Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study
Abstract
:1. Introduction
2. Results
2.1. Virucidal Activity of Tested Compounds on FCoVII and HCoV-OC43
2.2. Virucidal Activity of Individual Components of TTO on FCoVII and HCoV-OC43
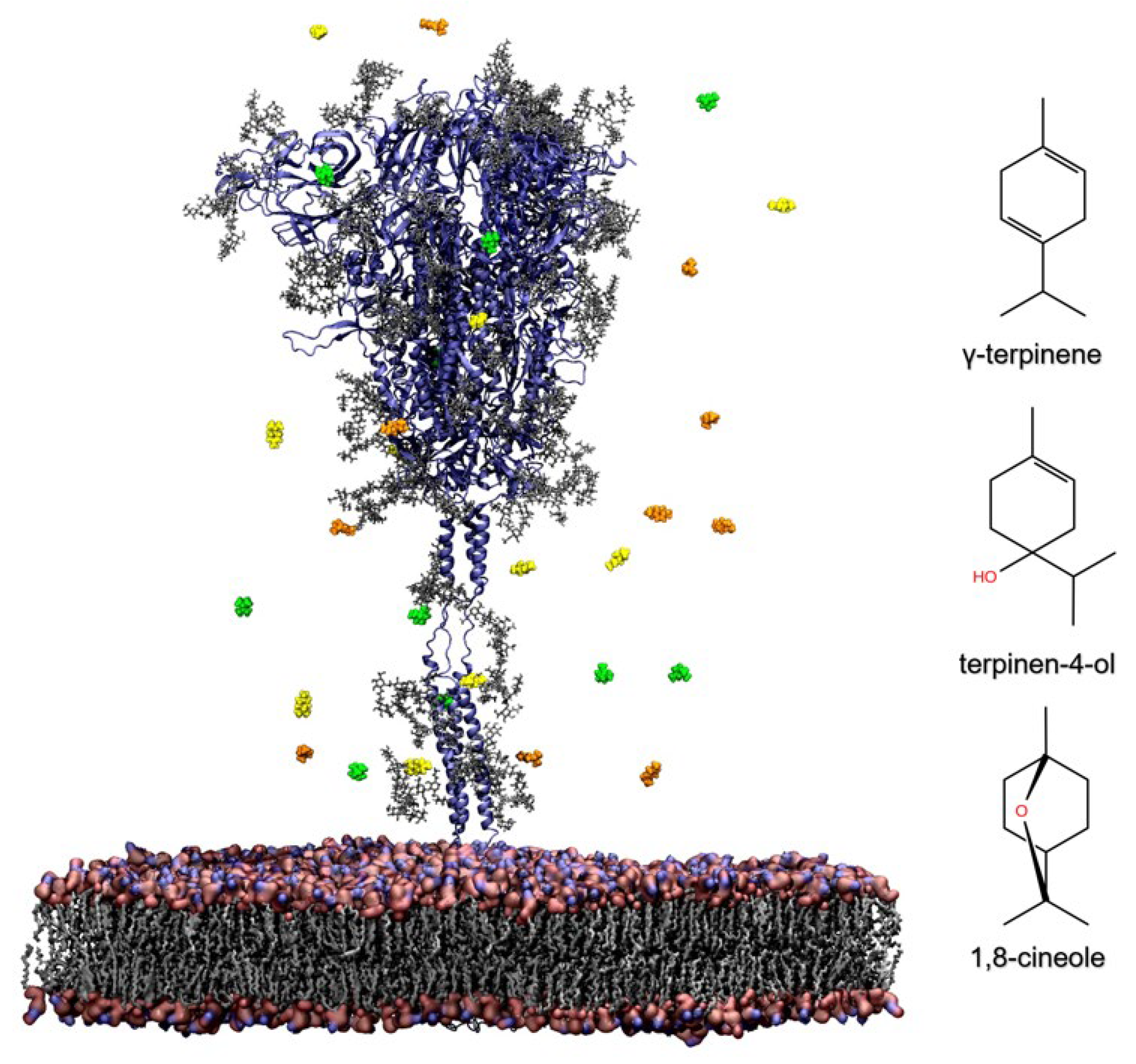
2.3. In Silico Evaluation of TTO Antiviral Activity on SARS-CoV-2
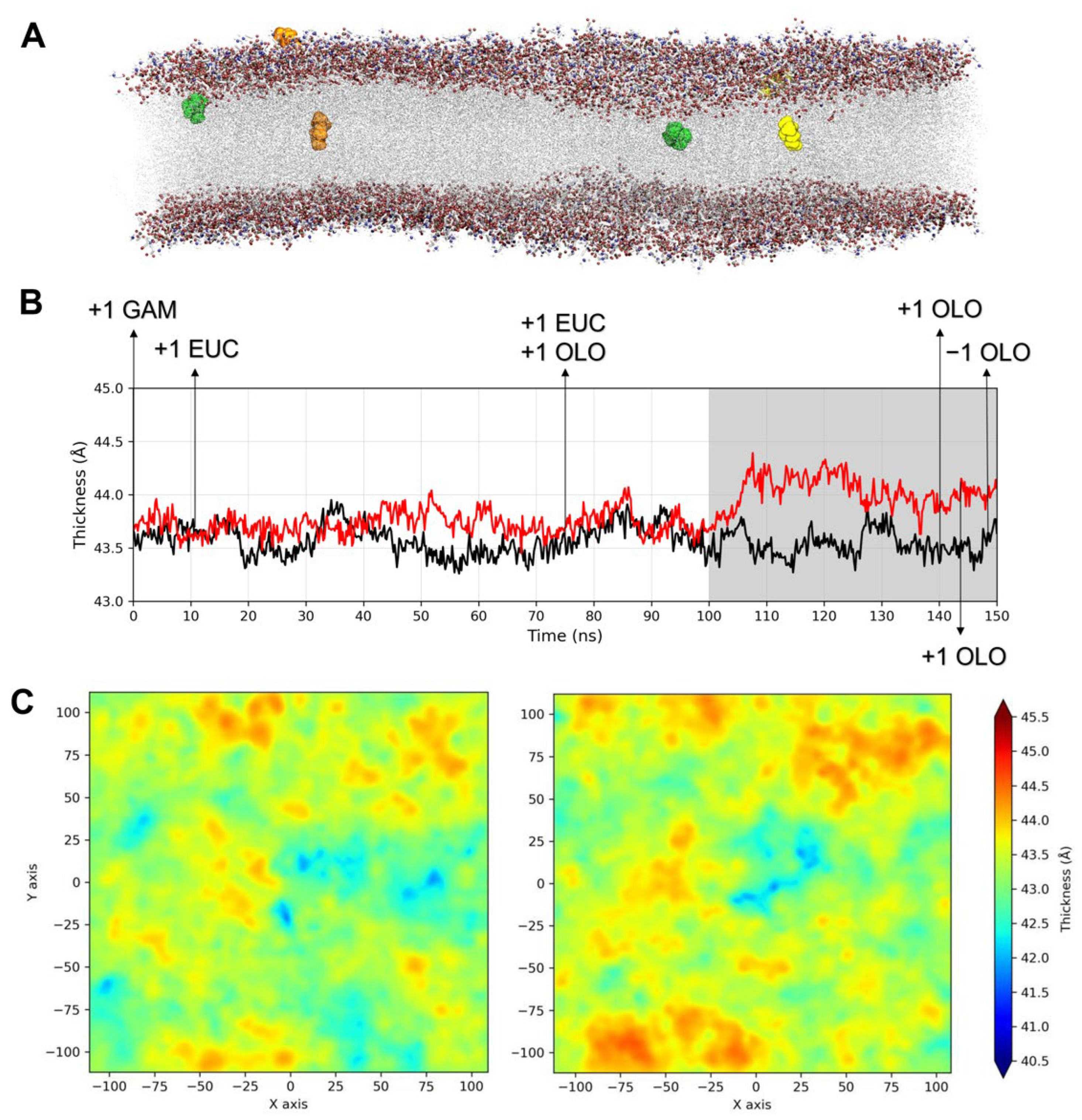
2.3.1. Influence of TTO on the Viral Envelope
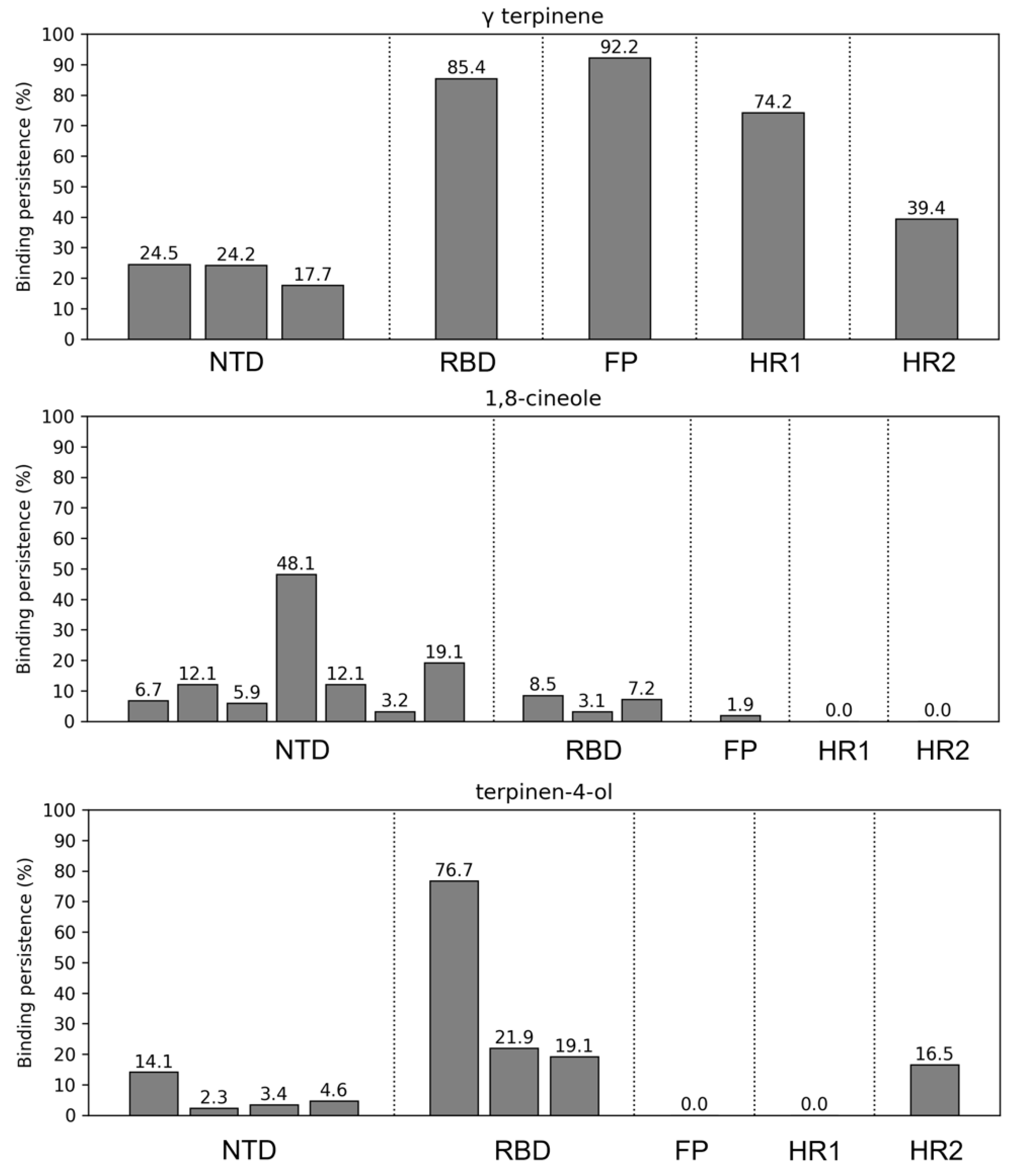
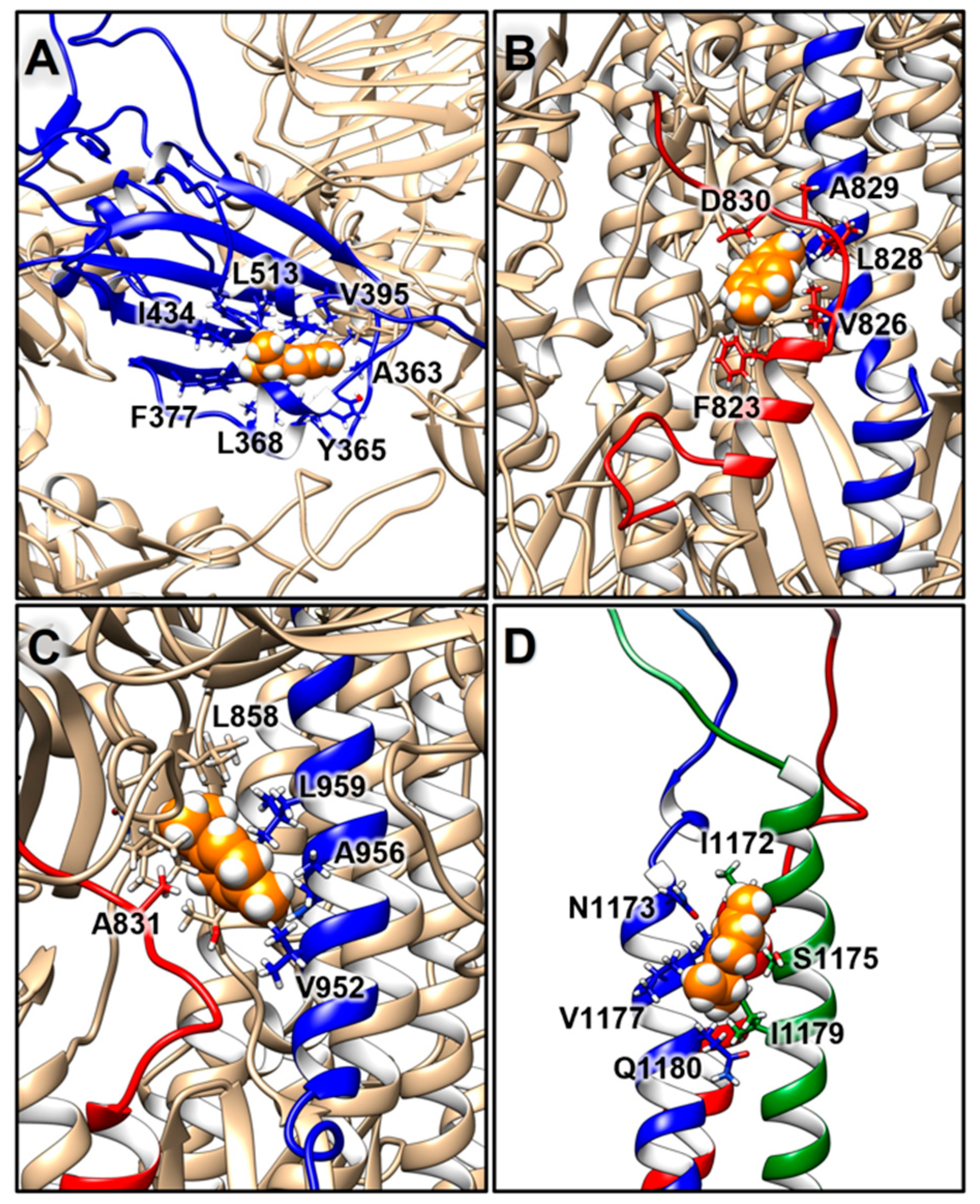
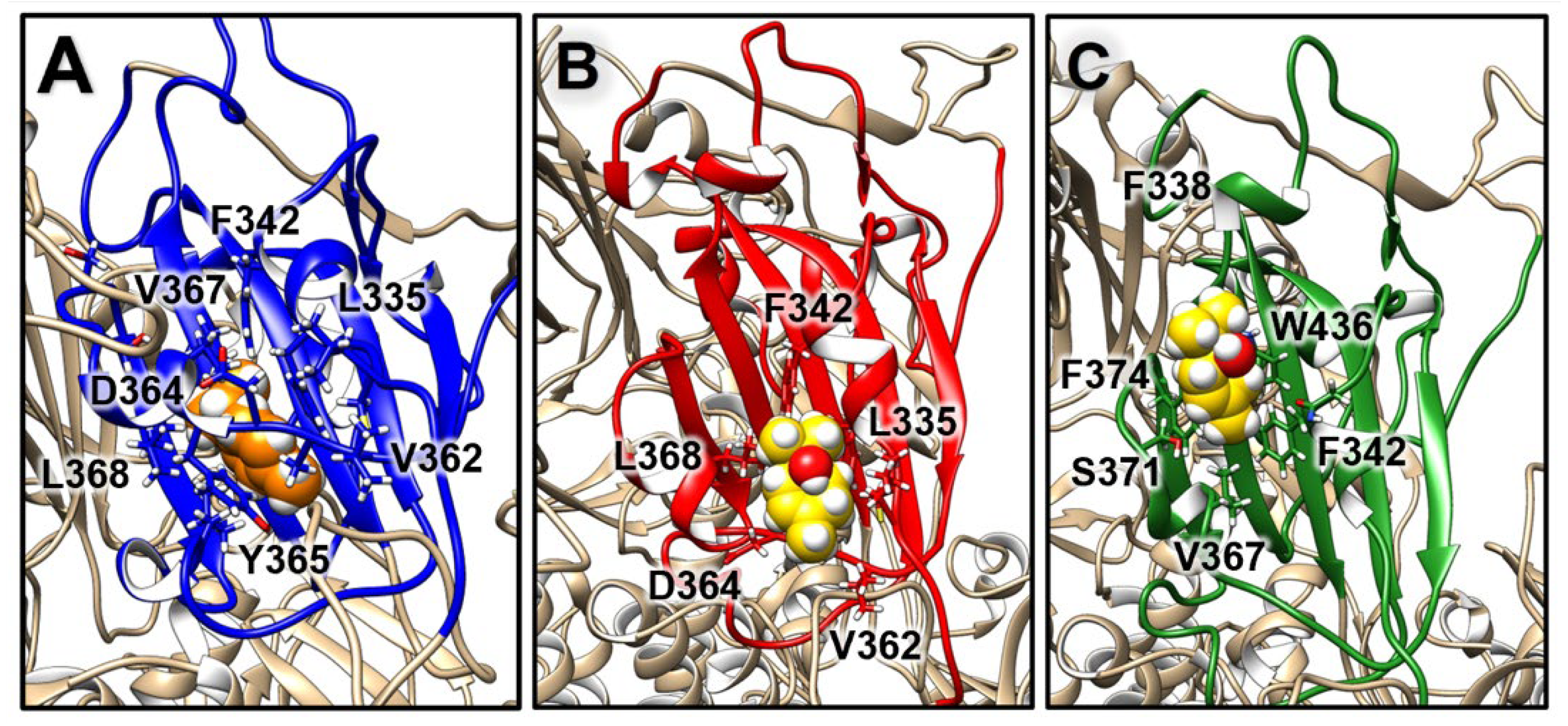
2.3.2. Structural Analysis of TTO Interactions with the S Glycoprotein
2.3.3. Influence of TTO on S Mobility
3. Discussion
4. Materials and Methods
4.1. Tea Tree Oil Formulations, Cells and Viruses
4.2. In Vitro Virucidal Assays
4.3. Real-Time, Quantitative Reverse Transcriptase PCR Assay for Absolute Quantitation of Human Coronaviruses OC43
| Forward | 5′-AGCAGACCTTCCTGAGCCTTCAAT-3′; |
| Reverse | 5′-AGCAACCAGGCTGATGTCAATACC-3′; |
| Probe | 5′-/56-FAM/TGACATTGTCGATCGGGACCCAAGTA/36-TAMSp/-3′. |
4.4. GaMD Simulations of the Spike Glycoprotein in a Viral Membrane
4.5. Trajectories Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020, 178, 104792. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 65, 193–292. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.M.; Rottier, P.J.M. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 21, eabd4251. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, H.; Mathieu, E.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Ortiz-Ospina, E.; Hasell, J.; Macdonald, B.; Beltekian, D.; Roser, M. Coronavirus Pandemic (COVID-19). Available online: https://ourworldindata.org/coronavirus (accessed on 26 February 2022).
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; García-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the pandemic: Perspectives on the future trajectory of COVID-19. Nature 2021, 596, 495–504. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Modes of Transmission of Virus Causing COVID-19: Implications for IPC Precaution Recommendations: Scientific Brief, 27 March 2020 (No. WHO/2019-nCoV/Sci_Brief/Transmission_modes/2020.1). Available online: https://apps.who.int/iris/handle/10665/331601 (accessed on 5 October 2021).
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Guide to Local Production: WHO-Recommended Handrub Formulations. 2020. Available online: https://www.who.int/gpsc/5may/Guide_to_Local_Production.pdf (accessed on 5 October 2021).
- Food and Drug Administration (FDA), Center for Drug Evaluation and Research. Policy for Temporary Compounding of Certain Alcohol-Based Hand Sanitizer Products during the Public Health Emergency. March 2020, Updated February 2021. Docket Number FDA-2020-D-1106. Available online: https://www.fda.gov/media/136118/download (accessed on 5 October 2021).
- Mahmood, A.; Eqan, M.; Pervez, S.; Alghamdi, H.A.; Tabinda, A.B.; Yasar, A.; Brindhadevi, K.; Pugazhendhi, A. COVID-19 and frequent use of hand sanitizers; human health and environmental hazards by exposure pathways. Sci. Total Environ. 2020, 742, 140561. [Google Scholar] [CrossRef]
- Jing, J.L.J.; Pei, Y.T.; Bose, R.J.C.; McCarthy, J.R.; Tharmalingam, N.; Madheswaran, T. Hand Sanitizers: A Review on Formulation Aspects, Adverse Effects, and Regulations. Int. J. Environ. Res. Public Health 2020, 17, 3326. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Lim, J.Y.C.; Xue, K.; Yew, P.Y.M.; Owh, C.; Chee, P.L.; Loh, X.J. Sanitizing agents for virus inactivation and disinfection. View 2020, 1, e16. [Google Scholar] [CrossRef]
- Daverey, A.; Dutta, K. COVID-19: Eco-friendly hand hygiene for human and environmental safety. J. Environ. Chem. Eng. 2021, 9, 104754. [Google Scholar] [CrossRef] [PubMed]
- Carson, C.F.; Hammer, K.A.; Riley, T.V. Melaleuca alternifolia (Tea Tree) Oil: A Review of Antimicrobial and Other Medicinal Properties. Clin. Microbiol. Rev. 2006, 19, 50–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penfold, A.R.; Grant, R. The germicidal values of the principal commercial Eucalyptus oils and their pure constituents, with observations on the value of concentrated disinfectants. J. R. Soc. N. S. W. 1923, 57, 80–89. [Google Scholar]
- Penfold, A.R.; Grant, R. The germicidal values of the pure constituents of Australian essential oils, together with those for some essential oil isolates and synthetics. Part II. J. R. Soc. N. S. W. 1924, 58, 117–123. [Google Scholar]
- Penfold, A.R.; Grant, R. The germicidal values of some Australian essential oils and their pure constituents, together with those for some essential oil isolates, and synthetics. Part III. J. R. Soc. N. S. W. 1925, 59, 346–349. [Google Scholar]
- Brun, P.; Bernabè, G.; Filippini, R.; Piovan, A. In Vitro Antimicrobial Activities of Commercially Available Tea Tree (Melaleuca alternifolia) Essential Oils. Curr. Microbiol. 2019, 76, 108–116. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Antifungal activity of the components of Melaleuca alternifolia (tea tree) oil. J. Appl. Microbiol. 2003, 95, 853–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Duan, S.; Chu, C.; Xu, J.; Zeng, G.; Lam, A.; Zhou, J.; Yin, Y.; Fang, D.; Reynolds, M.; et al. Melaleuca alternifolia Concentrate Inhibits In Vitro Entry of Influenza Virus into Host Cells. Molecules 2013, 18, 9550–9566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, B.; Piccirilli, E.; Ceddia, T.; Pontieri, E.; Aureli, P.; Ferrini, A.M. Antimycotic activity of Melaleuca alternifolia essential oil and its major components. Lett. Appl. Microbiol. 2003, 37, 185–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyankov, O.V.; Usachev, E.V.; Pyankova, O.; Agranovski, I.E. Inactivation of Airborne Influenza Virus by Tea Tree and Eucalyptus Oils. Aerosol. Sci. Technol. 2012, 46, 1295–1302. [Google Scholar] [CrossRef]
- Schnitzler, P.; Schön, K.; Reichling, J. Antiviral activity of Australian tea tree oil and eucalyptus oil against herpes simplex virus in cell culture. Pharmazie 2001, 56, 343–347. [Google Scholar] [PubMed]
- Usachev, E.V.; Pyankov, O.V.; Usacheva, O.V.; Agranovski, I.E. Antiviral activity of tea tree and eucalyptus oil aerosol and vapour. J. Aerosol. Sci. 2013, 59, 22–30. [Google Scholar] [CrossRef]
- Carson, C.F.; Ashton, L.; Dry, L.; Smith, D.W.; Riley, T.V. Melaleuca alternifolia (tea tree) oil gel (6%) for the treatment of recurrent herpes labialis. J. Antimicrob. Chemother. 2001, 48, 450–451. [Google Scholar] [CrossRef]
- Deyno, S.; Mtewa, A.G.; Abebe, A.; Hymete, A.; Makonnen, E.; Bazira, J.; Alele, P.E. Essential oils as topical anti-infective agents: A systematic review and meta-analysis. Complement. Ther. Med. 2019, 47, 102224. [Google Scholar] [CrossRef]
- Mertas, A.; Garbusińska, A.; Szliszka, E.; Jureczko, A.; Kowalska, M.; Król, W. The Influence of Tea Tree Oil (Melaleuca alternifolia) on Fluconazole Activity against Fluconazole-Resistant Candida albicans Strains. Biomed. Res. Int. 2015, 2015, 590470. [Google Scholar] [CrossRef] [Green Version]
- Giordani, C.; Molinari, A.; Toccacieli, L.; Calcabrini, A.; Stringaro, A.; Chistolini, P.; Arancia, G.; Diociaiuti, M. Interaction of Tea Tree Oil with Model and Cellular Membranes. J. Med. Chem. 2006, 49, 4581–4588. [Google Scholar] [CrossRef]
- Brophy, J.J.; Davies, N.W.; Southwell, I.A.; Stiff, I.A.; Williams, L.R. Gas chromatographic quality control for oil of Melaleuca terpinen-4-ol type (Australian tea tree). J. Agric. Food Chem. 1989, 37, 1330–1335. [Google Scholar] [CrossRef]
- Griffin, S.G.; Wyllie, S.G.; Markham, J.L.; Leach, D.N. The role of structure and molecular properties of terpenoids in determining their antimicrobial activity. Flavour Fragr. J. 1999, 14, 322–332. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Witzke, S.; Duelund, L.; Kongsted, J.; Petersen, M.; Mouritsen, O.G.; Khandelia, H. Inclusion of Terpenoid Plant Extracts in Lipid Bilayers Investigated by Molecular Dynamics Simulations. J. Phys. Chem. B 2010, 114, 15825–15831. [Google Scholar] [CrossRef] [PubMed]
- Astani, A.; Reichling, J.; Schnitzler, P. Comparative study on the antiviral activity of selected monoterpenes derived from essential oils. Phytother. Res. 2010, 24, 673–679. [Google Scholar] [CrossRef]
- Garozzo, A.; Timpanaro, R.; Stivala, A.; Bisignano, G.; Castro, A. Activity of Melaleuca alternifolia (tea tree) oil on Influenza virus A/PR/8: Study on the mechanism of action. Antiviral Res. 2011, 89, 83–88. [Google Scholar] [CrossRef]
- Hogue, B.G.; Brian, D.A. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986, 5, 131–144. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.J.; Bosch, B.J.; Rey, F.A.; de Groot, R.J.; et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Cueno, M.E.; Imai, K. Structural Comparison of the SARS-CoV-2 Spike Protein Relative to Other Human-Infecting Coronaviruses. Front. Med. 2021, 7, 594439. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Liang, X.Y.; Wang, Q.; Zhang, S.; Zhao, H.; Wang, K.; Hu, G.X.; Liu, W.J.; Gao, F.S. Mind the feline coronavirus: Comparison with SARS-CoV-2. Gene 2022, 825, 146443. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Woo, H.; Park, S.J.; Choi, Y.K.; Park, T.; Tanveer, M.; Cao, Y.; Kern, N.R.; Lee, J.; Yeom, M.S.; Croll, T.I.; et al. Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J. Phys. Chem. B 2020, 124, 7128–7137. [Google Scholar] [CrossRef] [PubMed]
- Toelzer, C.; Gupta, K.; Yadav, S.K.N.; Borucu, U.; Davidson, A.D.; Kavanagh Williamson, M.; Shoemark, D.K.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020, 370, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Shoemark, D.K.; Colenso, C.K.; Toelzer, C.; Gupta, K.; Sessions, R.B.; Davidson, A.D.; Berger, I.; Schaffitzel, C.; Spencer, J.; Mulholland, A.J. Molecular Simulations Suggest Vitamins, Retinoids and Steroids as Ligands of the Free Fatty Acid Pocket of the SARS-CoV-2 Spike Protein. Angew. Chem. Int. Ed. 2021, 60, 7098–7110. [Google Scholar] [CrossRef] [PubMed]
- Romeo, A.; Iacovelli, F.; Falconi, M. Targeting the SARS-CoV-2 spike glycoprotein prefusion conformation: Virtual screening and molecular dynamics simulations applied to the identification of potential fusion inhibitors. Virus Res. 2020, 286, 198068. [Google Scholar] [CrossRef]
- Battles, M.B.; Langedijk, J.P.; Furmanova-Hollenstein, P.; Chaiwatpongsakorn, S.; Costello, H.M.; Kwanten, L.; Vranckx, L.; Vink, P.; Jaensch, S.; Jonckers, T.H.M.; et al. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat. Chem. Biol. 2016, 12, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins Struct. Funct. Genet. 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. Chembiochem 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Warwicker, J. A model for pH coupling of the SARS-CoV-2 spike protein open/closed equilibrium. Brief. Bioinform. 2021, 22, 1499–1507. [Google Scholar] [CrossRef]
- Singh, D.; Joshi, K.; Samuel, A.; Patra, J.; Mahindroo, N. Alcohol-based hand sanitisers as first line of defence against SARS-CoV-2: A review of biology, chemistry and formulations. Epidemiol. Infect. 2020, 148, e229. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Chan, K.H.; Sridhar, S.; Zhang, R.R.; Chu, H.; Fung, A.Y.; Chan, G.; Chan, J.F.; To, K.K.; Hung, I.F.; Cheng, V.C.; et al. Factors affecting stability and infectivity of SARS-CoV-2. J. Hosp. Infect. 2020, 106, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.R.; Yadav, K.; Khursheed, A.; Rather, M.A. An updated and comprehensive review of the antiviral potential of essential oils and their chemical constituents with special focus on their mechanism of action against various influenza and coronaviruses. Microb. Pathog. 2021, 152, 104620. [Google Scholar] [CrossRef] [PubMed]
- Loizzo, M.R.; Saab, A.M.; Tundis, R.; Statti, G.A.; Menichini, F.; Lampronti, I.; Gambari, R.; Cinatl, J.; Doerr, H.W. Phytochemical Analysis andin Vitro Antiviral Activities of the Essential Oils of Seven Lebanon Species. Chem. Biodivers. 2008, 5, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [Green Version]
- Asif, M.; Saleem, M.; Saadullah, M.; Yaseen, H.S.; Al Zarzour, R. COVID-19 and therapy with essential oils having antiviral, anti-inflammatory, and immunomodulatory properties. Inflammopharmacology 2020, 28, 1153–1161. [Google Scholar] [CrossRef]
- Silveira, D.; Prieto-Garcia, J.M.; Boylan, F.; Estrada, O.; Fonseca-Bazzo, Y.M.; Jamal, C.M.; Magalhães, P.O.; Pereira, E.O.; Tomczyk, M.; Heinrich, M. COVID-19: Is There Evidence for the Use of Herbal Medicines as Adjuvant Symptomatic Therapy? Front. Pharmacol. 2020, 11, 1–44. [Google Scholar] [CrossRef]
- Wińska, K.; Mączka, W.; Łyczko, J.; Grabarczyk, M.; Czubaszek, A.; Szumny, A. Essential Oils as Antimicrobial Agents—Myth or Real Alternative? Molecules 2019, 24, 2130. [Google Scholar] [CrossRef] [Green Version]
- Messager, S.; Hammer, K.A.; Carson, C.F.; Riley, T.V. Effectiveness of hand-cleansing formulations containing tea tree oil assessed ex vivo on human skin and in vivo with volunteers using European standard EN 1499. J. Hosp. Infect. 2005, 59, 220–228. [Google Scholar] [CrossRef]
- Youn, B.H.; Kim, Y.S.; Yoo, S.; Hur, M.H. Antimicrobial and hand hygiene effects of Tea Tree Essential Oil disinfectant: A randomised control trial. Int. J. Clin. Pract. 2021, 75, e14206. [Google Scholar] [CrossRef]
- Theerawatanasirikul, S.; Kuo, C.J.; Phecharat, N.; Chootip, J.; Lekcharoensuk, C.; Lekcharoensuk, P. Structural-based virtual screening and in vitro assays for small molecules inhibiting the feline coronavirus 3CL protease as a surrogate platform for coronaviruses. Antiviral Res. 2020, 182, 104927. [Google Scholar] [CrossRef] [PubMed]
- Schirtzinger, E.E.; Kim, Y.; Davis, A.S. Improving human coronavirus OC43 (HCoV-OC43) research comparability in studies using HCoV-OC43 as a surrogate for SARS-CoV-2. J. Virol. Methods 2022, 299, 114317. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Kutter, J.S.; Spronken, M.I.; Fraaij, P.L.; Fouchier, R.A.; Herfst, S. Transmission routes of respiratory viruses among humans. Curr. Opin. Virol. 2018, 28, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Payne, D. The action of three antiseptics/disinfectants against enveloped and non-enveloped viruses. J. Hosp. Infect. 1998, 38, 283–295. [Google Scholar] [CrossRef]
- Pawnikar, S.; Bhattarai, A.; Wang, J.; Miao, Y. Binding Analysis Using Accelerated Molecular Dynamics Simulations and Future Perspectives. Adv. Appl. Bioinform. Chem. 2022, 15, 1–19. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Hirose, R.; Watanabe, N.; Bandou, R.; Yoshida, T.; Daidoji, T.; Naito, Y.; Itoh, Y.; Nakaya, T. A Cytopathic Effect-Based Tissue Culture Method for HCoV-OC43 Titration Using TMPRSS2-Expressing VeroE6 Cells. mSphere 2021, 6, e00159-21. [Google Scholar] [CrossRef]
- Maharjan, S.; Kang, M.; Kim, J.; Kim, D.; Park, S.; Kim, M.; Baek, K.; Lee, Y.; Kwon, H.J. Apoptosis Enhances the Replication of Human Coronavirus OC43. Viruses 2021, 13, 2199. [Google Scholar] [CrossRef]
- Pang, Y.T.; Miao, Y.; Wang, Y.; McCammon, J.A. Gaussian accelerated molecular dynamics in NAMD. J. Chem. Theory Comput. 2017, 13, 9–19. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W.; MacKerell, A.D. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate–Protein Modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Goga, N.; Rzepiela, A.J.; De Vries, A.H.; Marrink, S.J.; Berendsen, H.J.C. Efficient algorithms for langevin and DPD dynamics. J. Chem. Theory Comput. 2012, 8, 3637–3649. [Google Scholar] [CrossRef] [Green Version]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iannone, F.; Ambrosino, F.; Bracco, G.; De Rosa, M.; Funel, A.; Guarnieri, G.; Migliori, S.; Palombi, F.; Ponti, G.; Santomauro, G.; et al. CRESCO ENEA HPC clusters: A working example of a multifabric GPFS Spectrum Scale layout. In Proceedings of the 2019 International Conference on High Performance Computing and Simulation, HPCS 2019, Institute of Electrical and Electronics Engineers Inc., Dublin, Ireland, 15–19 July 2019; pp. 1051–1052. [Google Scholar] [CrossRef]
- Guixà-González, R.; Rodriguez-Espigares, I.; Ramírez-Anguita, J.M.; Carrió-Gaspar, P.; Martinez-Seara, H.; Giorgino, T.; Selent, J. MEMBPLUGIN: Studying membrane complexity in VMD. Bioinformatics 2014, 30, 1478–1480. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
| Formulation (TTO%, EtOH%) | Time of Exposure (min) | FCoVII (Log TCID50/mL) | HCoV-OC43 (Log RNA Copies/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Virus Exposed to TTO | Virus Control | RV | Virus Inactivation (%) | Virus Exposed to TTO | Virus Control | RV | Virus Inactivation (%) | ||
| A * (3.33, 5.33) | 5 | <0.50 | 4.00 | ≥3.50 | ≥99.97 * | 2.58 | 3.36 | 0.78 | 83.41 |
| 15 | <0.50 | 4.00 | ≥3.50 | ≥99.97 * | 2.56 | 3.28 | 0.72 | 80.96 | |
| 30 | <0.50 | 4.00 | ≥3.50 | ≥99.97 * | 2.78 | 4.16 | 1.38 | 95.83 | |
| B (0.66, 1.06) | 5 | 3.75 | 3.50 | 0.00 | 0.00 | 3.79 | 3.36 | 0.00 | 0.00 |
| 15 | 3.75 | 4.00 | 0.25 | 43.76 | 3.69 | 3.28 | 0.00 | 0.00 | |
| 30 | 4.00 | 4.50 | 0.50 | 68.36 | 4.78 | 5.13 | 0.35 | 55.36 | |
| C (0.13, 0.21) | 5 | 3.50 | 3.50 | 0.00 | 0.00 | 3.24 | 3.36 | 0.00 | 0.00 |
| 15 | 4.00 | 4.00 | 0.00 | 0.00 | 3.45 | 3.28 | 0.00 | 0.00 | |
| 30 | 4.50 | 4.50 | 0.00 | 0.00 | 4.80 | 5.13 | 0.33 | 53.28 | |
| D (0.00, 5.33) | 5 | ND | ND | ND | ND | ND | ND | ND | ND |
| 15 | ND | ND | ND | ND | ND | ND | ND | ND | |
| 30 | 4.00 | 4.00 | 0.00 | 0.00 | 4.12 | 4.05 | 0.00 | 0.00 | |
| Disinfectant ** | 5 | <0.50 | 3.50 | ≥3.00 | ≥99.90 * | 1.90 | 4.03 | 2.13 | 99.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, A.; Iacovelli, F.; Scagnolari, C.; Scordio, M.; Frasca, F.; Condò, R.; Ammendola, S.; Gaziano, R.; Anselmi, M.; Divizia, M.; et al. Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study. Molecules 2022, 27, 3786. https://doi.org/10.3390/molecules27123786
Romeo A, Iacovelli F, Scagnolari C, Scordio M, Frasca F, Condò R, Ammendola S, Gaziano R, Anselmi M, Divizia M, et al. Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study. Molecules. 2022; 27(12):3786. https://doi.org/10.3390/molecules27123786
Chicago/Turabian StyleRomeo, Alice, Federico Iacovelli, Carolina Scagnolari, Mirko Scordio, Federica Frasca, Roberta Condò, Serena Ammendola, Roberta Gaziano, Maurizio Anselmi, Maurizio Divizia, and et al. 2022. "Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study" Molecules 27, no. 12: 3786. https://doi.org/10.3390/molecules27123786
APA StyleRomeo, A., Iacovelli, F., Scagnolari, C., Scordio, M., Frasca, F., Condò, R., Ammendola, S., Gaziano, R., Anselmi, M., Divizia, M., & Falconi, M. (2022). Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study. Molecules, 27(12), 3786. https://doi.org/10.3390/molecules27123786