Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Experimental Samples
2.1.2. Chemicals and Reagents
2.2. Method
2.2.1. Preparation of Standard and Sample Solutions
2.2.2. Data Acquisition for E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis
E-Nose Data Acquisition
E-Tongue Data Acquisition
Multi-Component Quantitative Analysis
2.2.3. Chemometric Analysis and Data Fusion
3. Results and Discussion
3.1. Results of E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis
3.2. Comparison Results of E-Nose Measurements, E-Tongue Measurements, and Chemical Anylsis on Discrimination of the Two Origins
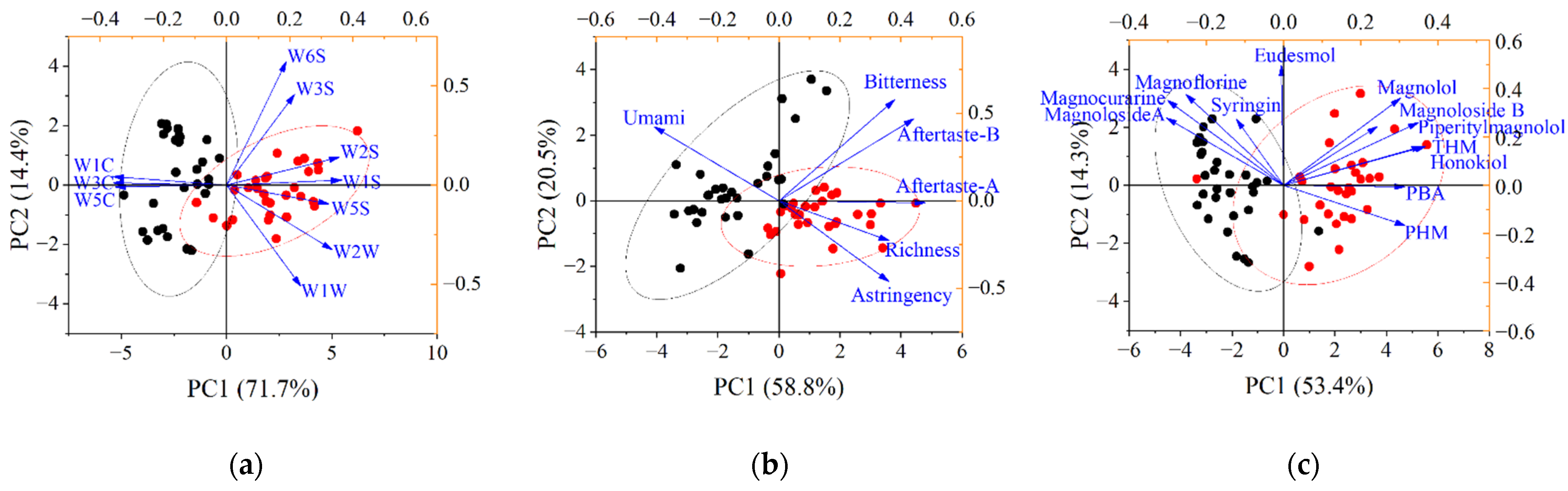
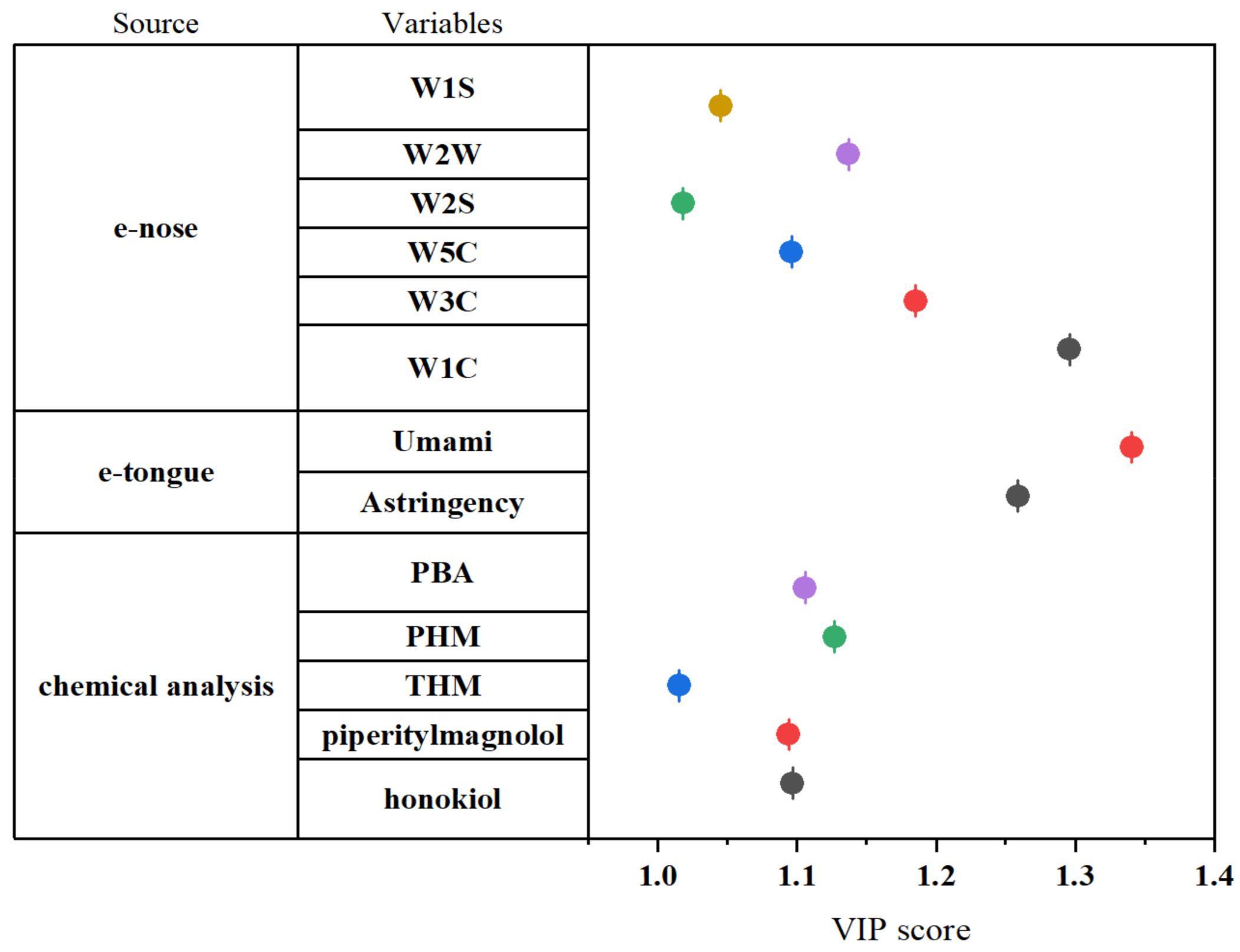
3.3. Extraction of Feature Variables Based on PCA and PLS-VIP
3.4. Multi-Source Data Fusion and Establishment of Discriminative Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Fang, D.L.; Yang, W.J.; Kimatu, B.M.; Zhao, L.; Hu, Q.H. Comparison of flavour qualities of mushrooms (Flammulina velutipes) packed with different packaging materials. Food Chem. 2017, 232, 1–9. [Google Scholar]
- Yin, X.Q.; Xu, X.X.; Zhang, Q.; Xu, J.G. Rapid Determination of the Geographical Origin of Chinese Red Peppers (Zanthoxylum Bungeanum Maxim.) Based on Sensory Characteristics and Chemometric Techniques. Molecules 2018, 23, 1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitra, M.; Angelica, R.; Hugo, W.; Bernd, S.; Markus, S. Sensory characteristics of human milk: Association between mothers’ diet and milk for bitter taste. J. Dairy Sci. 2018, 102, 1116–1130. [Google Scholar]
- Zhang, X.Y.; Jiang, D.; Li, D.M.; Yu, C.X.; Dong, X.F.; Qi, H. Characterization of a seafood-flavoring enzymatic hydrolysate from brown alga Laminaria japonica. J. Food Meas. Charact. 2019, 13, 1185–1194. [Google Scholar] [CrossRef]
- Li, B.; Kimatu, B.M.; Pei, F.; Chen, S.Y.; Zhao, L.Y. Non-volatile flavour components in Lentinus edodes after hot water blanching and microwave blanching. Int. J. Food Prop. 2018, 20, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mohamed-Ahmed, A.H.A.; Soto, J.; Ernest, T.; Tuleu, C. Non-human tools for the evaluation of bitter taste in the design and development of medicines: A systematic review. Drug Discov. Today 2016, 21, 1170–1180. [Google Scholar] [CrossRef]
- Petrovick, G.F.; Kleinebudde, P.; Breitkreutz, J. Orodispersible tablets containing taste-masked solid lipid pellets with metformin hydrochloride: Influence of process parameters on tablet properties. Eur. J. Pharm. Biopharm. 2017, 122, 137–145. [Google Scholar] [CrossRef]
- Immohr, L.I.; Dischinger, A.; Kuhl, P.; Kletzl, H.; Sturm, S.; Guenther, A.; Pein-Hackelbusch, M. Early pediatric formulation development with new chemical entities: Opportunities of e-tongue besides human taste assessment. Int. J. Pharm. 2017, 530, 201–212. [Google Scholar] [CrossRef]
- Immohr, L.I.; Turner, R.; Pein-Hackelbusch, M. Impact of sodium lauryl sulfate in oral liquids on e-tongue measurements. Int. J. Pharm. 2016, 515, 441–448. [Google Scholar] [CrossRef]
- Shiraishi, S.; Haraguchi, T.; Nakamura, S.; Kojima, H.; Kawasaki, I.; Yoshida, M.; Uchida, T. Suppression in Bitterness Intensity of Bitter Basic Drug by Chlorogenic Acid. Chem. Pharm. Bull. 2017, 65, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Phat, C.; Moon, B.K.; Lee, C. Evaluation of umami taste in mushroom extracts by chemical analysis, sensory evaluation, and an electronic tongue system. Food Chem. 2016, 192, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, M.; Bhandari, B.; Adhikari, B. Application of electronic tongue for fresh foods quality evaluation: A review. Food Rev. Int. 2018, 34, 746–769. [Google Scholar] [CrossRef]
- Tian, X.J.; Wang, J.; Cui, S.Q. Analysis of pork adulteration in minced mutton using electronic nose of metal oxide sensors. J. Food Eng. 2013, 119, 744–749. [Google Scholar] [CrossRef]
- Labrador, R.H.; Masot, R.; Alcaniz, M.; Baigts, D.; Soto, J.; Martinez-Manez, R.; Garcia-Breijo, E.; Gil, L.; Barat, J.M. Prediction of NaCl, nitrate and nitrite contents in minced meat by using a voltammetric electronic tongue and an impedimetric sensor. Food Chem. 2010, 122, 864–870. [Google Scholar] [CrossRef]
- Ritzer, J.; Lühmann, T.; Rode, C.; Pein-Hackelbusch, M.; Immohr, I.; Schedler, U.; Thiele, T.; Stübinger, S.; Rechenberg, B.V.; Waser-Althaus, J.; et al. Diagnosing peri-implant disease using the tongue as a 24/7 detector. Nat. Commun. 2017, 8, 264. [Google Scholar] [CrossRef]
- Sberveglieri, V.; Bhandari, M.P.; Pulvirenti, A.; Carmona, E.N. Grated Parmigiano Reggiano Cheese: Authenticity Determination and Characterization by a Novel Nanowire Device (S3) and GC-MS. In Sensors for Everyday Life: Environmental and Food Engineering; Mukhopadhyay, S.C., Postolache, O.A., Jayasundera, K.P., Swain, A.K., Eds.; Springer International Publishing: Cham, Germany, 2017; pp. 229–243. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.W.; Yu, X.K.; Luo, H.Y.; Lu, Y.Q.; Yang, H.J.; Li, X.; Li, Z.Y.; Tang, L.Y.; Wang, Z.J. Determination of Bitterness of Andrographis Herba Based on Electronic Tongue Technology and Discovery of the Key Compounds of Bitter Substances. Molecules 2018, 23, 3362. [Google Scholar] [CrossRef] [Green Version]
- Anjiki, N.; Hosoe, J.; Fuchino, H.; Kiuchi, F.; Sekita, S.; Ikezaki, H.; Mikage, M.; Kawahara, N.; Goda, Y. Evaluation of the taste of crude drug and Kampo formula by a taste-sensing system (4): Taste of Processed Aconite Root. J. Nat. Med. 2011, 65, 293–300. [Google Scholar] [CrossRef]
- Eckert, C.; Lutz, C.; Breitkreutz, J.R.; Woertz, K. Quality control of oral herbal products by an electronic tongue—Case study on sage lozenges. Sens. Actuators B Chem. 2011, 156, 204–212. [Google Scholar] [CrossRef]
- Buratti, S.; Malegori, C.; Benedetti, S.; Oliveri, P.; Giovanelli, G. E-nose, e-tongue and e-eye for edible olive oil characterization and shelf life assessment: A powerful data fusion approach. Talanta 2018, 182, 131–141. [Google Scholar] [CrossRef]
- Men, H.; Shi, Y.; Fu, S.L.; Jiao, Y.N.; Qiao, Y.; Liu, J.J. Mining Feature of Data Fusion in the Classification of Beer Flavor Information Using E-Tongue and E-Nose. Sensors 2017, 17, 1656. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Wang, J.; Zhu, L.Y. The qualitative and quantitative assessment of tea quality based on E-nose, E-tongue and E-eye combined with chemometrics. Food Chem. 2019, 289, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.; Covington, J.A.; Gardner, J.W. Combined electronic nose and tongue for a flavour sensing system. Sens. Actuators B Chem. 2011, 156, 832–839. [Google Scholar] [CrossRef] [Green Version]
- Banerjeeroy, R.; Tudu, B.; Shaw, L.; Jana, A.; Bhattacharyya, N.; Bandyopadhyay, R. Instrumental testing of tea by combining the responses of electronic nose and tongue. J. Food Eng. 2012, 110, 356–363. [Google Scholar] [CrossRef]
- Tian, X.; Wang, J.; Ma, Z.; Li, M.; Wei, Z. Combination of an E-Nose and an E-Tongue for Adulteration Detection of Minced Mutton Mixed with Pork. J Food Qual. 2019, 2019, 4342509. [Google Scholar] [CrossRef] [Green Version]
- Poivre, M.; Duez, P. Biological activity and toxicity of the Chinese herb Magnolia officinalis Rehder & E. Wilson (Houpo) and its constituents. J. Zhejiang Univ. Sci. B 2017, 18, 194–214. [Google Scholar]
- Youn, U.J.; Fatima, N.; Chen, Q.C.; Chae, S.; Hung, T.M.; Min, B.S. Apoptosis-inducing and antitumor activity of neolignans isolated from Magnolia officinalis in HeLa cancer cells. Phytother Res. 2013, 27, 1419–1422. [Google Scholar] [CrossRef]
- Youn, U.J.; Chen, Q.C.; Jin, W.Y.; Lee, I.S.; Kim, H.J.; Lee, J.P.; Chang, M.J.; Min, B.S.; Bae, K.H. Cytotoxic lignans from the stem bark of Magnolia officinalis. J. Nat. Prod. 2007, 70, 1687–1689. [Google Scholar] [CrossRef]
- Kuo, W.L.; Chung, C.Y.; Hwang, T.L.; Chen, J.J. Biphenyl-type neolignans from Magnolia officinalis and their anti-inflammatory activities. Phytochemistry 2013, 85, 153–160. [Google Scholar] [CrossRef]
- Shih, H.C.; Hwang, T.L.; Chen, H.C.; Kuo, P.C.; Lee, E.J.; Lee, K.H.; Wu, T.S. Honokiol dimers and magnolol derivatives with new carbon skeletons from the roots of Magnolia officinalis and their inhibitory effects on superoxide anion generation and elastase release. PLoS ONE 2013, 8, e59502. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.Y.; Liu, H.L.; Zhang, J.Y.; Yang, B. Phenolic glycosides and other constituents from the bark of Magnolia officinalis. J. Asian Nat. Prod. Res. 2014, 16, 400–405. [Google Scholar] [CrossRef]
- Yu, S.X.; Yan, R.Y.; Liang, R.X.; Wang, W.; Yang, B. Bioactive polar compounds from stem bark of Magnolia officinalis. Fitoterapia 2012, 83, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Yan, R.; Yang, B. Phenylethanoid glycosides and phenolic glycosides from stem bark of Magnolia officinalis. Phytochemistry 2016, 127, 50–62. [Google Scholar] [CrossRef]
- Porter, E.A.; Kite, G.C.; Veitch, N.C.; Geoghegan, I.A.; Larsson, S.; Simmonds, M.S.J. Phenylethanoid glycosides in tepals of Magnolia salicifolia and their occurrence in flowers of Magnoliaceae. Phytochemistry 2015, 117, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.Y.; Wang, W.H.; Guo, J.; Liu, H.L.; Zhang, J.Y.; Yang, B. Studies on the alkaloids of the bark of Magnolia officinalis: Isolation and on-line analysis by HPLC-ESI-MS(n). Molecules 2013, 18, 7739–7750. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.F.; Wang, X.B.; Luo, J.G.; Luo, J.; Wang, J.S.; Kong, L.Y. A novel aporphine alkaloid from Magnolia officinalis. Fitoterapia 2011, 82, 637–641. [Google Scholar] [CrossRef]
- Luo, H.a.; Wu, H.e.; Yu, X.u.; Zhang, X.; Lu, Y.Q. A review of the phytochemistry and pharmacological activities of Magnoliae officinalis cortex. J. Ethnopharmacol. 2019, 23, 412–442. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shao, Y.; Yao, Y.; Yu, Y.; Cao, G.; Zou, H.; Yan, Y. A novel quality evaluation method for magnolia bark using electronic nose and colorimeter data with multiple statistical algorithms. J. Trad. Chin. Med. Sci. 2020, 7, 221–227. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, Y.; Wang, C.C.; Zou, L.S.; Liu, X.H.; Chen, S.Y.; Shi, J.J. Comparison of Chemical Constituents in Magnoliae Officinalis Cortex Processed by “Sweating” and “Non Sweating” based on Ultra Fast Liquid Chromatography-Triple Quadrupole-Time of Flight Mass Spectrometry and Gas Chromatography-Triple Quadrupole Mass Spectrometry Combined with Multivariate Statistical Analysis. Nat. Prod. Commun. 2018, 13, 251–252. [Google Scholar]
- Peng, B.; He, R.; Yang, B.; Liu, Y.L.; Gao, J.; Li, J.R. Comparative study of effect of Magnolia officinalis and M. officinalis var. biloba on gastrointestinal motility dysfunction. China J. Chin. Mater. Med. 2010, 35, 2624–2627. [Google Scholar]
- He, J.S.; Chen, L.; Si, Y.; Huang, B.; Ban, X.Q.; Wang, Y.W. Population structure and genetic diversity distribution in wild and cultivated populations of the traditional Chinese medicinal plant Magnolia officinalis subsp. biloba (Magnoliaceae). Genetica 2009, 135, 233–243. [Google Scholar] [CrossRef]
- Yao, Y.Z.; Pan, S.Y.; Fan, G.; Dong, L.; Ren, J.N.; Zhu, Y. Evaluation of volatile profile of Sichuan dongcai, a traditional salted vegetable, by SPME–GC–MS and E-nose. LWT-Food Sci. Technol. 2015, 64, 528–535. [Google Scholar] [CrossRef]
- Jing, W.G.; Zhang, Q.; Deng, Z.; Wang, J.Y.; Sun, X.B. Quality evaluation of Magnoliae Officinalis Cortex based on combinative method of ingerprint, quantitative analysis of multicomponents and chemometrics. China J. Chin. Mater. Med. 2019, 44, 975–982. [Google Scholar]
- Anderson, K.A.; Hobbie, K.A.; Smith, B.W. Chemical Profiling with Modeling Differentiates Wild and Farm-Raised Salmon. J. Agric. Food Chem. 2010, 58, 11768–11774. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Ma, J.; Luo, S.Q.; Yang, Z.N.; Di, S.J.; Hu, J.; Zheng, Z. Comparative analysis of phenolics and volatiles in seeds of Magnolia officinalis and Magnolia officinalis var.biloba. Seed 2015, 34, 29–34. [Google Scholar]
- Xue, Z.Z.; Yan, R.Y.; Yu, S.X.; Cao, Y.Y.; Shao, A.J.; Yang, B. Determination of Six Active Compounds in Commercial Samples of Magnoliae Officinalis Cortex by HPLC-DAD. Chin. J. Exp. Trad. Med. Form. 2014, 20, 45–49. [Google Scholar]
- Xue, Z.Z.; Kotani, A.; Yang, B.; Hakamata, H. Discrimination of magnoliae officinalis cortex based on the quantitative profiles of magnolosides by two-channel liquid chromatography with electrochemical detection. J. Pharm. Biomed. Anal. 2018, 158, 166–173. [Google Scholar] [CrossRef]
- Zhang, M.H.; Feng, L.; Hu, S.Y.; Jia, X.B. Essence of material base in geoherbs: Specificality of constituent structure. China J. Chin. Mater. Med. 2013, 38, 136–140. [Google Scholar]
- Yeom, J.; Choe, A.; Lim, S.; Lee, Y.; Na, S.; Ko, H. Soft and ion-conducting hydrogel artificial tongue for astringency perception. Sci. Adv. 2020, 6, eaba5785. [Google Scholar] [CrossRef]
- Narukawa, M.; Kimata, H.; Noga, C.; Watanabe, T. Taste characterization of tea catechins. Int. J. Food Sci. Tech. 2010, 45, 1579–1589. [Google Scholar] [CrossRef]


| No | Sensor Name | Performance Description | Sensitive Substances and Threshold Values (mL·m−3) |
|---|---|---|---|
| 1 | W1C | Aromatic | Toluene, 10 |
| 2 | W5S | Hydrocarbon | Nitrogen dioxide, 1 |
| 3 | W3C | Aromatic | Benzene, 10 |
| 4 | W6S | Hydrogen | Hydrogen, 100 |
| 5 | W5C | Aromatic and aliphatic | Propane, 1 |
| 6 | W1S | Broad range and methane | Methane, 100 |
| 7 | W1W | Sulfur organic | Hydrogen sulfide, 1 |
| 8 | W2S | Broad range alcohol | Nitric oxide, 100 |
| 9 | W2W | Sulfur and chlorinate | Hydrogen sulfide, 1 |
| 10 | W3S | Methane and aliphatic | Methane, 10 |
| Model | Data Source | Resubstitution | Cross-Validation | Sample Dichotomy Strategy | |||
|---|---|---|---|---|---|---|---|
| NM 1 | PM 2 | NM | PM | NM | PM | ||
| LDA | E-nose | 1 | 1.67% | 1 | 1.67% | 0 | 0 |
| E-tongue | 1 | 1.67% | 2 | 3.33% | 1 | 1.67% | |
| Chemical analysis | 2 | 3.33% | 3 | 5.00% | T:2 | 5.00% | |
| QDA | E-nose | 0 | 0 | 0 | 0 | 0 | 0 |
| E-tongue | 0 | 0 | 1 | 1.67% | 1 | 1.67% | |
| Chemical analysis | 2 | 3.33% | 6 | 10.00% | T 3:1 P 4:2 | 2.50% 5.00% | |
| Fusion | Data Source | Model | Resubstitution | Cross-Validation | Sample Dichotomy Strategy | |||
|---|---|---|---|---|---|---|---|---|
| NM 1 | PM 2 | NM | PM | NM | PM | |||
| Low-level fusion | Original data with normalization | LDA | 0 | 0 | 0 | 0 | NA 3 | NA |
| QDA | 0 | 0 | 34 | 56.67% | NA | NA | ||
| Original data with PCA | LDA | 0 | 0 | 0 | 0 | 0 | 0 | |
| QDA | 0 | 0 | 1 | 1.67% | 0 | 0 | ||
| Mid-level fusion | Combination of 15 extracted PCs | LDA | 0 | 0 | 0 | 0 | 0 | 0 |
| QDA | 0 | 0 | 2 | 3.33% | 0 | 0 | ||
| Combination of 7 extracted PCs | LDA | 0 | 0 | 0 | 0 | 0 | 0 | |
| QDA | 0 | 0 | 1 | 1.67% | 0 | 0 | ||
| Combination of extracted variables | LDA | 0 | 0 | 0 | 0 | 0 | 0 | |
| QDA | 0 | 0 | 1 | 1.67% | 0 | 0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jing, W.; Zhao, X.; Li, M.; Hu, X.; Cheng, X.; Ma, S.; Wei, F. Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis. Molecules 2022, 27, 3892. https://doi.org/10.3390/molecules27123892
Jing W, Zhao X, Li M, Hu X, Cheng X, Ma S, Wei F. Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis. Molecules. 2022; 27(12):3892. https://doi.org/10.3390/molecules27123892
Chicago/Turabian StyleJing, Wenguang, Xiaoliang Zhao, Minghua Li, Xiaowen Hu, Xianlong Cheng, Shuangcheng Ma, and Feng Wei. 2022. "Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis" Molecules 27, no. 12: 3892. https://doi.org/10.3390/molecules27123892
APA StyleJing, W., Zhao, X., Li, M., Hu, X., Cheng, X., Ma, S., & Wei, F. (2022). Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis. Molecules, 27(12), 3892. https://doi.org/10.3390/molecules27123892






