Screening of Specific and Common Pathways in Breast Cancer Cell Lines MCF-7 and MDA-MB-231 Treated with Chlorophyllides Composites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Chlorophyll Extraction and Measurement
2.3. Preparation of Chlorophyllides Composites by Using Chlorophyllase
2.4. Total RNA Preparation for Sequencing
2.5. Preparation of cDNA Library and Sequencing
2.6. Microarray Gene Expression Profiling
2.7. Quantitative Reverse Transcription PCR (RT-qPCR)
2.8. Statistical Analysis
3. Results and Discussion
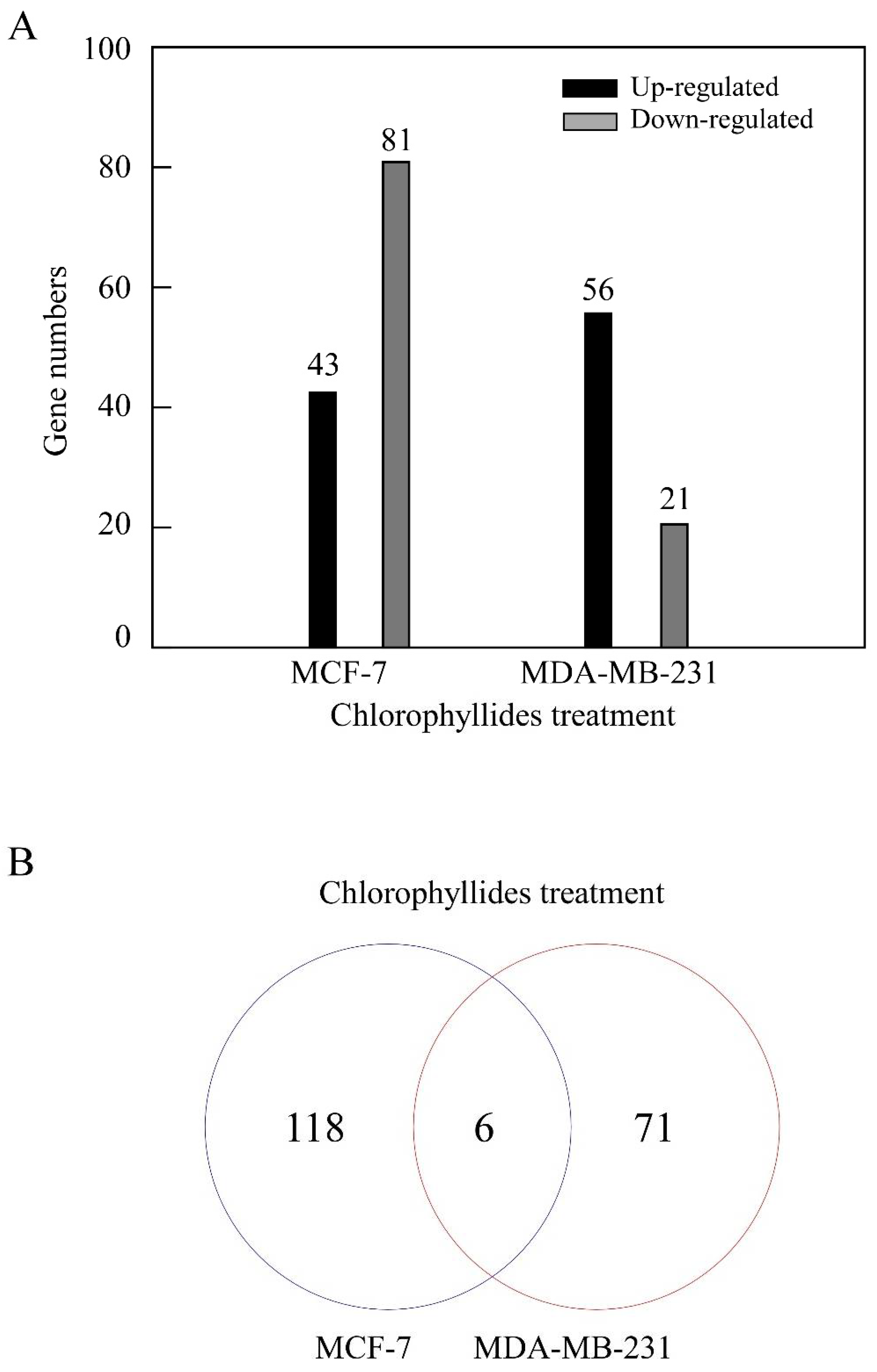
3.1. DEG Analysis in MCF-7 and MDA-MB-231 Cells
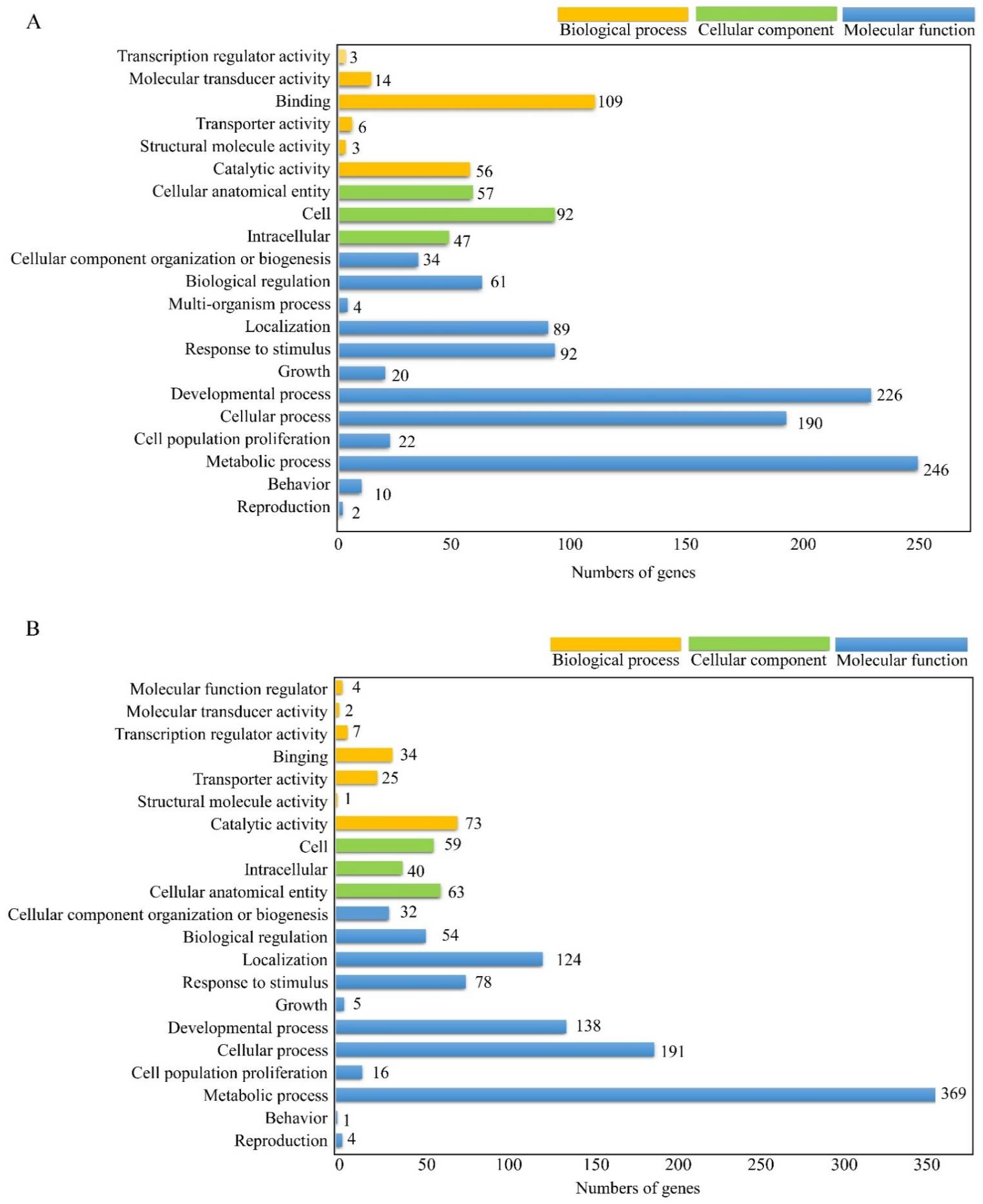
3.2. GO Annotation of Differential Expessed Genes
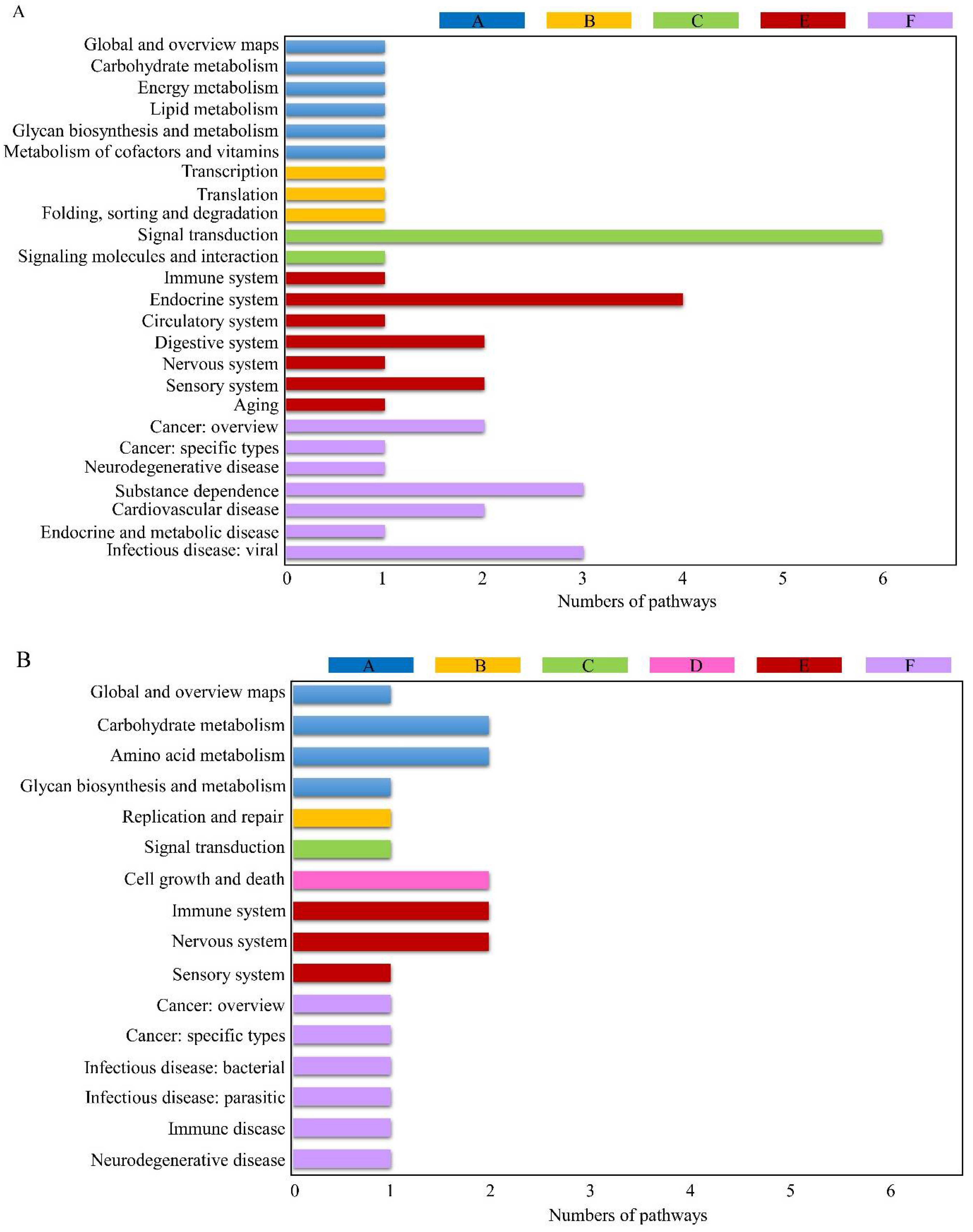
3.3. KEGG Pathway Analysis of DEGs
3.4. Analysis of Common KEGG Pathways
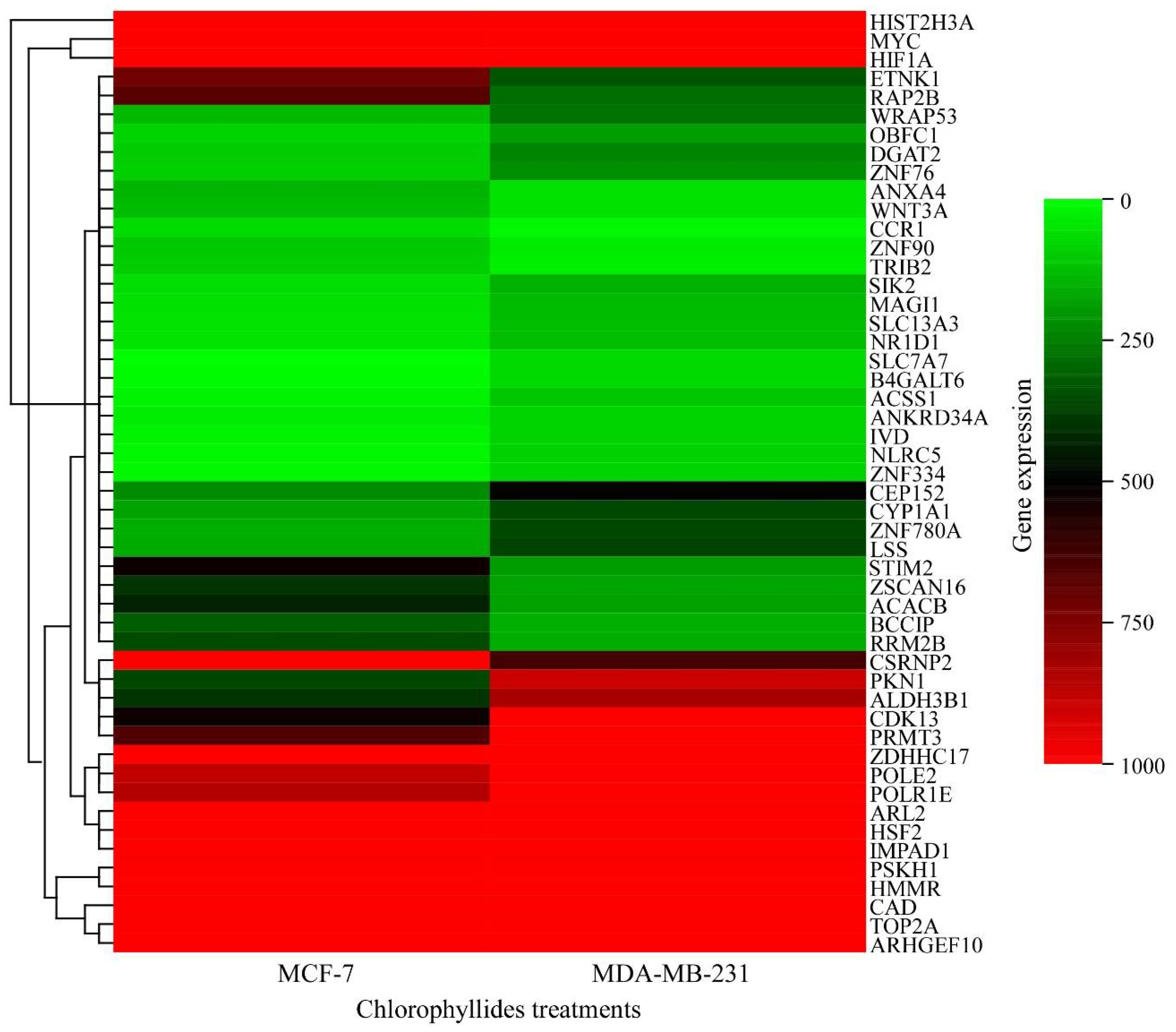
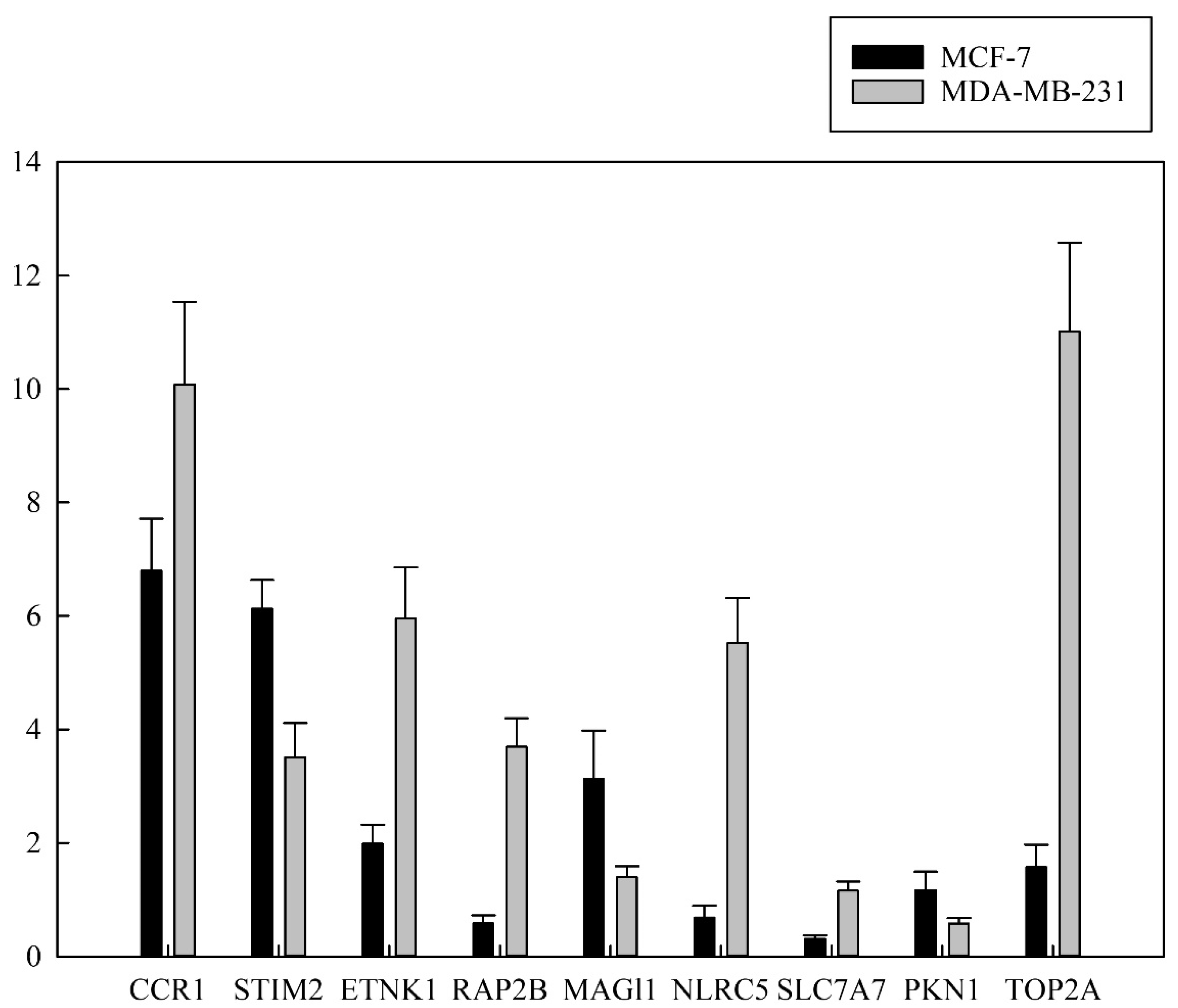
3.5. Validation of RNA Expression by RT-qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| (ANXA4) | annexin A4 |
| (BP) | biological process |
| (CC) | cellular component |
| (CCR1) | chemokine C-C motif receptor 1 |
| (TOP2A) | DNA topoisomerase II alpha 170 kDa |
| (ER) | estrogen receptor |
| (ETNK1) | ethanolamine kinase 1 |
| (FC) | fold change |
| (HER2) | human epidermal growth receptor 2 |
| (RAP2B) | member of RAS oncogene family |
| (MAGI1) | membrane associated guanylate kinase WW and PDZ domain containing 1 |
| (MF) | molecular function |
| (NLRC5) | NLR family CARD domain containing 5 |
| (SLC7A7) | solute carrier family 7 membrane 7 |
| (STIM2) | stromal interaction molecule 2 |
| (PR) | progesterone receptor |
| (PKN1) | protein kinase N1 |
| (TNBC) | triple negative breast cancer |
| (RT-qPCR) | quantitative reverse transcription PCR |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- McCann, K.E.; Hurvitz, S.A.; McAndrew, N. Advances in targeted therapies for triple-begative breast cancer. Drugs 2019, 79, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA A Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.P.; Rosato, R.R.; Li, X.; Moulder, S.; Piwnica-Worms, H.; Chang, J.C. A comprehensive overview of metaplastic breast cancer: Clinical features and molecular aberrations. Breast Cancer Res. 2020, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 10286, 1750–1769. [Google Scholar] [CrossRef]
- ComŞA, Ş.; CÎMpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147. [Google Scholar]
- Theodossiou, T.A.; Ali, M.; Grigalavicius, M.; Grallert, B.; Dillard, P.; Schink, K.O.; Olsen, C.E.; Wälchli, S.; Inderberg, E.M.; Kubin, A.; et al. Simultaneous defeat of MCF7 and MDA-MB-231 resistances by a hypericin PDT–tamoxifen hybrid therapy. NPJ Breast Cancer 2019, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- da Silva, J.L.; Cardoso Nunes, N.C.; Izetti, P.; de Mesquita, G.G.; de Melo, A.C. Triple negative breast cancer: A thorough review of biomarkers. Crit. Rev. Oncol. Hematol. 2020, 145, 102855. [Google Scholar] [CrossRef]
- Reis-Filho, J.S.; Pusztai, L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet 2011, 378, 1812–1823. [Google Scholar] [CrossRef]
- González-Martínez, S.; Pérez-Mies, B.; Carretero-Barrio, I.; Palacios-Berraquero, M.L.; Perez-García, J.; Cortés, J.; Palacios, J. Molecular Features of Metaplastic Breast Carcinoma: An Infrequent Subtype of Triple Negative Breast Carcinoma. Cancers 2020, 12, 1832. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast cancer treatment: A review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, F.; Nuzzolese, I.; Ponzone, R. Neoadjuvant or adjuvant chemotherapy in early breast cancer? Expert Opin Pharm. 2020, 21, 1071–1082. [Google Scholar] [CrossRef]
- Diaby, V.; Tawk, R.; Sanogo, V.; Xiao, H.; Montero, A.J. A review of systematic reviews of the cost-effectiveness of hormone therapy, chemotherapy, and targeted therapy for breast cancer. Breast Cancer Res. Treat. 2015, 151, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigematsu, H.; Fujisawa, T.; Shien, T.; Iwata, H. Omitting surgery for early breast cancer showing clinical complete response to primary systemic therapy. Jpn. J. Clin. Oncol. 2020, 50, 629–634. [Google Scholar] [CrossRef]
- Recht, A.; McArthur, H.; Solin, L.J.; Tendulkar, R.; Whitley, A.; Giuliano, A. Contemporary guidelines in whole-breast irradiation: An alternative perspective. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 567–573. [Google Scholar] [CrossRef]
- Castaneda, S.A.; Strasser, J. Updates in the treatment of breast cancer with radiotherapy. Surg. Oncol. Clin. 2017, 26, 371–382. [Google Scholar] [CrossRef]
- Lyseng-Williamson, K.A.; Fenton, C. Docetaxel: A review of its use in metastatic breast cancer. Drugs 2005, 65, 2513–2531. [Google Scholar] [CrossRef]
- Willson, M.L.; Burke, L.; Ferguson, T.; Ghersi, D.; Nowak, A.K.; Wilcken, N. Taxanes for adjuvant treatment of early breast cancer. Cochrane Database Syst. Rev. 2019, 9, CD004421. [Google Scholar] [CrossRef]
- Alqahtani, F.Y.; Aleanizy, F.S.; El Tahir, E.; Alkahtani, H.M.; AlQuadeib, B.T. Chapter Three—Paclitaxel. In Profiles of Drug Substances, Excipients and Related Methodology; Brittain, H.G., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 44, pp. 205–238. [Google Scholar]
- Gallego-Jara, J.; Lozano-Terol, G.; Sola-Martínez, R.A.; Cánovas-Díaz, M.; de Diego Puente, T. A compressive review about Taxol(®): History and future challenges. Molecules 2020, 25, 5986. [Google Scholar] [CrossRef]
- Dehelean, C.A.; Marcovici, I.; Soica, C.; Mioc, M.; Coricovac, D.; Iurciuc, S.; Cretu, O.M.; Pinzaru, I. Plant-derived anticancer compounds as new perspectives in drug discovery and alternative therapy. Molecules 2021, 26, 1109. [Google Scholar] [CrossRef] [PubMed]
- Nacht, M.; Ferguson, A.T.; Zhang, W.; Petroziello, J.M.; Cook, B.P.; Gao, Y.H.; Maguire, S.; Riley, D.; Coppola, G.; Landes, G.M.; et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999, 59, 5464–5470. [Google Scholar]
- Russo, G.; Zegar, C.; Giordano, A. Advantages and limitations of microarray technology in human cancer. Oncogene 2003, 22, 6497–6507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and Microarray Gene Expression Platforms for the Toxicogenomic Evaluation of Liver From Short-Term Rat Toxicity Studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brązert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, R.; Parker, J.S.; Jones, W.D.; Livasy, C.A.; Coleman, W.B. Microarray-Based Gene Expression Profiling for Molecular Classification of Breast Cancer and Identification of New Targets for Therapy. Lab. Med. 2010, 41, 364–372. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef]
- Malvia, S.; Bagadi, S.A.R.; Pradhan, D.; Chintamani, C.; Bhatnagar, A.; Arora, D.; Sarin, R.; Saxena, S. Study of Gene Expression Profiles of Breast Cancers in Indian Women. Sci. Rep. 2019, 9, 10018. [Google Scholar] [CrossRef]
- Glinsky, G.V.; Berezovska, O.; Glinskii, A.B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Investig. 2005, 115, 1503–1521. [Google Scholar] [CrossRef] [Green Version]
- Yoshimaru, T.; Nakamura, Y.; Katagiri, T. Functional genomics for breast cancer drug target discovery. J. Hum. Genet. 2021, 66, 927–935. [Google Scholar] [CrossRef]
- Kim, G.-E.; Kim, N.I.; Lee, J.S.; Park, M.H.; Kang, K. Differentially Expressed Genes in Matched Normal, Cancer, and Lymph Node Metastases Predict Clinical Outcomes in Patients With Breast Cancer. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 111. [Google Scholar] [CrossRef] [PubMed]
- Kuo, W.-H.; Chang, Y.-Y.; Lai, L.-C.; Tsai, M.-H.; Hsiao, C.K.; Chang, K.-J.; Chuang, E.Y. Molecular Characteristics and Metastasis Predictor Genes of Triple-Negative Breast Cancer: A Clinical Study of Triple-Negative Breast Carcinomas. PLoS ONE 2012, 7, e45831. [Google Scholar] [CrossRef] [PubMed]
- Bellon, J.R.; Burstein, H.J.; Frank, E.S.; Mittendorf, E.A.; King, T.A. Multidisciplinary considerations in the treatment of triple-negative breast cancer. CA A Cancer J. Clin. 2020, 70, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Pusztai, L. Gene-Expression Signatures in Breast Cancer. New Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef] [Green Version]
- Hortobagyi, G.N. Treatment of Breast Cancer. N. Engl. J. Med. 1998, 339, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Yang, C.H.; Huang, T.Y.; Tai, M.H.; Sie, R.H.; Shaw, J.F. Cytotoxic effects of chlorophyllides in ethanol crude extracts from plant leaves. Evid Based Complement Altern. Med. 2019, 2019, 9494328. [Google Scholar] [CrossRef]
- Hsiang, Y.P.; Wang, Y.T.; Huang, K.S.; Huang, T.Y.; Tai, M.H.; Lin, Y.M.; Yang, C.H.; Shaw, J.F. Facile production of chlorophyllides using recombinant CrCLH1 and their cytotoxicity towards multidrug resistant breast cancer cell lines. PLoS ONE 2021, 16, e0250565. [Google Scholar] [CrossRef]
- Qin, H.; Li, S.; Li, D. An improved method for determining phytoplankton chlorophyll a concentration without filtration. Hydrobiologia 2013, 707, 81–95. [Google Scholar] [CrossRef] [Green Version]
- Chou, Y.L.; Ko, C.Y.; Yen, C.C.; Chen, L.F.; Shaw, J.F. A novel recombinant chlorophyllase1 from Chlamydomonas reinhardtii for the production of chlorophyllide derivatives. J. Agric. Food Chem. 2015, 63, 9496–9503. [Google Scholar] [CrossRef]
- Hummon, A.B.; Lim, S.R.; Difilippantonio, M.J.; Ried, T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 2007, 42, 467–472. [Google Scholar] [CrossRef]
- Sheu, C.C.; Tsai, M.J.; Chen, F.W.; Chang, K.F.; Chang, W.A.; Chong, I.W.; Kuo, P.L.; Hsu, Y.L. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget 2017, 8, 82674–82688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, J.; Cui, R.; Lin, J.; Ding, X. Curcumin in Treating Breast Cancer: A Review. J. Lab. Autom. 2016, 21, 723–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yang, Y.; An, Y.; Fang, G. The mechanism of anticancer action and potential clinical use of kaempferol in the treatment of breast cancer. Biomed. Pharm. 2019, 117, 109086. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Li, T.; Tian, J.X.; Xi, P.; Liu, R.H. Ursolic acid, a potential anticancer compound for breast cancer therapy. Crit. Rev. Food Sci. Nutr. 2018, 58, 568–574. [Google Scholar] [CrossRef]
- Ezzati, M.; Yousefi, B.; Velaei, K.; Safa, A. A review on anti-cancer properties of Quercetin in breast cancer. Life Sci. 2020, 248, 117463. [Google Scholar] [CrossRef]
- Guo, W.; Xu, B.; Wang, X.; Zheng, B.; Du, J.; Liu, S. The Analysis of the Anti-Tumor Mechanism of Ursolic Acid Using Connectively Map Approach in Breast Cancer Cells Line MCF-7. Cancer Manag. Res. 2020, 12, 3469–3476. [Google Scholar] [CrossRef]
- Bachmeier, B.E.; Mohrenz, I.V.; Mirisola, V.; Schleicher, E.; Romeo, F.; Höhneke, C.; Jochum, M.; Nerlich, A.G.; Pfeffer, U. Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFkappaB. Carcinogenesis 2008, 29, 779–789. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Zhang, B.; Bie, Q.; Qian, H.; Xu, W. Transcriptome Analysis Reveals Key Genes and Pathways Associated with Metastasis in Breast Cancer. OncoTargets Ther. 2020, 13, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Sun, N.; Gao, P.; Li, Y.; Yan, Z.; Peng, Z.; Zhang, Y.; Han, F.; Qi, X. Screening and Identification of Key Common and Specific Genes and Their Prognostic Roles in Different Molecular Subtypes of Breast Cancer. Front. Mol. Biosci. 2021, 8, 619110. [Google Scholar] [CrossRef]
- Kim, C.; Kim, B. Anti-Cancer Natural Products and Their Bioactive Compounds Inducing ER Stress-Mediated Apoptosis: A Review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akram, I.G.; Georges, R.; Hielscher, T.; Adwan, H.; Berger, M.R. The chemokines CCR1 and CCRL2 have a role in colorectal cancer liver metastasis. Tumor Biol. 2016, 37, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Kojder, K.; Barczak, K.; Simińska, D.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Hypoxia alters the expression of CC chemokines and CC chemokine receptors in a tumor-a literature review. Int. J. Mol. Sci. 2020, 21, 5647. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Tallapragada, S.; Schaar, B.; Kamat, K.; Chanana, A.M.; Zhang, Y.; Patel, S.; Parkash, V.; Rinker-Schaeffer, C.; Folkins, A.K.; et al. Omental macrophages secrete chemokine ligands that promote ovarian cancer colonization of the omentum via CCR1. Commun. Biol. 2020, 3, 524. [Google Scholar] [CrossRef]
- Shin, S.Y.; Lee, D.H.; Lee, J.; Choi, C.; Kim, J.Y.; Nam, J.S.; Lim, Y.; Lee, Y.H. C-C motif chemokine receptor 1 (CCR1) is a target of the EGF-AKT-mTOR-STAT3 signaling axis in breast cancer cells. Oncotarget 2017, 8, 94591–94605. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Wu, H. CC chemokine receptors in lung adenocarcinoma: The inflammation-related prognostic biomarkers and immunotherapeutic targets. J. Inflamm. Res. 2021, 14, 267–285. [Google Scholar] [CrossRef]
- Emrich, S.M.; Yoast, R.E.; Xin, P.; Zhang, X.; Pathak, T.; Nwokonko, R.; Gueguinou, M.F.; Subedi, K.P.; Zhou, Y.; Ambudkar, I.S.; et al. Cross-talk between N-terminal and C-terminal domains in stromal interaction molecule 2 (STIM2) determines enhanced STIM2 sensitivity. J. Biol. Chem. 2019, 294, 6318–6332. [Google Scholar] [CrossRef] [Green Version]
- Novello, M.J.; Zhu, J.; Feng, Q.; Ikura, M.; Stathopulos, P.B. Structural elements of stromal interaction molecule function. Cell Calcium 2018, 73, 88–94. [Google Scholar] [CrossRef]
- Nelson, H.A.; Roe, M.W. Molecular physiology and pathophysiology of stromal interaction molecules. Exp. Biol. Med. 2018, 243, 451–472. [Google Scholar] [CrossRef]
- Berna-Erro, A.; Jardin, I.; Salido, G.M.; Rosado, J.A. Role of STIM2 in cell function and physiopathology. J. Physiol. 2017, 595, 3111–3128. [Google Scholar] [CrossRef] [Green Version]
- López, E.; Salido, G.M.; Rosado, J.A.; Berna-Erro, A. Unraveling STIM2 function. J. Physiol. Biochem. 2012, 68, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Lasho, T.L.; Finke, C.M.; Zblewski, D.; Patnaik, M.; Ketterling, R.P.; Chen, D.; Hanson, C.A.; Tefferi, A.; Pardanani, A. Novel recurrent mutations in ethanolamine kinase 1 (ETNK1) gene in systemic mastocytosis with eosinophilia and chronic myelomonocytic leukemia. Blood Cancer J. 2015, 5, e275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, D.; Mauri, M.; Renso, R.; Docci, M.; Crespiatico, I.; Røst, L.M.; Jang, M.; Niro, A.; D’Aliberti, D.; Massimino, L.; et al. ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine. Nat. Commun. 2020, 11, 5938. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Huang, H.; Di, J.; Gao, K.; Lu, Z.; Zheng, J. Structure, functional regulation and signaling properties of Rap2B. Oncol. Lett. 2016, 11, 2339–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.Y.; Li, X.H.; Tian, G.W.; Zhang, D.Y.; Gao, H.; Wang, Z.Y. MAGI1 inhibits the proliferation, migration and invasion of glioma cells. OncoTargets Ther. 2019, 12, 11281–11290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Jia, S.; Martin, T.A.; Jiang, W.G. Regulation and involvement in cancer and pathological conditions of MAGI1, a tight junction protein. Anticancer Res. 2014, 34, 3251–3256. [Google Scholar] [PubMed]
- Shukla, A.; Cloutier, M.; Appiya Santharam, M.; Ramanathan, S.; Ilangumaran, S. The MHC Class-I Transactivator NLRC5: Implications to Cancer Immunology and Potential Applications to Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 1964. [Google Scholar] [CrossRef]
- Cho, S.X.; Vijayan, S.; Yoo, J.S.; Watanabe, T.; Ouda, R.; An, N.; Kobayashi, K.S. MHC class I transactivator NLRC5 in host immunity, cancer and beyond. Immunology 2021, 162, 252–261. [Google Scholar] [CrossRef]
- Noguchi, A.; Takahashi, T. Overview of symptoms and treatment for lysinuric protein intolerance. J. Hum. Genet. 2019, 64, 849–858. [Google Scholar] [CrossRef]
- Bodoy, S.; Sotillo, F.; Espino-Guarch, M.; Sperandeo, M.P.; Ormazabal, A.; Zorzano, A.; Sebastio, G.; Artuch, R.; Palacín, M. Inducible Slc7a7 knockout mouse model recapitulates lysinuric protein intolerance disease. Int. J. Mol. Sci. 2019, 20, 5294. [Google Scholar] [CrossRef] [Green Version]
- Zeng, R.; Wang, Z.; Li, X.; Chen, Y.; Yang, S.; Dong, J. Cyclin-dependent kinase 1-mediated phosphorylation of protein kinase N1 promotes anchorage-independent growth and migration. Cell. Signal. 2020, 69, 109546. [Google Scholar] [CrossRef] [PubMed]
- Venkadakrishnan, V.B.; DePriest, A.D.; Kumari, S.; Senapati, D.; Ben-Salem, S.; Su, Y.; Mudduluru, G.; Hu, Q.; Cortes, E.; Pop, E.; et al. Protein Kinase N1 control of androgen-responsive serum response factor action provides rationale for novel prostate cancer treatment strategy. Oncogene 2019, 38, 4496–4511. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Shao, B.; Zhou, Y.; Chen, Z. High expression of TOP2A in hepatocellular carcinoma is associated with disease progression and poor prognosis. Oncol. Lett. 2020, 20, 232. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.A.; Kirkland, J.G.; Caswell-Jin, J.L.; Crabtree, G.R.; Curtis, C. Chromatin regulators mediate anthracycline sensitivity in breast cancer. Nat. Med. 2019, 25, 1721–1727. [Google Scholar] [CrossRef]
- Wu, Y.M.; Hu, W.; Wang, Y.; Wang, N.; Gao, L.; Chen, Z.Z.; Zheng, W.Q. Exploring novel targets of basal-like breast carcinoma by comparative gene profiling and mechanism analysis. Breast Cancer Res. Treat. 2013, 141, 23–32. [Google Scholar] [CrossRef]
- Weisman, P.S.; Ng, C.K.Y.; Brogi, E.; Eisenberg, R.E.; Won, H.H.; Piscuoglio, S.; De Filippo, M.R.; Ioris, R.; Akram, M.; Norton, L.; et al. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod. Pathol. 2016, 29, 476–488. [Google Scholar] [CrossRef] [Green Version]
- Dillon, J.L.; Mockus, S.M.; Ananda, G.; Spotlow, V.; Wells, W.A.; Tsongalis, G.J.; Marotti, J.D. Somatic gene mutation analysis of triple negative breast cancers. Breast 2016, 29, 202–207. [Google Scholar] [CrossRef]

| Name | Sequence | Target Gene | Product Size (bp) |
|---|---|---|---|
| GAPDH-F | ATCACTGCCACCCAGA AGAC | GAPDH | 460 |
| GAPDH-R | ATGAGGTCCACCACCCTGTT | ||
| CCR1-F | AGAAGCCGGGATGGAAACTC | CCR1 | 165 |
| CCR1-R | TTCCAACCAGGCCAATGACA | ||
| STIM2-F | AGTCTTTGGGACTCTGCACG | STIM2 | 129 |
| STIM2-R | TGTTGCCAGCGAAAAAGTCG | ||
| ETNK1-F | CCAAAGCATGTCTGCAACCC | ETNK1 | 114 |
| ETNK1-R | AAGCAGAAGCCTTGACCCTC | ||
| RAP2B-F | AGCTTCCAGGACATCAAGCC | RAP2B | 190 |
| RAP2B-R | AGGCTTTGTTTTTGGCCGAC | ||
| MAGIL-F | GCCTTGCACAACCCGATCT | MAGIL | 150 |
| MAGIL-R | GGCTTGGGTGTCCCATAATAG | ||
| NLRC5-F | ACCTTAAGCCTGTGTCCACG | NLRC5 | 115 |
| NLRC5-R | CTGTGAACCTGCCACAGCA | ||
| SLC7A7-F | CTCACTGCTTAACGGCGTGT | SLC7A7 | 170 |
| SLC7A7-R | CCAGTTCCGCATAACAAAGG | ||
| PKN1-F | GCCATCAAGGCTCTGAAGAA | PKN1 | 136 |
| PKN1-R | GTCTGGAAACAGCCGAAGAG | ||
| TOP2A-F | CTTTGGCTCGATTGTTATTTCC | TOP2A | 142 |
| TOP2A-R | CCCAGTACCGATTCCTTCAG |
| Pathway ID | Pathway Description | Number of DEGs | All Genes with Pathway Annotation | q-Value | ||
|---|---|---|---|---|---|---|
| Up | Down | Total DEGs | ||||
| hsa05202 | Transcriptional misregulation in cancer | 47 | 42 | 89 (3.749%) | 186 (2.347%) | 1.296 × 10−5 |
| hsa05203 | Viral carcinogenesis | 34 | 40 | 74 (3.117%) | 201 (2.536%) | 0.0428029 |
| hsa05205 | Proteoglycans in cancer | 35 | 55 | 90 (3.791%) | 204 (2.574%) | 0.0002796 |
| hsa05210 | Colorectal cancer | 22 | 26 | 48 (2.022%) | 86 (1.085%) | 2.367 × 10−5 |
| hsa05211 | Renal cell carcinoma | 17 | 12 | 29 (1.222%) | 69 (0.871%) | 0.0433838 |
| hsa05212 | Pancreatic cancer | 15 | 22 | 37 (1.559%) | 69 (0.871%) | 0.0021288 |
| hsa05213 | Endometrial cancer | 13 | 14 | 27 (1.137%) | 69 (0.871%) | 0.0159683 |
| hsa05214 | Glioma | 17 | 17 | 34 (1.432%) | 69 (0.871%) | 0.0118167 |
| hsa05215 | Prostate cancer | 25 | 23 | 48 (2.022%) | 97 (1.224%) | 0.0005016 |
| hsa05216 | Thyroid cancer | 10 | 11 | 21 (0.885%) | 37 (0.467%) | 0.0032991 |
| hsa05217 | Basal cell carcinoma | 9 | 15 | 24 (1.011%) | 63 (0.795%) | 0.1293504 |
| hsa05218 | Melanoma | 15 | 16 | 31 (1.306%) | 72 (0.909%) | 0.0298569 |
| hsa05219 | Bladder cancer | 9 | 9 | 18 (0.758%) | 41 (0.517%) | 0.0669118 |
| hsa05220 | Chronic myeloid leukemia | 14 | 22 | 36 (1.516%) | 76 (0.959%) | 0.0045089 |
| hsa05221 | Acute myeloid leukemia | 15 | 16 | 31 (1.306%) | 67 (0.845%) | 0.0118167 |
| hsa05222 | Small cell lung cancer | 13 | 33 | 46 (1.938%) | 92 (1.161%) | 0.0005016 |
| hsa05223 | Non-small cell lung cancer | 13 | 20 | 33 (1.390%) | 66 (0.833%) | 0.0029084 |
| hsa05224 | Breast cancer | 34 | 27 | 61 (2.570%) | 147 (1.855%) | 0.0066776 |
| hsa05225 | Hepatocellular carcinoma | 32 | 40 | 72 (3.033%) | 168 (2.120%) | 0.0016521 |
| hsa05226 | Gastric cancer | 30 | 28 | 58 (2.443%) | 149 (1.880%) | 0.0283904 |
| hsa05230 | Central carbon metabolism in cancer | 13 | 17 | 30 (1.264%) | 69 (0.871%) | 0.0286728 |
| hsa05231 | Choline metabolism in cancer | 20 | 16 | 36 (1.516%) | 98 (1.237%) | 0.1137391 |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 14 | 21 | 35 (1.474%) | 89 (1.123%) | 0.0624114 |
| Pathway ID | Pathway Description | Number of DEGs | All Genes with Pathway Annotation | q-Value | ||
|---|---|---|---|---|---|---|
| Up | Down | Total DEGs | ||||
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 22 | 20 | 42 (1.769%) | 79 (0.997%) | 0.0002796 |
| hsa01522 | Endocrine resistance | 24 | 21 | 45 (1.896%) | 98 (1.237%) | 0.0032168 |
| hsa01523 | Antifolate resistance | 5 | 10 | 15 (0.632%) | 31 (0.391%) | 0.0464471 |
| hsa01524 | Platinum drug resistance | 15 | 23 | 38 (1.601%) | 73 (0.921%) | 0.0006735 |
| Description | Gene Name | Log2 FC * | KEGG Pathway |
|---|---|---|---|
| Up regulation (MCF-7-chlorophyllides/MDA-MB-231-chlorophyllides) | |||
| annexin A4 | ANXA4 | 1.3495564 | hsa04974 |
| C-C motif chemokine receptor 1 | CCR1 | 2.573958 | ko04060, ko04061, ko04062, ko05163, ko05167 |
| stromal interaction molecule 2 | STIM2 | 1.4764014 | hsa04020 |
| ethanolamine kinase 1 | ETNK1 | 1.1246655 | hsa00564, hsa01100 |
| RAP2B, member of RAS oncogene family | RAP2B | 1.2477774 | NA |
| BRCA2 and CDKN1A interacting protein | BCCIP | 1.0360939 | NA |
| ribonucleotide reductase M2 B | RRM2B | 1.1502474 | hsa00230, hsa00240, hsa00480, hsa00983, hsa01100, hsa04115 |
| cysteine-serine-rich nuclear protein 2 | CSRNP2 | 1.155312 | NA |
| serine kinase H1 | PSKH1 | 1.1608988 | NA |
| zinc finger and SCAN domain containing 16 | ZSCAN16 | 1.175633 | NA |
| histone cluster 2, H3a | HIST2H3A | 1.2060455 | hsa04613, hsa05034, hsa05131, hsa05202, hsa05322 |
| wingless-type MMTV integration site family, member 3A | WNT3A | 1.2382799 | hsa04150, hsa04310, hsa04390, hsa04550, hsa04916, hsa04934, hsa05010, hsa05022, hsa05165, hsa05200, hsa05205, hsa05206, hsa05217, hsa05224, hsa05225, hsa05226 |
| acetyl-CoA carboxylase beta | ACACB | 1.2477973 | hsa00061, hsa00620, hsa00640, hsa01100, hsa04152, hsa04910, hsa04920, hsa04922, hsa04931 |
| zinc finger protein 90 | ZNF90 | 1.4318171 | hsa05168 |
| hyaluronan-mediated motility receptor | HMMR | 1.4742341 | ko04512 |
| tribbles pseudokinase 2 | TRIB2 | 1.5311222 | NA |
| Down- regulation (MCF-7-chlorophyllides/MDA-MB-231-chlorophyllides) | |||
| membrane associated guanylate kinase, WW and PDZ domain containing 1 | MAGI1 | −1.2064317 | hsa04015, hsa04151, hsa04530, hsa05165 |
| NLR family, CARD domain containing 5 | NLRC5 | −2.5420052 | NA |
| solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 | SLC7A7 | −4.4729806 | hsa04974 |
| protein kinase N1 | PKN1 | −1.3322328 | hsa04151, hsa04621, hsa05132, hsa05135 |
| topoisomerase (DNA) II alpha 170kDa | TOP2A | −1.1590858 | hsa01524 |
| UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 | B4GALT6 | −3.2967566 | ko00600, ko01100 |
| zinc finger protein 334 | ZNF334 | −2.4680017 | hsa05168 |
| acyl-CoA synthetase short-chain family member 1 | ACSS1 | −2.1925126 | hsa00010, hsa00620, hsa00630, hsa00640, hsa01100, hsa01200 |
| isovaleryl-CoA dehydrogenase | IVD | −1.8816549 | ko00280, ko01100 |
| ADP-ribosylation factor-like 2 | ARL2 | −1.8091316 | NA |
| Rho guanine nucleotide exchange factor 10 | ARHGEF10 | −1.9429478 | ko04270, ko04611, ko04810, ko04928, ko05130, ko05135, ko05163, ko05200, ko05205, ko05417 |
| cyclin-dependent kinase 13 | CDK13 | −1.3298924 | NA |
| diacylglycerol O-acyltransferase 2 | DGAT2 | −1.3269336 | ko00561, ko01100, ko04975 |
| solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | SLC13A3 | −1.3249987 | NA |
| nuclear receptor subfamily 1, group D, member 1 | NR1D1 | −1.2920206 | ko04710 |
| zinc finger protein 76 | ZNF76 | −1.2438793 | hsa05168 |
| ankyrin repeat domain 34A | ANKRD34A | −1.2280619 | NA |
| salt-inducible kinase 2 | SIK2 | −1.2129109 | ko04922 |
| v-myc avian myelocytomatosis viral oncogene homolog | MYC | −1.2045108 | ko04010, ko04012, ko04110, ko04151, ko04218, ko04310, ko04350, ko04390, ko04391, ko04550, ko04630, ko04919, ko05132, ko05160, ko05161, ko05163, ko05166, ko05167, ko05169, ko05200, ko05202, ko05205, ko05206, ko05207, ko05210, ko05213, ko05216, ko05219, ko05220, ko05221, ko05222, ko05224, ko05225, ko05226, ko05230 |
| zinc finger protein 780A | ZNF780A | −1.1839843 | hsa05168 |
| oligonucleotide/oligosaccharide-binding fold containing 1 | OBFC1 | −1.1801386 | NA |
| lanosterol synthase | LSS | −1.163355 | ko00100, ko01100, ko01110, ko01130 |
| zinc finger, DHHC-type containing 17 | ZDHHC17 | −1.1437738 | NA |
| carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | CAD | −1.1388409 | hsa00240, hsa00250, hsa01100, hsa01240 |
| centrosomal protein 152kDa | CEP152 | −1.1385807 | NA |
| hypoxia inducible factor 1, alpha subunit | HIF1A | −1.0661852 | ko04066, ko04137, ko04140, ko04212, ko04361 Axon regeneration ko04659, ko04919, ko05167, ko05200, ko05205, ko05211, ko05230, ko05231, ko05235 |
| aldehyde dehydrogenase 3 family, member B1 | ALDH3B1 | −1.063386 | hsa00010, hsa00340, hsa00350, hsa00360, hsa00410, hsa00980, hsa00982, hsa01100 |
| polymerase (DNA directed), epsilon 2, accessory subunit | POLE2 | −1.0376518 | ko03030, ko03410, ko03420 |
| arginine methyltransferase 3 | PRMT3 | −1.0357205 | NA |
| polymerase (RNA) I polypeptide E, 53kDa | POLR1E | −1.0155994 | ko03020 |
| cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | −1.0129887 | ko00140, ko00380, ko00830, ko00980, ko01100, ko04913, ko05204 |
| WD repeat containing, antisense to TP53 | WRAP53 | −1.0114268 | NA |
| heat shock transcription factor 2 | HSF2 | −1.0091325 | ko03000 |
| inositol monophosphatase domain containing 1 | IMPAD1 | −1.0071035 | ko00562, ko00920, ko01100, ko01120, ko01130, ko04070 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.-S.; Wang, Y.-T.; Byadgi, O.; Huang, T.-Y.; Tai, M.-H.; Shaw, J.-F.; Yang, C.-H. Screening of Specific and Common Pathways in Breast Cancer Cell Lines MCF-7 and MDA-MB-231 Treated with Chlorophyllides Composites. Molecules 2022, 27, 3950. https://doi.org/10.3390/molecules27123950
Huang K-S, Wang Y-T, Byadgi O, Huang T-Y, Tai M-H, Shaw J-F, Yang C-H. Screening of Specific and Common Pathways in Breast Cancer Cell Lines MCF-7 and MDA-MB-231 Treated with Chlorophyllides Composites. Molecules. 2022; 27(12):3950. https://doi.org/10.3390/molecules27123950
Chicago/Turabian StyleHuang, Keng-Shiang, Yi-Ting Wang, Omkar Byadgi, Ting-Yu Huang, Mi-Hsueh Tai, Jei-Fu Shaw, and Chih-Hui Yang. 2022. "Screening of Specific and Common Pathways in Breast Cancer Cell Lines MCF-7 and MDA-MB-231 Treated with Chlorophyllides Composites" Molecules 27, no. 12: 3950. https://doi.org/10.3390/molecules27123950
APA StyleHuang, K.-S., Wang, Y.-T., Byadgi, O., Huang, T.-Y., Tai, M.-H., Shaw, J.-F., & Yang, C.-H. (2022). Screening of Specific and Common Pathways in Breast Cancer Cell Lines MCF-7 and MDA-MB-231 Treated with Chlorophyllides Composites. Molecules, 27(12), 3950. https://doi.org/10.3390/molecules27123950









