Transcriptome Analyses Revealed the Key Metabolic Genes and Transcription Factors Involved in Terpenoid Biosynthesis in Sacred Lotus. Molecules 2022, 27, 4599. https://doi.org/10.3390/molecules27144599
Qin L, Du F, Yang N, Zhang C, Wang Z, Zheng X, Tang J, Yang L, Dong C. Transcriptome Analyses Revealed the Key Metabolic Genes and Transcription Factors Involved in Terpenoid Biosynthesis in Sacred Lotus. Molecules. 2022; 27(14):4599. https://doi.org/10.3390/molecules27144599
Chicago/Turabian StyleQin, Lili, Fei Du, Ningning Yang, Chen Zhang, Zhiwen Wang, Xingwen Zheng, Jiawei Tang, Liangbo Yang, and Chen Dong. 2022. "Transcriptome Analyses Revealed the Key Metabolic Genes and Transcription Factors Involved in Terpenoid Biosynthesis in Sacred Lotus" Molecules 27, no. 14: 4599. https://doi.org/10.3390/molecules27144599
APA StyleQin, L., Du, F., Yang, N., Zhang, C., Wang, Z., Zheng, X., Tang, J., Yang, L., & Dong, C. (2022). Transcriptome Analyses Revealed the Key Metabolic Genes and Transcription Factors Involved in Terpenoid Biosynthesis in Sacred Lotus. Molecules, 27(14), 4599. https://doi.org/10.3390/molecules27144599






