Abstract
Hydrogen/deuterium (H/D) exchange combined with two-dimensional (2D) NMR spectroscopy has been widely used for studying the structure, stability, and dynamics of proteins. When we apply the H/D-exchange method to investigate non-native states of proteins such as equilibrium and kinetic folding intermediates, H/D-exchange quenching techniques are indispensable, because the exchange reaction is usually too fast to follow by 2D NMR. In this article, we will describe the dimethylsulfoxide (DMSO)-quenched H/D-exchange method and its applications in protein science. In this method, the H/D-exchange buffer is replaced by an aprotic DMSO solution, which quenches the exchange reaction. We have improved the DMSO-quenched method by using spin desalting columns, which are used for medium exchange from the H/D-exchange buffer to the DMSO solution. This improvement has allowed us to monitor the H/D exchange of proteins at a high concentration of salts or denaturants. We describe methodological details of the improved DMSO-quenched method and present a case study using the improved method on the H/D-exchange behavior of unfolded human ubiquitin in 6 M guanidinium chloride.
1. Introduction
Hydrogen/deuterium (H/D) exchange is a powerful tool to investigate the structure, stability, dynamics, and interactions of proteins [1,2,3,4]. The advancements in two-dimensional (2D) nuclear magnetic resonance (NMR) techniques have made it possible to monitor the H/D-exchange kinetics of the individually identified peptide amide (NH) protons of proteins at amino-acid residue resolution [5,6]. Recent H/D-exchange studies also employ a mass spectrometric (MS) technique combined with rapid proteolysis and HPLC separation, which allows us to obtain the exchange kinetics at a nearly single-residue resolution [4,7,8]. In H/D-exchange experiments, the exchange reaction of each NH group of a protein to ND takes place in D2O. The exchange kinetics of the NH proton are determined by a structural opening reaction of the NH group and the intrinsic exchange rate constant, kint, of the NH proton, and are represented by:
where NHcl and NHop denote the closed and open states of the NH group, ND indicates the amide group after H/D exchange, and kop and kcl are the opening and closing rate constants [1,2,3,4]. Under the steady-state condition, the observed H/D-exchange rate constant, kobs, is thus given by:
The kint of each NH proton of a protein depends on solution conditions (pH, temperature, and salt concentration), neighboring residues (i.e., the amino-acid sequence), and isotope effects. Englander and his colleagues [9,10,11,12] accurately calibrated these effects on kint and reported the calibration parameters, from which we can quantitatively estimate the kint value of each NH proton of the protein.
The protection factor, P, which represents the degree of protection of the NH group against H/D exchange is given by the ratio of kint to kobs as:
The P value of native globular proteins is as large as 106–109, which corresponds to the structural opening free energy of 8–13 kcal/mol. This free energy change is equivalent to the free energy change of the global unfolding for each protein, indicating that the H/D-exchange reactions of the most stable NH groups are brought about by global unfolding [13,14].
A native-state H/D-exchange method was developed by Bai et al. [15] in 1995, and the H/D-exchange behavior at low concentrations of denaturants was characterized by this method. Temperature or pressure perturbation was also used in the native-state H/D exchange [16,17,18]. The method is effective for studying different kinds of protein dynamic behavior, ranging from local fluctuations up to sub-global and whole-molecule global unfolding reactions [15,19,20,21]. The method has been applied to a large number of proteins (reviewed in [4,19,20,21]).
The H/D-exchange techniques have also been used effectively for studying non-native states, including equilibrium unfolding intermediates and transient intermediates in kinetic refolding reactions of proteins. The molten globule (MG) state, which has a substantial amount of secondary structure but lacks the specific tertiary side-chain packing characteristics of native proteins, is an equilibrium intermediate state under mildly denaturing conditions for numerous globular proteins, many with more than ~100 residues [22,23,24]. In the 1990s, the structural characterizations of the MG state by H/D-exchange 2D NMR were carried out for a number of globular proteins, including apomyoglobin [25], cytochrome c [26,27], α-lactalbumin [28,29,30,31,32], Ca2+-binding milk lysozyme [33,34], and other proteins [35,36,37,38,39,40,41]. The P values of slowly exchanging NH protons in the MG state range from 102 to 103, which is more than three orders of magnitude smaller than the values in the native (N) state. The H/D-exchange rate in the MG state and in other non-native unfolded states [42,43,44,45,46,47,48,49,50,51,52] is usually too fast to follow by 2D NMR spectroscopy. Therefore, the H/D exchange was quenched after the desired period of H/D exchange by rapid refolding, and the NMR spectra were measured in the N state.
The use of a hydrogen-exchange method to detect and characterize a transient folding intermediate of proteins was first reported by Schmid and Baldwin [53] in 1979. They used a tritium-exchange technique and carried out a kinetic competition between folding and hydrogen exchange to investigate the folding kinetics of ribonuclease A. Subsequently, Roder and Wüthrich [54] proposed an extension of this technique by combined use of rapid mixing and NMR analysis. In 1988, two seminal papers, one by Udgaonkar and Baldwin [55] for ribonuclease A and the other by Roder et al. [56] for oxidized cytochrome c, appeared and reported the structural characterization of kinetic folding intermediates by hydrogen-exchange labeling and 2D NMR spectroscopy. In both studies, the NH groups of proteins were first fully deuterated in the fully unfolded (U) state in D2O, and the nonprotected amide ND groups in folding intermediates were proton-labeled in H2O by a short alkaline pH pulse (pulse-labeling hydrogen exchange), followed by quenching the D/H exchange by rapid refolding to the N state and NMR measurements in the N state. Since then, the pulse-labeling or competition hydrogen-exchange studies combined with 2D NMR to detect and characterize transient folding intermediates have been reported for many globular proteins, including barnase by the Fersht group [57,58], the lysozyme–α-lactalbumin family proteins by the Chris Dobson group [59,60,61,62,63], apomyoglobin and apoleghemoglobin by the Wright group [64,65,66,67], and other proteins by other groups [41,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91]. The early transient folding intermediates thus characterized are very similar in structure and stability to the equilibrium MG state, indicating that the MG state is the equilibrium counterpart of the kinetic folding intermediate formed early during refolding from the U state [23]. However, there are exceptions to this rule. The kinetic folding intermediate of the plant globin apoleghemoglobin under the refolding condition is significantly different in structure from its equilibrium MG state [66]. The early kinetic folding intermediate of ribonuclease H from Escherichia coli (E. coli) has a well-folded region with a closely packed tertiary structure, which is absent in its equilibrium MG state [92].
In this article, we describe the dimethylsulfoxide (DMSO)-quenched H/D-exchange method, in which the H/D-exchange reaction is quenched by the DMSO solution [93]. As shown above, the hydrogen-exchange reactions in the MG state, other non-native states, and transient kinetic folding intermediates of proteins need to be quenched before measurements of 2D NMR spectra. The rapid refolding was used for quenching the exchange reactions in the above studies, and hence, only NH protons stably protected in the N state are available for analysis. The DMSO-quenched H/D-exchange method is more versatile, because the exchange reactions of all NH groups, including nonprotected NH groups in the N state, are effectively quenched in an aprotic solvent DMSO and are available for the NMR analysis [93]. In the following, we will give a brief historical summary of the DMSO-quenched H/D-exchange method in proteins. In addition, the DMSO-quenched H/D-exchange method has recently been improved by the use of spin desalting columns [94]. This improvement has made it possible to apply the DMSO-quenched method to the exchange reactions of proteins in the presence of a high concentration of salt or denaturant. We thus describe the methodological details of the use of spin desalting columns in the DMSO-quenched H/D-exchange method and present a study on the H/D-exchange behavior of unfolded ubiquitin in 6 M guanidinium chloride (GdmCl), in which the spin desalting column was used in the DMSO-quenched H/D-exchange 2D NMR experiments.
2. DMSO-Quenched H/D-Exchange Method
The DMSO-quenched H/D-exchange method was developed by Zhang et al. [93] in 1995. They investigated various solution conditions to minimize the H/D-exchange rate of NH protons of proteins, and the presence of 95% (v/v) DMSO-d6 in a DMSO-d6/D2O mixture at pH* 5–6 was the best condition, where the H/D-exchange rate was ~100 fold slower than the minimum exchange rate in D2O. Here, pH* is the uncorrected pH-meter reading, and the pH* was adjusted by dichloroacetic acid-d2 (DCA-d2), whose pKa is 5.72 in the DMSO/D2O mixture [93]. To use the DMSO-quenched H/D-exchange method to investigate the H/D-exchange reaction of a protein, aliquots of the reaction mixture at various exchange times are first quenched by rapid freezing in liquid nitrogen, and then lyophilized. The lyophilized powder is dissolved in the quenching DMSO solution, and the NMR spectra of the protein are measured. Because proteins are unfolded in the DMSO solution, the NMR peaks may be distributed in a very narrow spectral region. However, the use of isotope (15N and 13C)-enriched proteins and the triple-resonance multi-dimensional NMR techniques [95] have made it feasible to identify most of the cross peaks in the 2D 1H–15N heteronuclear single-quantum coherence (HSQC) spectrum [96] of a small protein and to follow the H/D-exchange kinetics of individually identified NH protons.
2.1. Applications to Folding Intermediates and Amyloid Fibrils
Nishimura et al. [65,66,97] applied the DMSO-quenched H/D-exchange method to elucidate the structure in the equilibrium and transient folding intermediates of apomyoglobin and apoleghemoglobin; they used 99.4% DMSO instead of the 95% DMSO solution as a quenching solvent, however. Using the DMSO-quenched H/D-exchange method, they acquired data for the NH protons of 94 residues for the 153 residues of apomyoglobin, as compared with the 52 residues probed by the conventional pulse-labeling hydrogen-exchange method, in which the exchange was quenched by rapid refolding [97]. The DMSO-quenched method could be applied only for pH-jump refolding experiments because it was difficult to dissolve the lyophilized protein in DMSO in the presence of residual denaturant (urea or GdmCl) after denaturant-jump experiments [65,66]. Sakamoto et al. [98] studied the H/D-exchange kinetics of disulfide-deficient lysozyme in glycerol solution by the DMSO-quenched H/D-exchange method. They removed glycerol from the reaction mixture by reversed-phase HPLC, and the fractionated portion was lyophilized before dissolving in the DMSO solution.
DMSO effectively dissolves amyloid fibrils in vivo and in vitro [99,100,101]. Therefore, since the early 2000s, the DMSO-quenched H/D-exchange method has been widely used in studies on the H/D-exchange kinetics of amyloid fibrils [102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124] and other protein aggregates, including protein supermolecular complexes [125,126]. These studies have demonstrated the presence of a hydrogen-bonded (H-bonded) core highly resistant to H/D exchange in the amyloid fibrils and the other protein complexes. In the amyloid experiments, insoluble amyloid fibrils were first suspended in D2O to carry out the H/D-exchange reaction for the desired exchange periods, followed by separation of fibrils by centrifugation and lyophilization. The lyophilized fibrils were dissolved and dissociated into monomers in the DMSO solution, and the unfolded monomeric form was subjected to 2D NMR analysis. To characterize transient kinetic intermediates during the formation of amyloid fibrils, Carulla et al. [127] and Konuma et al. [90] combined the pulse-labeling hydrogen-exchange strategy and the DMSO-quenched method. After a short labeling pH pulse, aliquots of the reaction mixture were frozen and lyophilized, followed by dissolution in the DMSO solution for the subsequent 2D NMR or MS analysis. These studies gave us information about the molecular mechanisms of amyloid formation. Although the DMSO solution based on the standard protocol is composed of 95% DMSO-d6 at pH* 5–6 adjusted by DCA-d2, slightly modified compositions, e.g., dry DMSO-d6 [103,125] and DMSO-d6/trifluoroacetic acid-d1 (0.01–1%) mixture [107,110,114,115,116,117,119,122,126,128,129] were also used as an H/D-exchange quenching solution. Several excellent review articles on the DMSO-quenched H/D-exchange method have been published and cover more details about the method [130,131,132,133,134].
2.2. Use of Spin Desalting Columns
We improved the DMSO-quenched H/D-exchange NMR method by the use of spin desalting columns for medium exchange from the H/D-exchange buffer to the DMSO solution [94]. As shown above, the conventional DMSO-quenched H/D-exchange experiments had used lyophilization for the medium exchange. Therefore, it was difficult to carry out the DMSO-quenched experiments at a high concentration of salt or denaturant (urea or GdmCl), because the presence of residual salt or denaturant after lyophilization interferes with the 2D NMR analysis of proteins dissolved in DMSO. This is a drawback of the conventional DMSO-quenched method, and it prevents us from using the method to characterize the H/D-exchange behaviors of proteins in the intermediate or unfolded state in denaturant and native proteins under physiological conditions at 0.15 M NaCl (or KCl).
To prepare the quenching DMSO solution, we adjusted the pH* of the 94.5% (v/v) DMSO-d6/5% (v/v) D2O/0.5% (v/v) DCA-d2 solution to between 5 and 6 by adding 10 M NaOD. The addition of NaOD was accompanied by crystalline sodium dichloroacetate-d1, which was dissolved by stirring with an increase in pH*. Before starting the H/D-exchange reaction of a sample protein, we first prepared a 10-fold concentrated stock solution of the protein in H2O. The H/D-exchange reaction was started by a 10-fold dilution of the stock solution with a D2O buffer. At appropriate time points in the H/D-exchange, we collected 1.0 mL aliquots of the reaction solution, which had been pre-dispensed into 2 mL polypropylene cryo-tubes in advance, and froze them by immersing the cryo-tubes in liquid nitrogen to stop the reaction. The frozen aliquots were kept at −85 °C to −80 °C until the medium exchange by a spin desalting column and the subsequent 2D NMR measurements. The frozen aliquots were thawed at an appropriate temperature, at or below room temperature, just before the medium exchange. The dead time of the H/D-exchange measurement depends on how the experiment is conducted. If the experiment is carried out with the cooperation of two experimenters, one quenching the reaction mixture quickly in liquid nitrogen and the other recording the reaction time of the H/D exchange, it is rather easy to realize the dead time of 30 s (see Figure 3).
Figure 1 shows a schematic of the medium exchange procedure from the D2O buffer of the H/D-exchange reaction mixture to the DMSO solution with the use of spin desalting columns [94]. We used 5 mL columns (ZebaTM Spin Desalting Column 89891, Thermo Fisher Scientific K.K., Tokyo, Japan) and 15 mL polypropylene centrifuge tubes as collection tubes. The procedure consists of the following five steps:
- Remove the bottom plug of a spin desalting column, and place the column in a collection tube.
- Centrifuge for 2 min at 1000× g to pack the column bed and remove storage buffer.
- Apply 2.5 mL of the DMSO solution to the column, centrifuge for 2–3 min at 1000× g, and discard the flow through. Repeat this process two to three times, but centrifuge for 3 min in the second and third runs, because the DMSO solution is more viscous than water.
- Apply 1.0 mL of the H/D-exchange sample solution to the column, and centrifuge for 2 min at 20 °C. The application volume should not exceed 1.0 mL, because the application of more than 1.0 mL results in leakage of the H/D-exchange sample solution into the collection tube.
- Recover the desalted protein sample in the DMSO solution; the protein sample can now be applied to 2D NMR analysis.
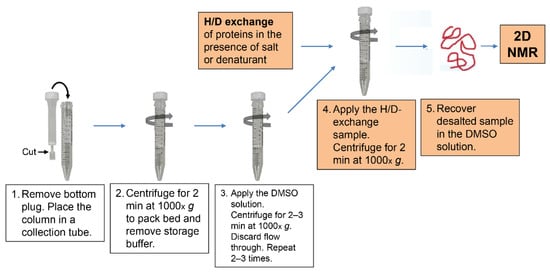
Figure 1.
A schematic procedure of the medium exchange by a spin desalting column.
We used 2D 1H–15N HSQC spectra for the NMR analysis, and hence the sample protein was 15N-labeled, and the assignment of the HSQC cross peaks was carried out by three-dimensional (3D) HN(CA)NNH, HNCA, HN(CO)CA, HNCO, CBCA(CO)NH, and HNCAHA experiments using 13C/15N-double-labeled proteins. We applied the improved DMSO-quenched 2D NMR method to investigate the H/D-exchange behavior of the E. coli co-chaperonin GroES at pH* 6.5 (or 7.5) and 25 °C [135] and unfolded ubiquitin at pH* 3.2 and 15.0 °C in 6 M GdmCl [136], In the following, we will describe the study on the H/D-exchange behavior of unfolded ubiquitin as a case study.
3. A Case Study: Unfolded Ubiquitin in 6 M GdmCl
The characterization of residual structures persistent in unfolded proteins in concentrated denaturant (6 M GdmCl or 8 M urea) is an important issue in studies of protein folding. The problem of protein folding has been described with reference to the Levinthal paradox, in which the initial unfolded state is assumed to be a random coil, and hence, there may exist an astronomically large number of conformations, inaccessible in a reasonable time by a random search, at the beginning of the folding reactions [137,138,139]. Solving the Levinthal paradox is a fundamental problem in folding studies [140,141,142,143,144,145,146]. The presence of the residual structure, if any, in the unfolded state thus invalidates the Levinthal paradox, because such residual structure may form a folding initiation site and guide the subsequent folding reactions. We therefore studied the H/D-exchange behavior of unfolded human ubiquitin in 6 M GdmCl by the DMSO-quenched H/D-exchange 2D NMR method with the use of spin desalting columns [136]. Although the persistence of residual structures in unfolded proteins in concentrated denaturant has been reported for several different proteins [50,147,148,149,150,151,152,153], the present method enabled us to estimate the p values of individually identified NH protons, including nonprotected NH protons in the N state [136].
Ubiquitin is a 76-residue α/β protein, composed of a mixed parallel–anti-parallel β-sheet packing against a middle α-helix to form the hydrophobic core (Figure 2) [154]. Ubiquitin is a typical model protein for protein folding studies, and thus its folding reactions have been studied by a variety of biophysical techniques, including stopped-flow [155,156,157,158] and continuous-flow [159] kinetic refolding experiments, pulse-labeling hydrogen-exchange experiments combined with 2D NMR spectroscopy [72,160] and electrospray ionization mass spectrometry [161], mutational ϕ-value analysis [162,163], and other techniques [164,165,166]. These results may be compared with the present results of the H/D-exchange behavior of unfolded ubiquitin in 6 M GdmCl.
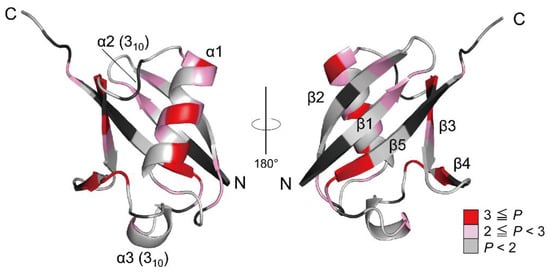
Figure 2.
The 3D structure of native ubiquitin (PDB code: 1UBQ). The residues are colored according to the P values of the NH protons, and the red gradient indicates the scale of the P value. The proline residues and the residues that could not be used as probes due to severe broadening or overlapping are shown in black. Adapted with permission from Ref. [136]. Copyright 2020 Biophysical Society.
To analyze the H/D-exchange kinetics of individually identified NH protons of ubiquitin, we first made their spectral assignments in the DMSO solution [136]. Using a combination of 3D spectral measurements, we successfully assigned all the peaks observed in the HSQC spectrum. We then investigated the H/D-exchange kinetics of all the individual NH groups. Excluding NH groups whose residues could not be used as probes due to severe broadening or overlapping, we successfully followed the H/D-exchange kinetics of 60 NH protons [136]. These 60 NH protons include not only the protons stably protected in the native structure but also nonprotected NH protons. The observed kinetic exchange curve, given by the volume, Y(t), of cross peaks in 2D NMR spectra as a function of the H/D-exchange time, t, was a single exponential fitted to the equation:
where ΔY and Y(∞) are the kinetic amplitude and the final value of the peak volume, respectively. Figure 3 shows the kinetic progress curves of the Val5, Asn25, Gln40, and Glu51 NH protons measured by the DMSO-quenched method. The protection factor P was calculated by Equation (3) for the 60 NH protons, resulting in the protection profile shown in Figure 4, in which the P value is plotted as a function of the residue number.
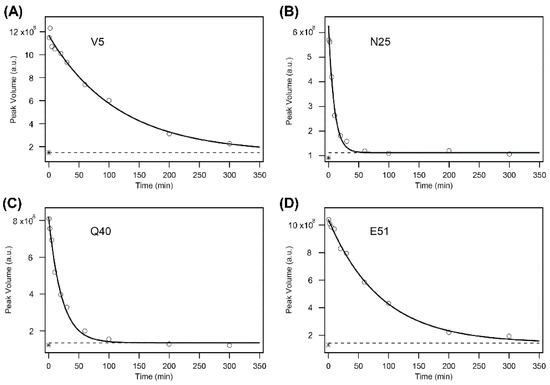
Figure 3.
The H/D-exchange curves for Val5 (V5) (A), Asn25 (N25) (B), Gln40 (Q40) (C), and Glu51 (E51) (D) of human ubiquitin in 6 M GdmCl at pH* 3.3 and 15 °C. The solid lines are the theoretical curves best fitted to a single-exponential function (Equation (4)). A broken line in each panel indicates the theoretically estimated peak volume after the complete exchange (i.e., Y(∞) in Equation (4)), and an asterisk “*” in each panel, located between (1–2) × 108 of the peak volume, indicates the experimentally observed value after heating the sample at 50 °C for 30 min. Because the reaction mixtures contained 10% H2O, the final peak volumes did not reach zero. The kobs values for the four residues are: (A) (8.7 ± 0.8) × 10−3 min−1; (B) (10.5 ± 1.2) × 10−2 min−1; (C) (4.6 ± 0.4) × 10−2 min−1; (D) (1.2 ± 0.1) × 10−2 min−1. The first and last time points are at 0.62 and 300.09 min, respectively, for all panels. Adapted with permission from Ref. [136]. Copyright 2020 Biophysical Society.
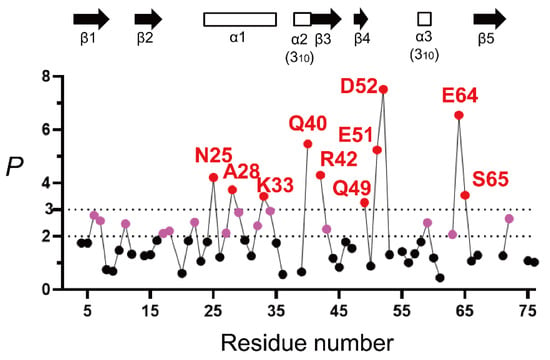
Figure 4.
The H/D-exchange protection profile of unfolded ubiquitin in 6 M GdmCl, represented by P as a function of the residue number (pH* 3.3 and 15 °C). The dashed lines indicate the P values of 2 and 3. The amino-acid residues with P values larger than 2 and 3 are indicated in pink and red, respectively, and the other residues in black. The locations of the secondary structures in native ubiquitin (PDB code: 1UBQ) are shown by arrows (β-strands) and open rectangles (helices). The kobs values for the majority of NH protons were obtained by three independent H/D-exchange experiments, and the percent standard error estimate of kobs was ~8%, indicating that the P value of 2.0 can be written as P = 2.0 ± 0.16 (see [136] for the complete list of the standard error estimates of kobs values). Adapted with permission from Ref. [136]. Copyright 2020 Biophysical Society.
From Figure 4, a majority of the NH protons have a P value below 2 and larger than or equal to 0.8, indicating that these peptide NH protons are almost fully exposed to solvent water in 6 M GdmCl. This is consistent with the previous reports that ubiquitin in concentrated denaturant (6 M GdmCl or 8 M urea) at acidic pH is almost fully unfolded [155,167,168,169,170]. However, the 10 NH protons of Asn25, Ala28, Lys33, Gln40, Arg42, Gln49, Glu51, Asp52, Glu64, and Ser65 were significantly protected with a p value larger than 3, and the additional 14 NH protons of Lys6, Thr7, Lys11, Val17, Glu18, Thr22, Lys27, Lys29, Asp32, Glu34, Leu43, Tyr59, Lys63, and Arg72 showed p values between 2 and 3. These results thus clearly indicate the presence of residual structures in unfolded ubiquitin. Because the protein was unfolded in 6 M GdmCl, it is most likely that these residues were protected by the formation of an H-bond with a certain acceptor group.
When the H/D-exchange protection is brought about by the formation of the H-bond with a specific acceptor group, we can estimate the fraction of H-bonding (fHbond) for the protected NH groups. Because only the non-H-bonded form of the NH proton is available for H/D exchange, (1 − fHbond) is equal to kobs/kint (= 1/P). Therefore, it follows that:
When NH protons have P values larger than 3 and 2, the fHbond values are larger than 0.67 and 0.50, respectively, from Equation (5). The free energy of the H-bond breakage (0.0–0.41 kcal/mol) estimated from the fHbond values is thus negligibly small as compared with the unfolding free energy (7.5 kcal/mol) of ubiquitin [155]. Nevertheless, the fHbond values larger than 0.5 should be significant when we consider the kinetic folding mechanisms of the protein, and some of these weakly protected residues may play an important role in the formation of folding initiation sites at an initial stage of kinetic refolding of the protein from the GdmCl-induced unfolded state.
To understand the relationships between the residual structure in unfolded ubiquitin and the H-bonds formed in native ubiquitin, the H-bonding network in the native structure is shown in Figure 5. From Figure 4 and Figure 5, the NH protons of Asn25, Lys27, Ala28, Lys29, Asp32, Lys33, and Glu34, which are all significantly protected with a P value larger than 2, are involved in the middle α-helix (Ile23–E34) in native ubiquitin, and each NH proton of these residues forms an α-helical H-bond with the peptide CO group of the four amino-acid residues earlier, except for Asn25 (Figure 5B). The Asn25 NH proton forms a more local H-bond with the side-chain Oγ atom of Thr22, which acts as an N-cap residue of the helix in native ubiquitin. These results thus clearly demonstrate the presence of a residual structure in this α-helix of ubiquitin in 6 M GdmCl at pH* 3.3 and 15 °C, and Thr22 may also function as a helix stop signal by forming the N-cap conformation as in the native ubiquitin structure [171]. The NH groups of Thr7 and Val17, which form H-bonds with the CO groups of Lys11 and Met1, respectively, in the N-terminal β-hairpin, also have P values larger than 2 in the U state (Figure 5C), suggesting that the N-terminal β-hairpin may also be partially preserved in unfolded ubiquitin. The other NH groups having a P value larger than 2 in the N-terminal β-hairpin include those of Lys6 and Lys11. Although the NH proton of Lys11 does not form a backbone H-bond in native ubiquitin, it forms a local backbone to the side-chain H-bond with the Oγ of Thr7, and hence such a local backbone to the side-chain H-bond may be at least partially preserved in unfolded ubiquitin and may stabilize the residual structure of the N-terminal β-hairpin (Figure 5C). Previous NMR studies on HN–NN residual dipolar couplings, chemical shifts, 3JHNHA couplings, relaxation rates, and h3JNC′ couplings have also shown that the native-like first β-hairpin conformation was populated to at most 25% in unfolded ubiquitin in 8 M urea [172,173,174,175]. From these results, we conclude that there are native-like residual structures in the middle helix and the N-terminal β-hairpin in unfolded ubiquitin in 6 M GdmCl and that these residual structures may play an important role at an initial stage of kinetic refolding from the unfolded state.
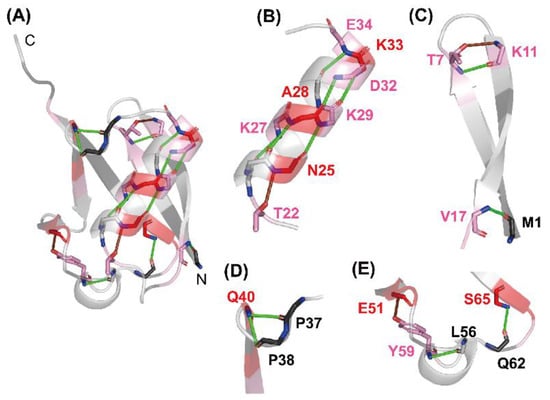
Figure 5.
The H-bonding network observed in native ubiquitin (PDB code: 1UBQ). (A) A whole view, and (B–E) closer views of (B) the middle α-helix, (C) the N-terminal β-hairpin, (D) the one-turn 310 helix (Pro38–Gln40), and (E) the Type II β-turn (Gln62–Ser65) and the one-turn 310 helix (Ser57–Tyr59) are shown. The H-bonds of the NH protons of Thr7, Val17, Lys27, Ala28, Lys29, Asp32, Lys33, Glu34, Gln40, Tyr59, and Ser65 with the CO groups of their counterparts are shown as green lines. The local H-bonds formed by the NH protons of Lys11, Asn25, and Glu51 with the side-chain atoms of Thr7, Thr22, and Tyr59, respectively, are shown as brown lines. The red gradient indicates the same P value scale as shown in Figure 2. Adapted with permission from Ref. [136]. Copyright 2020 Biophysical Society.
In support of this conclusion, a pulsed H/D-exchange study with rapid mixing methods and 2D NMR analysis by Briggs and Roder [72] has shown that the NH protons in the α-helix and the β-hairpin of the fast-folding species (ca. 80%) of unfolded ubiquitin become protected in an initial 8 ms folding phase from the GdmCl-induced unfolded state of ubiquitin. Went and Jackson [163] performed a comprehensive ϕ-value analysis on the structure of the transition state ensemble of the ubiquitin folding and showed that medium and high ϕ values were found only in the N-terminal β-hairpin and the middle helix, which was also consistent with the above conclusion. The α-helical residual structure as detected by the unfolded-state H/D-exchange NMR spectroscopy in a concentrated denaturant solution was also observed in cytochrome c in 6 M urea [91], suggesting that the present observation of the residual structure may not be a rare example.
Three locally stabilized H-bonds, which form two one-turn 310 helices (Gln40–Pro37 and Tyr59–Leu56) and a type II β-turn (Ser65–Gln62) in native ubiquitin, are also partially preserved in 6 M GdmCl. The NH proton of Gln40, which has a P value of 5.5 (fHbond = 0.81), forms H-bonds with the CO groups of Pro37 and Pro38 (Figure 5D). The NH proton of Tyr59, having a P value of 2.5 (fHbond = 0.60), forms an H-bond with the CO group of Leu56. The NH proton of Ser65, having a P value of 3.5 (fHbond = 0.71), forms an H-bond with the CO group of Gln62 (Figure 5E). Therefore, these locally stabilized H-bonds may not be fully disrupted even in 6 M GdmCl. However, it is not yet clear whether these local H-bonding interactions are important for the kinetic folding mechanisms of ubiquitin, because there is a dearth of experimental data concerning the effects of these local H-bonds on the kinetics of the ubiquitin folding.
The NH protons of the three residues of Arg42, Glu64, and Arg72 form native H-bonds with the backbone CO groups of Val70, Gln2, and Gln40, respectively [154], and have P values of 2.7 to 6.5 in unfolded ubiquitin (Figure 4). However, these are nonlocal H-bonds formed between residues at least 28 residues apart from each other, and such nonlocal H-bonds in native ubiquitin may not be stably formed in the U state in 6 M GdmCl. These NH protons may thus be protected by non-native H-bonding interactions. In fact, certain other NH protons, including those of Leu43, Gln49, Asp52, and Lys63, which have P values of 2.1 to 7.5 (Figure 4), are protected by non-native H-bonding interactions in 6 M GdmCl, because these NH protons do in fact lack backbone H-bonds in the native structure. The majority of residues with a P value larger than 3 are either charged or contain a side-chain amide or guanidinium group (Arg42, Gln49, Glu51, and Glu64). It is thus also possible that the protection might be afforded by the H-bonding between the backbone NH and its own side-chain atoms.
4. Conclusions
- We described the DMSO-quenched H/D-exchange 2D NMR spectroscopy and its applications in protein science. The DMSO-quenched method is superior to the conventionally used refolding-quenched method, because the former allows us to monitor the H/D-exchange kinetics of not only the protected NH protons but also the nonprotected NH protons in the N state.
- We described the improvement of the DMSO-quenched H/D-exchange method by the use of spin desalting columns and gave the methodological details of the use of spin desalting columns.
- We presented a case study in which we characterized the H/D-exchange kinetics of 60 peptide NH protons in unfolded ubiquitin in 6 M GdmCl by the improved DMSO-quenched method. The residual structures were preserved in the middle α-helix and the N-terminal β-hairpin in unfolded ubiquitin with a protection factor P larger than 2 (i.e., a fraction of H-bonding fHbond of larger than 0.5), and these residual structures may play an important role in the folding process as nucleation sites and guide the subsequent folding reactions to the N state.
Author Contributions
Writing—original draft preparation, K.K. (Kunihiro Kuwajima); writing—review and editing, M.Y.-U., S.Y. and K.K. (Koichi Kato). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by JSPS (Japan Society for the Promotion of Science) KAKENHI Grant Number JP20K06574 and by Joint Research of the Exploratory Research Center on Life and Living Systems (ExCELLS) Program Number 22EXC315. A part of this work was conducted in Institute for Molecular Science, and supported by the Nanotechnology Platform Program <Molecule and Material Synthesis> (JPMXP09S21MS0021) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hvidt, A.; Nielsen, S.O. Hydrogen exchange in proteins. Adv. Protein Chem. 1966, 21, 287–386. [Google Scholar] [CrossRef] [PubMed]
- Woodward, C.; Carulla, N.; Barany, G. Native state hydrogen-exchange analysis of protein folding and protein motional domains. Methods Enzymol. 2004, 380, 379–400. [Google Scholar] [PubMed]
- Englander, S.W.; Mayne, L.; Krishna, M.M. Protein folding and misfolding: Mechanism and principles. Q. Rev. Biophys. 2007, 40, 287–326. [Google Scholar] [CrossRef] [PubMed]
- Englander, S.W.; Mayne, L.; Kan, Z.Y.; Hu, W. Protein folding—How and why: By hydrogen exchange, fragment separation, and mass spectrometry. Annu. Rev. Biophys. 2016, 45, 135–152. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.; Wüthrich, K. Amide protein exchange and surface conformation of the basic pancreatic trypsin inhibitor in solution. Studies with two-dimensional nuclear magnetic resonance. J. Mol. Biol. 1982, 160, 343–361. [Google Scholar] [CrossRef]
- Wand, A.J.; Roder, H.; Englander, S.W. Two-dimensional 1H NMR studies of cytochrome c: Hydrogen exchange in the N-terminal helix. Biochemistry 1986, 25, 1107–1114. [Google Scholar] [CrossRef]
- Konermann, L.; Simmons, D.A. Protein-folding kinetics and mechanisms studied by pulse-labeling and mass spectrometry. Mass Spectrom. Rev. 2003, 22, 1–26. [Google Scholar] [CrossRef]
- Vadas, O.; Jenkins, M.L.; Dornan, G.L.; Burke, J.E. Using hydrogen-deuterium exchange mass spectrometry to examine protein-membrane interactions. Methods Enzymol. 2017, 583, 143–172. [Google Scholar] [CrossRef]
- Molday, R.S.; Englander, S.W.; Kallen, R.G. Primary structure effects on peptide group hydrogen exchange. Biochemistry 1972, 11, 150–158. [Google Scholar] [CrossRef]
- Bai, Y.; Milne, J.S.; Mayne, L.; Englander, S.W. Primary structure effects on peptide group hydrogen exchange. Proteins 1993, 17, 75–86. [Google Scholar] [CrossRef]
- Connelly, G.P.; Bai, Y.; Jeng, M.F.; Englander, S.W. Isotope effects in peptide group hydrogen exchange. Proteins 1993, 17, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Mayne, L.; Phillips, M.C.; Walter Englander, S. Reference parameters for protein hydrogen exchange rates. J. Am. Soc. Mass Spectrom. 2018, 29, 1936–1939. [Google Scholar] [CrossRef] [PubMed]
- Huyghues-Despointes, B.M.P.; Scholtz, J.M.; Pace, C.N. Protein conformational stabilities can be determined from hydrogen exchange rates. Nat. Struct. Biol. 1999, 6, 910–912. [Google Scholar] [PubMed]
- Huyghues-Despointes, B.M.; Pace, C.N.; Englander, S.W.; Scholtz, J.M. Measuring the conformational stability of a protein by hydrogen exchange. Methods Mol. Biol. 2001, 168, 69–92. [Google Scholar]
- Bai, Y.; Sosnick, T.R.; Mayne, L.; Englander, S.W. Protein folding intermediates: Native-state hydrogen exchange. Science 1995, 269, 192–197. [Google Scholar] [CrossRef]
- Brorsson, A.C.; Kjellson, A.; Aronsson, G.; Sethson, I.; Hambraeus, C.; Jonsson, B.H. The “two-state folder” MerP forms partially unfolded structures that show temperature dependent hydrogen exchange. J. Mol. Biol. 2004, 340, 333–344. [Google Scholar] [CrossRef]
- Rennella, E.; Corazza, A.; Fogolari, F.; Viglino, P.; Giorgetti, S.; Stoppini, M.; Bellotti, V.; Esposito, G. Equilibrium unfolding thermodynamics of β2-microglobulin analyzed through native-state H/D exchange. Biophys. J. 2009, 96, 169–179. [Google Scholar] [CrossRef]
- Fuentes, E.J.; Wand, A.J. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry 1998, 37, 9877–9883. [Google Scholar] [CrossRef]
- Bai, Y. Hidden intermediates and Levinthal paradox in the folding of small proteins. Biochem. Biophys. Res. Commun. 2003, 305, 785–788. [Google Scholar] [CrossRef]
- Bai, Y. Protein folding pathways studied by pulsed- and native-state hydrogen exchange. Chem. Rev. 2006, 106, 1757–1768. [Google Scholar] [CrossRef]
- Malhotra, P.; Udgaonkar, J.B. Native state hydrogen exchange-mass spectrometry methods to probe protein folding and unfolding. Methods Mol. Biol. 2022, 2376, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Ptitsyn, O.B. Molten globule and protein folding. Adv. Protein Chem. 1995, 47, 83–229. [Google Scholar] [PubMed]
- Arai, M.; Kuwajima, K. Role of the molten globule state in protein folding. Adv. Protein Chem. 2000, 53, 209–282. [Google Scholar] [PubMed]
- Kuwajima, K. The molten globule, and two-state vs. non-two-state folding of globular proteins. Biomolecules 2020, 10, 407. [Google Scholar] [CrossRef]
- Hughson, F.M.; Wright, P.E.; Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 1990, 249, 1544–1548. [Google Scholar] [CrossRef]
- Jeng, M.F.; Englander, S.W.; Gülnur, A.; Elöve, G.A.; Wand, A.J.; Roder, H. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry 1990, 29, 10433–10437. [Google Scholar] [CrossRef]
- Kuroda, Y.; Endo, S.; Nagayama, K.; Wada, A. Stability of α-helices in a molten globule state of cytochrome c by hydrogen-deuterium exchange and two-dimensional NMR spectroscopy. J. Mol. Biol. 1995, 247, 682–688. [Google Scholar] [CrossRef]
- Alexandrescu, A.T.; Evans, P.A.; Pitkeathly, M.; Baum, J.; Dobson, C.M. Structure and dynamics of the acid-denatured molten globule state of α-lactalbumin: A two-dimensional NMR study. Biochemistry 1993, 32, 1707–1718. [Google Scholar] [CrossRef]
- Chyan, C.-L.; Wormald, C.; Dobson, C.M.; Evans, P.A.; Baum, J. Structure and stability of the molten globule state of guinea-pig α-lactalbumin: A hydrogen exchange study. Biochemistry 1993, 32, 5681–5691. [Google Scholar] [CrossRef]
- Schulman, B.A.; Redfield, C.; Peng, Z.Y.; Dobson, C.M.; Kim, P.S. Different subdomains are most protected from hydrogen exchange in the molten globule and native states of human α-lactalbumin. J. Mol. Biol. 1995, 253, 651–657. [Google Scholar] [CrossRef]
- Engel, M.F.; Visser, A.J.; van Mierlo, C.P. Conformation and orientation of a protein folding intermediate trapped by adsorption. Proc. Natl. Acad. Sci. USA 2004, 101, 11316–11321. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Makabe, K.; Tomoyori, K.; Maki, K.; Mukaiyama, A.; Kuwajima, K. Different folding pathways taken by highly homologous proteins, goat α-lactalbumin and canine milk lysozyme. J. Mol. Biol. 2010, 396, 1361–1378. [Google Scholar] [CrossRef] [PubMed]
- Morozova-Roche, L.A.; Arico-Muendel, C.C.; Haynie, D.T.; Emelyanenko, V.I.; Van Dael, H.; Dobson, C.M. Structural characterisation and comparison of the native and A-states of equine lysozyme. J. Mol. Biol. 1997, 268, 903–921. [Google Scholar] [CrossRef] [PubMed]
- Kobashigawa, Y.; Demura, M.; Koshiba, T.; Kumaki, Y.; Kuwajima, K.; Nitta, K. Hydrogen exchange study of canine milk lysozyme: Stabilization mechanism of the molten globule. Proteins 2000, 40, 579–589. [Google Scholar] [CrossRef]
- Pan, Y.; Briggs, M.S. Hydrogen exchange in native and alcohol forms of ubiquitin. Biochemistry 1992, 31, 11405–11412. [Google Scholar] [CrossRef] [PubMed]
- Dabora, J.M.; Pelton, J.G.; Marqusee, S. Structure of the acid state of Escherichia coli ribonuclease HI. Biochemistry 1996, 35, 11951–11958. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, A.K.; Marqusee, S. Molten globule unfolding monitored by hydrogen exchange in urea. Biochemistry 1998, 37, 1736–1742. [Google Scholar] [CrossRef]
- Yamasaki, K.; Yamasaki, T.; Kanaya, S.; Oobatake, M. Acid-induced denaturation of Escherichia coli ribonuclease HI analyzed by CD and NMR spectroscopies. Biopolymers 2003, 69, 176–188. [Google Scholar] [CrossRef]
- Davis-Searles, P.R.; Morar, A.S.; Saunders, A.J.; Erie, D.A.; Pielak, G.J. Sugar-induced molten-globule model. Biochemistry 1998, 37, 17048–17053. [Google Scholar] [CrossRef]
- Mazon, H.; Marcillat, O.; Forest, E.; Smith, D.L.; Vial, C. Conformational dynamics of the GdmHCl-induced molten globule state of creatine kinase monitored by hydrogen exchange and mass spectrometry. Biochemistry 2004, 43, 5045–5054. [Google Scholar] [CrossRef]
- Nabuurs, S.M.; van Mierlo, C.P.M. Interrupted hydrogen/deuterium exchange reveals the stable core of the remarkably helical molten globule of α-β parallel protein flavodoxin. J. Biol. Chem. 2010, 285, 4165–4172. [Google Scholar] [CrossRef] [PubMed]
- Roder, H.; Wagner, G.; Wüthrich, K. Individual amide proton-exchange rates in thermally unfolded basic pancreatic trypsin-inhibitor. Biochemistry 1985, 24, 7407–7411. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.D.; Baldwin, R.L. Hydrogen exchange in thermally denatured ribonuclease A. Biochemistry 1991, 30, 9907–9914. [Google Scholar] [CrossRef]
- Radford, S.E.; Buck, M.; Topping, K.D.; Dobson, C.M.; Evans, P.A. Hydrogen exchange in native and denatured states of hen egg-white lysozyme. Proteins 1992, 14, 237–248. [Google Scholar] [CrossRef]
- Buck, M.; Radford, S.E.; Dobson, C.M. A partially folded state of hen egg white lysozyme in trifluoroethanol: Structural characterization and implications for protein folding. Biochemistry 1993, 32, 669–678. [Google Scholar] [CrossRef]
- Fan, P.; Bracken, C.; Baum, J. Structural characterization of monellin in the alcohol-denatured state by NMR: Evidence for β-sheet to α-helix conversion. Biochemistry 1993, 32, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, X.; Jonas, A.; Jonas, J. NMR study of the cold, heat, and pressure unfolding of ribonuclease A. Biochemistry 1995, 34, 8631–8641. [Google Scholar] [CrossRef]
- Nash, D.P.; Jonas, J. Structure of pressure-assisted cold denatured lysozyme and comparison with lysozyme folding intermediates. Biochemistry 1997, 36, 14375–14383. [Google Scholar] [CrossRef]
- Nash, D.P.; Jonas, J. Structure of the pressure-assisted cold denatured state of ubiquitin. Biochem. Biophys. Res. Commun. 1997, 238, 289–291. [Google Scholar] [CrossRef]
- Wang, H.M.; Yu, C. Investigating the refolding pathway of human acidic fibroblast growth factor (hFGF-1) from the residual structure(s) obtained by denatured-state hydrogen/deuterium exchange. Biophys. J. 2011, 100, 154–164. [Google Scholar] [CrossRef][Green Version]
- Buck, M.; Radford, S.E.; Dobson, C.M. Amide hydrogen exchange in a highly denatured state. Hen egg-white lysozyme in urea. J. Mol. Biol. 1994, 237, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Katou, H.; Hoshino, M.; Kamikubo, H.; Batt, C.A.; Goto, Y. Native-like β-hairpin retained in the cold-denatured state of bovine β-lactoglobulin. J. Mol. Biol. 2001, 310, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Schmid, F.X.; Baldwin, R.L. Detection of an early intermediate in the folding of ribonuclease A by protection of amide protons against exchange. J. Mol. Biol. 1979, 135, 199–215. [Google Scholar] [CrossRef]
- Roder, H.; Wüthrich, K. Protein folding kinetics by combined use of rapid mixing techniques and NMR observation of individual amide protons. Proteins 1986, 1, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Udgaonkar, J.B.; Baldwin, R.L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature 1988, 335, 694–699. [Google Scholar] [CrossRef]
- Roder, H.; Elöve, G.A.; Englander, S.W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature 1988, 335, 700–704. [Google Scholar] [CrossRef]
- Bycroft, M.; Matouschek, A.; Kellis, J.J.; Serrano, L.; Fersht, A.R. Detection and characterization of a folding intermediate in barnase by NMR. Nature 1990, 346, 488–490. [Google Scholar] [CrossRef]
- Matouschek, A.; Serrano, L.; Meiering, E.M.; Bycroft, M.; Fersht, A.R. The folding of an enzyme V. H/2H exchange–Nuclear magnetic resonance studies on the folding pathway of barnase: Complementarity to and agreement with protein engineering studies. J. Mol. Biol. 1992, 224, 837–845. [Google Scholar] [CrossRef]
- Miranker, A.; Radford, S.E.; Karplus, M.; Dobson, C.M. Demonstration by NMR of folding domains in lysozyme. Nature 1991, 349, 633–636. [Google Scholar] [CrossRef]
- Hooke, S.D.; Radford, S.E.; Dobson, C.M. The refolding of human lysozyme: A comparison with the structurally homologous hen lysozyme. Biochemistry 1994, 33, 5867–5876. [Google Scholar] [CrossRef]
- Eyles, S.J.; Radford, S.E.; Robinson, C.V.; Dobson, C.M. Kinetic consequences of the removal of a disulfide bridge on the folding of hen lysozyme. Biochemistry 1994, 33, 13038–13048. [Google Scholar] [CrossRef] [PubMed]
- Forge, V.; Wijesinha, R.T.; Balbach, J.; Brew, K.; Robinson, C.V.; Redfield, C.; Dobson, C.M. Rapid collapse and slow structural reorganisation during the refolding of bovine α-lactalbumin. J. Mol. Biol. 1999, 288, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Morozova-Roche, L.A.; Jones, J.A.; Noppe, W.; Dobson, C.M. Independent nucleation and heterogeneous assembly of structure during folding of equine lysozyme. J. Mol. Biol. 1999, 289, 1055–1073. [Google Scholar] [CrossRef] [PubMed]
- Jennings, P.A.; Wright, P.E. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science 1993, 262, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Dyson, H.J.; Wright, P.E. Enhanced picture of protein-folding intermediates using organic solvents in H/D exchange and quench-flow experiments. Proc. Natl. Acad. Sci. USA 2005, 102, 4765–4770. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Dyson, H.J.; Wright, P.E. The kinetic and equilibrium molten globule intermediates of apoleghemoglobin differ in structure. J. Mol. Biol. 2008, 378, 715–725. [Google Scholar] [CrossRef][Green Version]
- Uzawa, T.; Nishimura, C.; Akiyama, S.; Ishimori, K.; Takahashi, S.; Dyson, H.J.; Wright, P.E. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc. Natl. Acad. Sci. USA 2008, 105, 13859–13864. [Google Scholar] [CrossRef]
- Udgaonkar, J.B.; Baldwin, R.L. Nature of the early folding intermediate of ribonuclease A. Biochemistry 1995, 34, 4088–4096. [Google Scholar] [CrossRef]
- Houry, W.A.; Scheraga, H.A. Structure of a hydrophobically collapsed intermediate on the conformational folding pathway of ribonuclease A probed by hydrogen-deuterium exchange. Biochemistry 1996, 35, 11734–11746. [Google Scholar] [CrossRef]
- Krishna, M.M.; Lin, Y.; Mayne, L.; Englander, S.W. Intimate view of a kinetic protein folding intermediate: Residue-resolved structure, interactions, stability, folding and unfolding rates, homogeneity. J. Mol. Biol. 2003, 334, 501–513. [Google Scholar] [CrossRef]
- Lu, J.; Dahlquist, F.W. Detection and characterization of an early folding intermediate of T4 lysozyme using pulsed hydrogen exchange and two-dimensional NMR. Biochemistry 1992, 31, 4749–4756. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.S.; Roder, H. Early hydrogen-bonding events in the folding reaction of ubiquitin. Proc. Natl. Acad. Sci. USA 1992, 89, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Mullins, L.S.; Pace, C.N.; Raushel, F.M. Investigation of ribonuclease T1 folding intermediates by hydrogen-deuterium amide exchange-two-dimensional NMR spectroscopy. Biochemistry 1993, 32, 6152–6156. [Google Scholar] [CrossRef]
- Koide, S.; Dyson, H.J.; Wright, P.E. Characterization of a folding intermediate of apoplastocyanin trapped by proline isomerization. Biochemistry 1993, 32, 12299–12310. [Google Scholar] [CrossRef] [PubMed]
- Varley, P.; Gronenborn, A.M.; Christensen, H.; Wingfield, P.T.; Pain, R.H.; Clore, G.M. Kinetics of folding of the all β-sheet protein interleukin-1β. Science 1993, 260, 1110–1113. [Google Scholar] [CrossRef] [PubMed]
- Kuszewski, J.; Clore, G.M.; Gronenborn, A.M. Fast folding of a prototypic polypeptide: The immunoglobulin binding domain of streptococcal protein G. Protein Sci. 1994, 3, 1945–1952. [Google Scholar] [CrossRef]
- Heidary, D.K.; Gross, L.A.; Roy, M.; Jennings, P.A. Evidence for an obligatory intermediate in the folding of interleukin-1β. Nat. Struct. Biol. 1997, 4, 725–731. [Google Scholar] [CrossRef]
- Jacobs, M.D.; Fox, R.O. Staphylococcal nuclease folding intermediate characterized by hydrogen exchange and NMR spectroscopy. Proc. Natl. Acad. Sci. USA 1994, 91, 449–453. [Google Scholar] [CrossRef]
- Walkenhorst, W.F.; Edwards, J.A.; Markley, J.L.; Roder, H. Early formation of a beta hairpin during folding of staphylococcal nuclease H124L as detected by pulsed hydrogen exchange. Protein Sci. 2002, 11, 82–91. [Google Scholar] [CrossRef]
- Jones, B.E.; Matthews, C.R. Early intermediates in the folding of dihydrofolate reductase from Escherichia coli detected by hydrogen exchange and NMR. Protein Sci. 1995, 4, 167–177. [Google Scholar] [CrossRef]
- Raschke, T.M.; Marqusee, S. The kinetic folding intermediate of ribonuclease H resembles the acid molten globule and partially unfolded molecules detected under native conditions. Nat. Struct. Biol. 1997, 4, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.J.; Dempsey, C.E.; Lorch, M.; Clarke, A.R. Acquisition of native β-strand topology during the rapid collapse phase of protein folding. Biochemistry 1997, 36, 13396–13405. [Google Scholar] [CrossRef] [PubMed]
- Hosszu, L.L.P.; Craven, C.J.; Parker, M.J.; Lorch, M.; Spencer, J.; Clarke, A.R.; Waltho, J.P. Structure of a kinetic protein folding intermediate by equilibrium amide exchange. Nat. Struct. Biol. 1997, 4, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Freund, C.; Gehrig, P.; Holak, T.A.; Plückthun, A. Comparison of the amide proton exchange behavior of the rapidly formed folding intermediate and the native state of an antibody scFv fragment. FEBS Lett. 1997, 407, 42–46. [Google Scholar] [CrossRef]
- Teilum, K.; Kragelund, B.B.; Knudsen, J.; Poulsen, F.M. Formation of hydrogen bonds precedes the rate-limiting formation of persistent structure in the folding of ACBP. J. Mol. Biol. 2000, 301, 1307–1314. [Google Scholar] [CrossRef]
- Forge, V.; Hoshino, M.; Kuwata, K.; Arai, M.; Kuwajima, K.; Batt, C.A.; Goto, Y. Is folding of β-Lactoglobulin non-hierarchic? Intermediate with native-like β-sheet and non-native α-helix. J. Mol. Biol. 2000, 296, 1039–1051. [Google Scholar] [CrossRef]
- Kuwata, K.; Shastry, R.; Cheng, H.; Hoshino, M.; Batt, C.A.; Goto, Y.; Roder, H. Structural and kinetic characterization of early folding events in β-lactoglobulin. Nat. Struct. Biol. 2001, 8, 151–155. [Google Scholar] [CrossRef]
- Schulenburg, C.; Löw, C.; Weininger, U.; Mrestani-Klaus, C.; Hofmann, H.; Balbach, J.; Ulbrich-Hofmann, R.; Arnold, U. The folding pathway of onconase is directed by a conserved intermediate. Biochemistry 2009, 48, 8449–8457. [Google Scholar] [CrossRef]
- Di Paolo, A.; Balbeur, D.; De Pauw, E.; Redfield, C.; Matagne, A. Rapid collapse into a molten globule is followed by simple two-state kinetics in the folding of lysozyme from bacteriophage λ. Biochemistry 2010, 49, 8646–8657. [Google Scholar] [CrossRef]
- Konuma, T.; Chatani, E.; Yagi, M.; Sakurai, K.; Ikegami, T.; Naiki, H.; Goto, Y. Kinetic intermediates of β2-microglobulin fibril elongation probed by pulse-labeling H/D exchange combined with NMR analysis. J. Mol. Biol. 2011, 405, 851–862. [Google Scholar] [CrossRef]
- Fazelinia, H.; Xu, M.; Cheng, H.; Roder, H. Ultrafast hydrogen exchange reveals specific structural events during the initial stages of folding of cytochrome c. J. Am. Chem. Soc. 2014, 136, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.E.; Connell, K.B.; Marqusee, S. Evidence for close side-chain packing in an early protein folding intermediate previously assumed to be a molten globule. Proc. Natl. Acad. Sci. USA 2014, 111, 14746–14751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Paterson, Y.; Roder, H. Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci. 1995, 4, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Chandak, M.S.; Nakamura, T.; Takenaka, T.; Chaudhuri, T.K.; Yagi-Utsumi, M.; Chen, J.; Kato, K.; Kuwajima, K. The use of spin desalting columns in DMSO-quenched H/D-exchange NMR experiments. Protein Sci. 2013, 22, 486–491. [Google Scholar] [CrossRef]
- Bax, A.; Grzesiek, S. Methodological advances in protein NMR. Acc. Chem. Res. 1993, 26, 131–138. [Google Scholar] [CrossRef]
- Bax, A.; Ikura, M.; Kay, L.E.; Torchia, D.A.; Tschudin, R. Comparison of different modes of two-dimensional reverse-correlation NMR for the study of proteins. J. Magn. Reson. 1990, 86, 304–318. [Google Scholar] [CrossRef]
- Nishimura, C. Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 10–27. [Google Scholar] [CrossRef]
- Sakamoto, K.; Hirai, K.; Kitamura, Y.; Yamazaki, K.; Yusa, M.; Tokunaga, N.; Doi, G.; Noda, Y.; Tachibana, H.; Segawa, S. Glycerol-induced folding of unstructured disulfide-deficient lysozyme into a native-like conformation. Biopolymers 2009, 91, 665–675. [Google Scholar] [CrossRef]
- van Rijswijk, M.H.; Donker, A.J.; Ruinen, L. Dimethylsulphoxide in amyloidosis. Lancet 1979, 313, 207–208. [Google Scholar] [CrossRef]
- Regelson, W.; Harkins, S.W. “Amyloid is not a tombstone”—A summation. The primary role for cerebrovascular and CSF dynamics as factors in Alzheimer’s disease (AD): DMSO, fluorocarbon oxygen carriers, thyroid hormonal, and other suggested therapeutic measures. Ann. N. Y. Acad. Sci. 1997, 826, 348–374. [Google Scholar] [CrossRef]
- Hirota-Nakaoka, N.; Hasegawa, K.; Naiki, H.; Goto, Y. Dissolution of β2-microglobulin amyloid fibrils by dimethylsulfoxide. J. Biochem. 2003, 134, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, A.T. An NMR-based quenched hydrogen exchange investigation of model amyloid fibrils formed by cold shock protein A. Pac. Symp. Biocomput. 2001, 2000, 67–78. [Google Scholar]
- Ippel, J.H.; Olofsson, A.; Schleucher, J.; Lundgren, E.; Wijmenga, S.S. Probing solvent accessibility of amyloid fibrils by solution NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2002, 99, 8648–8653. [Google Scholar] [CrossRef]
- Hoshino, M.; Katou, H.; Hagihara, Y.; Hasegawa, K.; Naiki, H.; Goto, Y. Mapping the core of the β2-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 2002, 9, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, K.; Matumoto, T.; Cheng, H.; Nagayama, K.; James, T.L.; Roder, H. NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106–126. Proc. Natl. Acad. Sci. USA 2003, 100, 14790–14795. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.I.; Katou, H.; Hoshino, M.; Hasegawa, K.; Naiki, H.; Goto, Y. Core and heterogeneity of β2-microglobulin amyloid fibrils as revealed by H/D exchange. J. Mol. Biol. 2004, 338, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Ritter, C.; Maddelein, M.L.; Siemer, A.B.; Lührs, T.; Ernst, M.; Meier, B.H.; Saupe, S.J.; Riek, R. Correlation of structural elements and infectivity of the HET-s prion. Nature 2005, 435, 844–848. [Google Scholar] [CrossRef]
- Carulla, N.; Caddy, G.L.; Hall, D.R.; Zurdo, J.; Gairi, M.; Feliz, M.; Giralt, E.; Robinson, C.V.; Dobson, C.M. Molecular recycling within amyloid fibrils. Nature 2005, 436, 554–558. [Google Scholar] [CrossRef]
- Whittemore, N.A.; Mishra, R.; Kheterpal, I.; Williams, A.D.; Wetzel, R.; Serpersu, E.H. Hydrogen-deuterium (H/D) exchange mapping of Aβ1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry 2005, 44, 4434–4441. [Google Scholar] [CrossRef]
- Lührs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Döbeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef]
- Wilson, L.M.; Mok, Y.F.; Binger, K.J.; Griffin, M.D.; Mertens, H.D.; Lin, F.; Wade, J.D.; Gooley, P.R.; Howlett, G.J. A structural core within apolipoprotein C-II amyloid fibrils identified using hydrogen exchange and proteolysis. J. Mol. Biol. 2007, 366, 1639–1651. [Google Scholar] [CrossRef] [PubMed]
- Toyama, B.H.; Kelly, M.J.; Gross, J.D.; Weissman, J.S. The structural basis of yeast prion strain variants. Nature 2007, 449, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Morgan, G.J.; Giannini, S.; Hounslow, A.M.; Craven, C.J.; Zerovnik, E.; Turk, V.; Waltho, J.P.; Staniforth, R.A. Exclusion of the native α-helix from the amyloid fibrils of a mixed α/β protein. J. Mol. Biol. 2008, 375, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Maji, S.K.; Sawaya, M.R.; Eisenberg, D.; Riek, R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008, 6, e195. [Google Scholar] [CrossRef]
- Vilar, M.; Chou, H.T.; Lührs, T.; Maji, S.K.; Riek-Loher, D.; Verel, R.; Manning, G.; Stahlberg, H.; Riek, R. The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 8637–8642. [Google Scholar] [CrossRef]
- Wasmer, C.; Zimmer, A.; Sabaté, R.; Soragni, A.; Saupe, S.J.; Ritter, C.; Meier, B.H. Structural similarity between the prion domain of HET-s and a homologue can explain amyloid cross-seeding in spite of limited sequence identity. J. Mol. Biol. 2010, 402, 311–325. [Google Scholar] [CrossRef]
- Kurouski, D.; Washington, J.; Ozbil, M.; Prabhakar, R.; Shekhtman, A.; Lednev, I.K. Disulfide bridges remain intact while native insulin converts into amyloid fibrils. PLoS ONE 2012, 7, e36989. [Google Scholar] [CrossRef]
- Alexandrescu, A.T. Amide proton solvent protection in amylin fibrils probed by quenched hydrogen exchange NMR. PLoS ONE 2013, 8, e56467. [Google Scholar] [CrossRef]
- Garvey, M.; Baumann, M.; Wulff, M.; Kumar, S.T.; Markx, D.; Morgado, I.; Knüpfer, U.; Horn, U.; Mawrin, C.; Fändrich, M.; et al. Molecular architecture of Aβ fibrils grown in cerebrospinal fluid solution and in a cell culture model of Aβ plaque formation. Amyloid 2016, 23, 76–85. [Google Scholar] [CrossRef]
- Rezaei-Ghaleh, N.; Kumar, S.; Walter, J.; Zweckstetter, M. Phosphorylation interferes with maturation of amyloid-β fibrillar structure in the N Terminus. J. Biol. Chem. 2016, 291, 16059–16067. [Google Scholar] [CrossRef]
- Wälti, M.A.; Orts, J.; Riek, R. Quenched hydrogen-deuterium exchange NMR of a disease-relevant Aβ(1-42) amyloid polymorph. PLoS ONE 2017, 12, e0172862. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Nishizawa, R.; Walinda, E.; Takashima, S.; Sugase, K.; Shirakawa, M. Hydrogen-deuterium exchange profiles of polyubiquitin fibrils. Polymers 2018, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Zlatic, C.O.; Mao, Y.; Todorova, N.; Mok, Y.F.; Howlett, G.J.; Yarovsky, I.; Gooley, P.R.; Griffin, M.D.W. Polymorphism in disease-related apolipoprotein C-II amyloid fibrils: A structural model for rod-like fibrils. FEBS J. 2018, 285, 2799–2812. [Google Scholar] [CrossRef] [PubMed]
- Aucoin, D.; Xia, Y.; Theint, T.; Nadaud, P.S.; Surewicz, K.; Surewicz, W.K.; Jaroniec, C.P. Protein-solvent interfaces in human Y145Stop prion protein amyloid fibrils probed by paramagnetic solid-state NMR spectroscopy. J. Struct. Biol. 2019, 206, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Kostic, M.; Liu, J.; Martinez-Yamout, M.A. Hydrogen-deuterium exchange strategy for delineation of contact sites in protein complexes. FEBS Lett. 2008, 582, 1495–1500. [Google Scholar] [CrossRef]
- Chakraborty, S.; Hosur, R.V. NMR insights into the core of GED assembly by H/D exchange coupled with DMSO dissociation and analysis of the denatured state. J. Mol. Biol. 2011, 405, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Carulla, N.; Zhou, M.; Arimon, M.; Gairí, M.; Giralt, E.; Robinson, C.V.; Dobson, C.M. Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation. Proc. Natl. Acad. Sci. USA 2009, 106, 7828–7833. [Google Scholar] [CrossRef]
- Kulminskaya, N.V.; Yoshimura, Y.; Runager, K.; Sørensen, C.S.; Bjerring, M.; Andreasen, M.; Otzen, D.E.; Enghild, J.J.; Nielsen, N.C.; Mulder, F.A. Near-complete 1H, 13C, 15N resonance assignments of dimethylsulfoxide-denatured TGFBIp FAS1-4 A546T. Biomol. NMR Assign. 2016, 10, 25–29. [Google Scholar] [CrossRef]
- Karsisiotis, A.I.; Deacon, O.M.; Macdonald, C.; Blumenschein, T.M.A.; Moore, G.R.; Worrall, J.A.R. Near-complete backbone resonance assignments of acid-denatured human cytochrome c in dimethylsulfoxide: A prelude to studying interactions with phospholipids. Biomol. NMR Assign. 2017, 11, 165–168. [Google Scholar] [CrossRef]
- Hoshino, M.; Katou, H.; Yamaguchi, K.I.; Goto, Y. Dimethylsulfoxide-quenched hydrogen/deuterium exchange method to study amyloid fibril structure. Biochim. Biophys. Acta 2007, 1768, 1886–1899. [Google Scholar] [CrossRef]
- Carulla, N.; Zhou, M.; Giralt, E.; Robinson, C.V.; Dobson, C.M. Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange. Acc. Chem. Res. 2010, 43, 1072–1079. [Google Scholar] [CrossRef]
- Vilar, M.; Wang, L.; Riek, R. Structural studies of amyloids by quenched hydrogen-deuterium exchange by NMR. Methods Mol. Biol. 2012, 849, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Goto, Y. Kinetic intermediates of amyloid fibrillation studied by hydrogen exchange methods with nuclear magnetic resonance. Biochim. Biophys. Acta 2012, 1824, 1307–1323. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, A.T. Quenched hydrogen exchange NMR of amyloid fibrils. Methods Mol. Biol. 2016, 1345, 211–222. [Google Scholar] [PubMed]
- Chandak, M.S.; Nakamura, T.; Makabe, K.; Takenaka, T.; Mukaiyama, A.; Chaudhuri, T.K.; Kato, K.; Kuwajima, K. The H/D-exchange kinetics of the Escherichia coli co-chaperonin GroES studied by 2D NMR and DMSO-quenched exchange methods. J. Mol. Biol. 2013, 425, 2541–2560. [Google Scholar] [CrossRef]
- Yagi-Utsumi, M.; Chandak, M.S.; Yanaka, S.; Hiranyakorn, M.; Nakamura, T.; Kato, K.; Kuwajima, K. Residual structure of unfolded ubiquitin as revealed by hydrogen/deuterium-exchange 2D NMR. Biophys. J. 2020, 119, 2029–2038. [Google Scholar] [CrossRef]
- Levinthal, C. Are there pathways for protein folding? J. Chim. Phys. 1968, 65, 44–45. [Google Scholar] [CrossRef]
- Levinthal, C. How to fold graciously. Mössbauer Spectrosc. Biol. Syst. 1969, 67, 22–24. [Google Scholar]
- Wetlaufer, D.B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 697–701. [Google Scholar] [CrossRef]
- Zwanzig, R.; Szabo, A.; Bagchi, B. Levinthal’s paradox. Proc. Natl. Acad. Sci. USA 1992, 89, 20–22. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 1995, 21, 167–195. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Chan, H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997, 4, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Sali, A.; Karplus, M. Protein folding: A perspective from theory and experiment. Angew. Chem. Int. Ed. Engl. 1998, 37, 868–893. [Google Scholar] [CrossRef]
- Englander, S.W.; Mayne, L. The nature of protein folding pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 15873–15880. [Google Scholar] [CrossRef] [PubMed]
- Ivankov, D.N.; Finkelstein, A.V. Solution of Levinthal’s paradox and a physical theory of protein folding times. Biomolecules 2020, 10, 250. [Google Scholar] [CrossRef]
- Rose, G.D. Protein folding—Seeing is deceiving. Protein Sci. 2021, 30, 1606–1616. [Google Scholar] [CrossRef]
- Neri, D.; Billeter, M.; Wider, G.; Wüthrich, K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science 1992, 257, 1559–1563. [Google Scholar] [CrossRef]
- Shortle, D.; Ackerman, M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science 2001, 293, 487–489. [Google Scholar] [CrossRef]
- Klein-Seetharaman, J.; Oikawa, M.; Grimshaw, S.B.; Wirmer, J.; Duchardt, E.; Ueda, T.; Imoto, T.; Smith, L.J.; Dobson, C.M.; Schwalbe, H. Long-range interactions within a nonnative protein. Science 2002, 295, 1719–1722. [Google Scholar] [CrossRef]
- Wirmer, J.; Berk, H.; Ugolini, R.; Redfield, C.; Schwalbe, H. Characterization of the unfolded state of bovine α-lactalbumin and comparison with unfolded states of homologous proteins. Protein Sci. 2006, 15, 1397–1407. [Google Scholar] [CrossRef]
- Danielson, T.A.; Stine, J.M.; Dar, T.A.; Briknarova, K.; Bowler, B.E. Effect of an imposed contact on secondary structure in the denatured state of yeast Iso-1-cytochrome c. Biochemistry 2017, 56, 6662–6676. [Google Scholar] [CrossRef] [PubMed]
- Leavens, M.J.; Spang, L.E.; Cherney, M.M.; Bowler, B.E. Denatured state conformational biases in three-helix bundles containing divergent sequences localize near turns and helix capping residues. Biochemistry 2021, 60, 3071–3085. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Udgaonkar, J.B. Heterogeneity in protein folding and unfolding reactions. Chem. Rev. 2022, 122, 8911–8935. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 1987, 194, 531–544. [Google Scholar] [CrossRef]
- Khorasanizadeh, S.; Peters, I.D.; Butt, T.R.; Roder, H. Folding and stability of a tryptophan-containing mutant of ubiquitin. Biochemistry 1993, 32, 7054–7063. [Google Scholar] [CrossRef] [PubMed]
- Khorasanizadeh, S.; Peters, I.D.; Roder, H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat. Struct. Biol. 1996, 3, 193–205. [Google Scholar] [CrossRef]
- Went, H.M.; Benitez-Cardoza, C.G.; Jackson, S.E. Is an intermediate state populated on the folding pathway of ubiquitin? FEBS Lett. 2004, 567, 333–338. [Google Scholar] [CrossRef]
- Larios, E.; Li, J.S.; Schulten, K.; Kihara, H.; Gruebele, M. Multiple probes reveal a native-like intermediate during low-temperature refolding of ubiquitin. J. Mol. Biol. 2004, 340, 115–125. [Google Scholar] [CrossRef]
- Roder, H.; Maki, K.; Cheng, H. Early events in protein folding explored by rapid mixing methods. Chem. Rev. 2006, 106, 1836–1861. [Google Scholar] [CrossRef]
- Gladwin, S.T.; Evans, P.A. Structure of very early protein folding intermediates: New insights through a variant of hydrogen exchange labelling. Fold. Des. 1996, 1, 407–417. [Google Scholar] [CrossRef]
- Pan, J.; Wilson, D.J.; Konermann, L. Pulsed hydrogen exchange and electrospray charge-state distribution as complementary probes of protein structure in kinetic experiments: Implications for ubiquitin folding. Biochemistry 2005, 44, 8627–8633. [Google Scholar] [CrossRef] [PubMed]
- Sosnick, T.R.; Dothager, R.S.; Krantz, B.A. Differences in the folding transition state of ubiquitin indicated by ϕ and ψ analyses. Proc. Natl. Acad. Sci. USA 2004, 101, 17377–17382. [Google Scholar] [CrossRef]
- Went, H.M.; Jackson, S.E. Ubiquitin folds through a highly polarized transition state. Protein Eng. Des. Sel. 2005, 18, 229–237. [Google Scholar] [CrossRef]
- Kitahara, R.; Akasaka, K. Close identity of a pressure-stabilized intermediate with a kinetic intermediate in protein folding. Proc. Natl. Acad. Sci. USA 2003, 100, 3167–3172. [Google Scholar] [CrossRef]
- Jackson, S.E. Ubiquitin: A small protein folding paradigm. Org. Biomol. Chem. 2006, 4, 1845–1853. [Google Scholar] [CrossRef]
- Wirtz, H.; Schäfer, S.; Hoberg, C.; Reid, K.M.; Leitner, D.M.; Havenith, M. Hydrophobic collapse of ubiquitin generates rapid protein-water motions. Biochemistry 2018, 57, 3650–3657. [Google Scholar] [CrossRef]
- Ibarra-Molero, B.; Loladze, V.V.; Makhatadze, G.I.; Sanchez-Ruiz, J.M. Thermal versus guanidine-induced unfolding of ubiquitin. An analysis in terms of the contributions from charge-charge interactions to protein stability. Biochemistry 1999, 38, 8138–8149. [Google Scholar] [CrossRef]
- Peti, W.; Hennig, M.; Smith, L.J.; Schwalbe, H. NMR spectroscopic investigation of ψ torsion angle distribution in unfolded ubiquitin from analysis of 3J(Cα, Cα) coupling constants and cross-correlated ΓCHNN,CαHα relaxation rates. J. Am. Chem. Soc. 2000, 122, 12017–12018. [Google Scholar] [CrossRef]
- Peti, W.; Smith, L.J.; Redfield, C.; Schwalbe, H. Chemical shifts in denatured proteins: Resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J. Biomol. NMR 2001, 19, 153–165. [Google Scholar] [CrossRef]
- Wirmer, J.; Peti, W.; Schwalbe, H. Motional properties of unfolded ubiquitin: A model for a random coil protein. J. Biomol. NMR 2006, 35, 175–186. [Google Scholar] [CrossRef]
- Harper, E.T.; Rose, G.D. Helix stop signals in proteins and peptides: The capping box. Biochemistry 1993, 32, 7605–7609. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Strohmeier, M.; Blackledge, M.; Grzesiek, S. Direct observation of dipolar couplings and hydrogen bonds across a β-hairpin in 8 M urea. J. Am. Chem. Soc. 2007, 129, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.R.; Grzesiek, S. Ensemble calculations of unstructured proteins constrained by RDC and PRE data: A case study of urea-denatured ubiquitin. J. Am. Chem. Soc. 2010, 132, 694–705. [Google Scholar] [CrossRef]
- Candotti, M.; Esteban-Martin, S.; Salvatella, X.; Orozco, M. Toward an atomistic description of the urea-denatured state of proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 5933–5938. [Google Scholar] [CrossRef]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.R.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA 2016, 113, E5389–E5398. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).