Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1
Abstract
1. Introduction
2. Results and Discussion
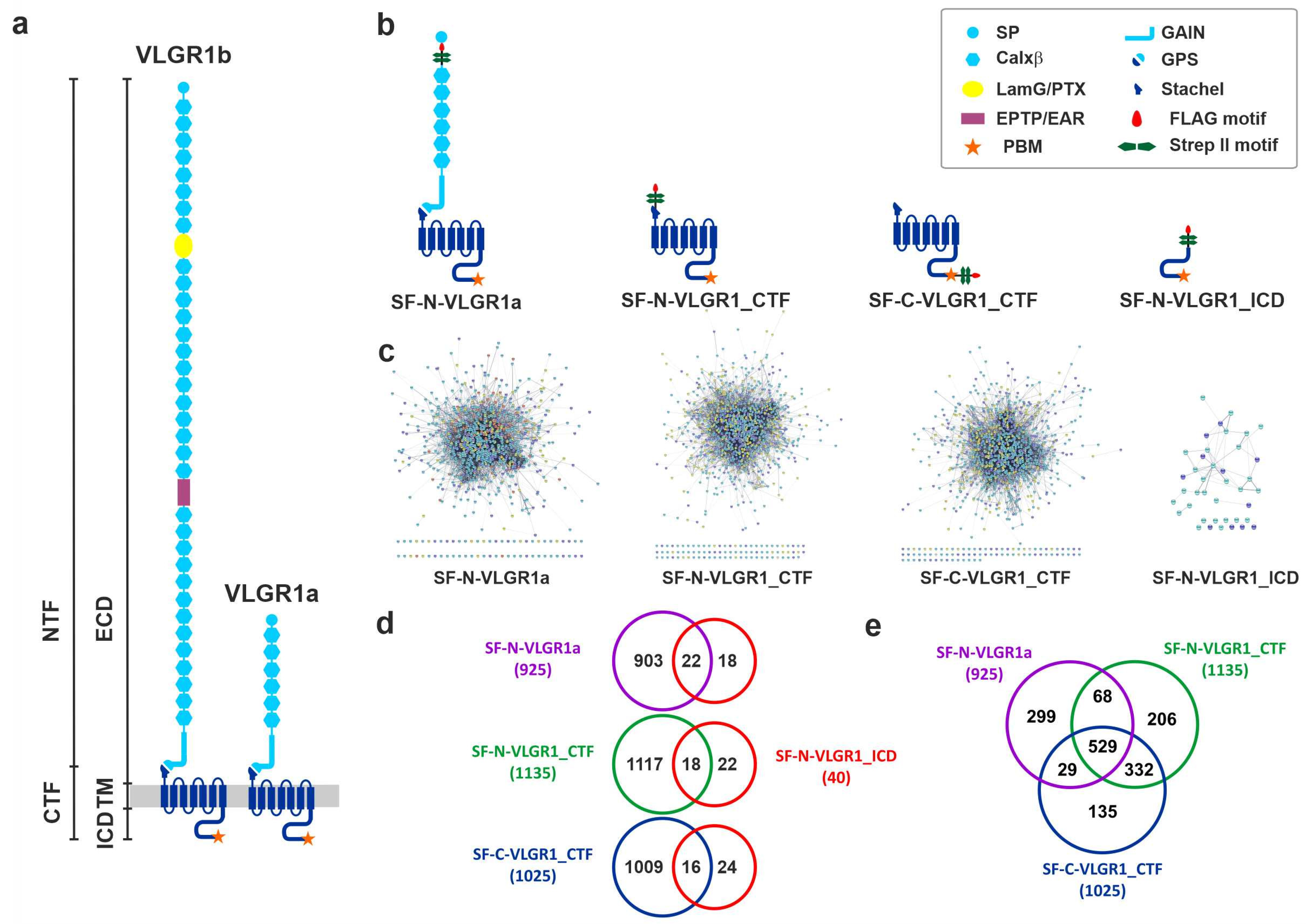
2.1. Identification of Novel VLGR1 Protein Complex Partners by Tandem Affinity Purification (TAP)
2.1.1. Specificity of TAP Hits
2.1.2. Grouping and Categorizing of TAP Hits
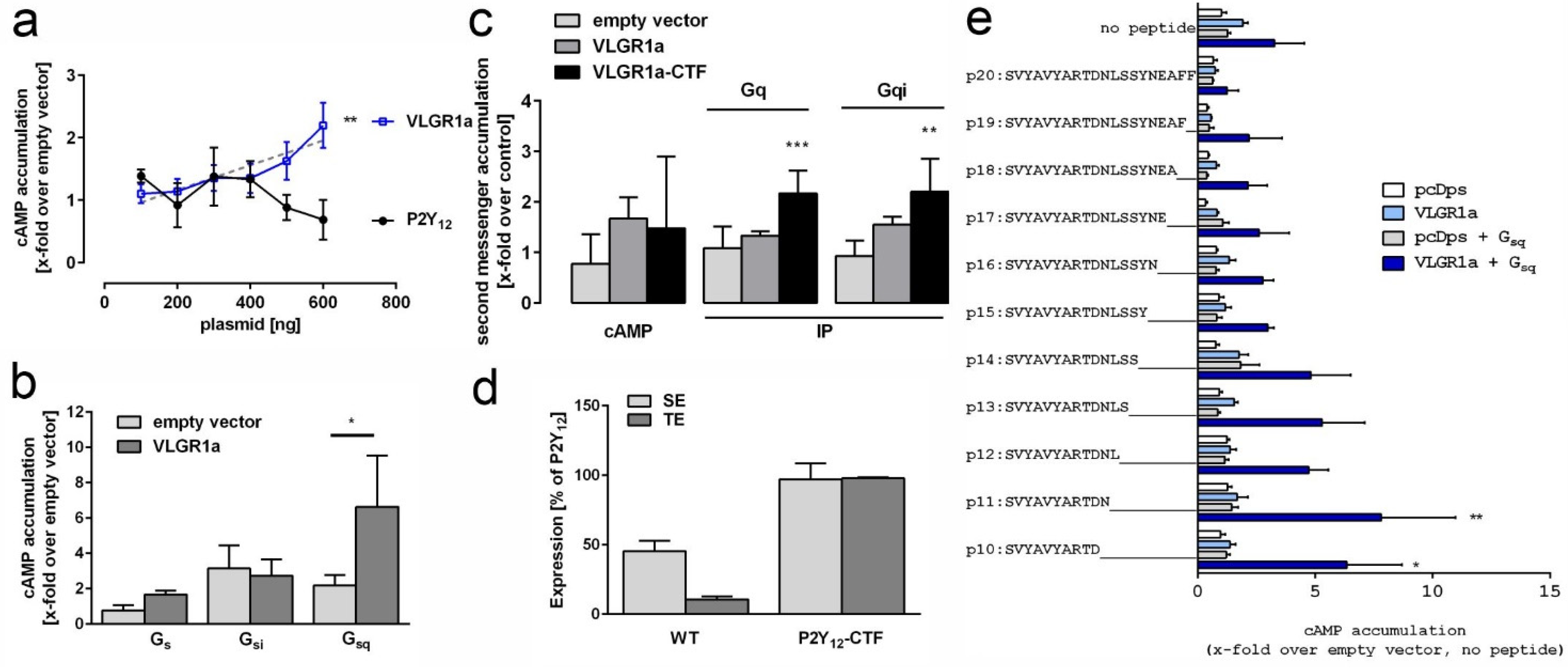
2.2. VLGR1 G Protein Coupling Switches between Gαs/Gαq and Gαi
2.3. TAP Data Analysis Indicates Coupling of VLGR1 to Various Downstream Signaling Pathways
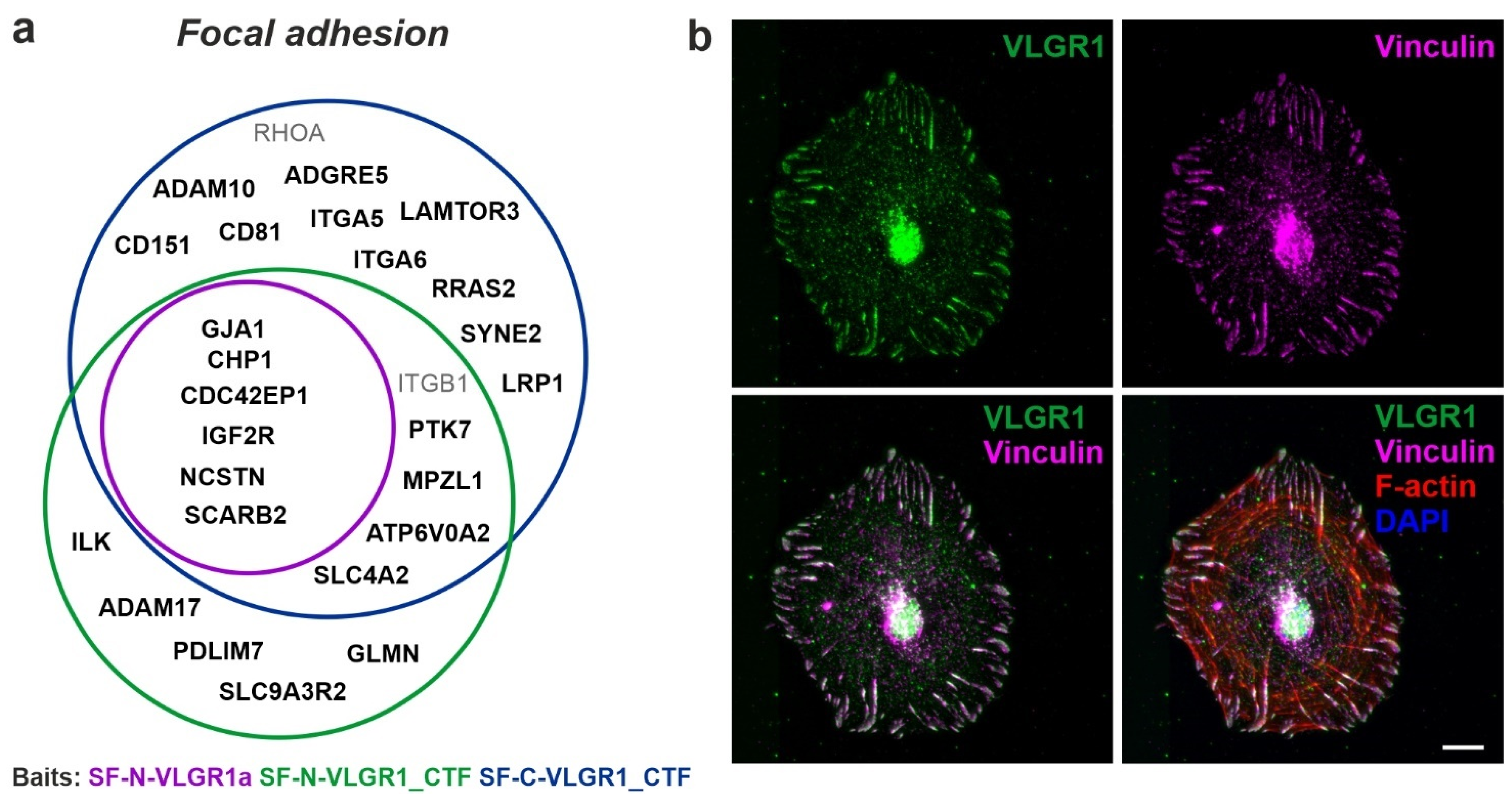
2.4. VLGR1 Participates in Signaling at Focal Adhesions
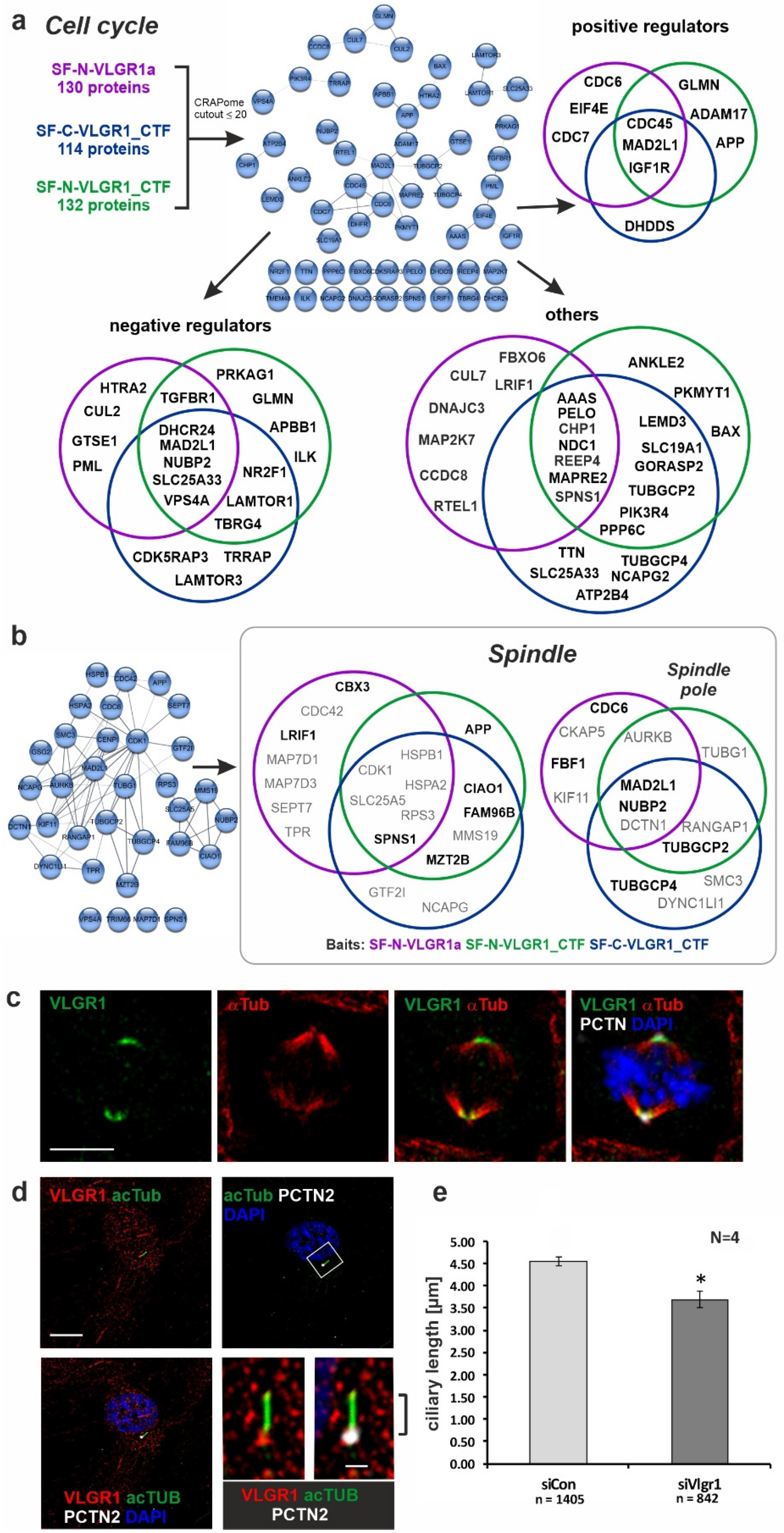
2.5. VLGR1 Partners Participate in Cell Cycle Regulation, Cell Division, and Ciliogenesis
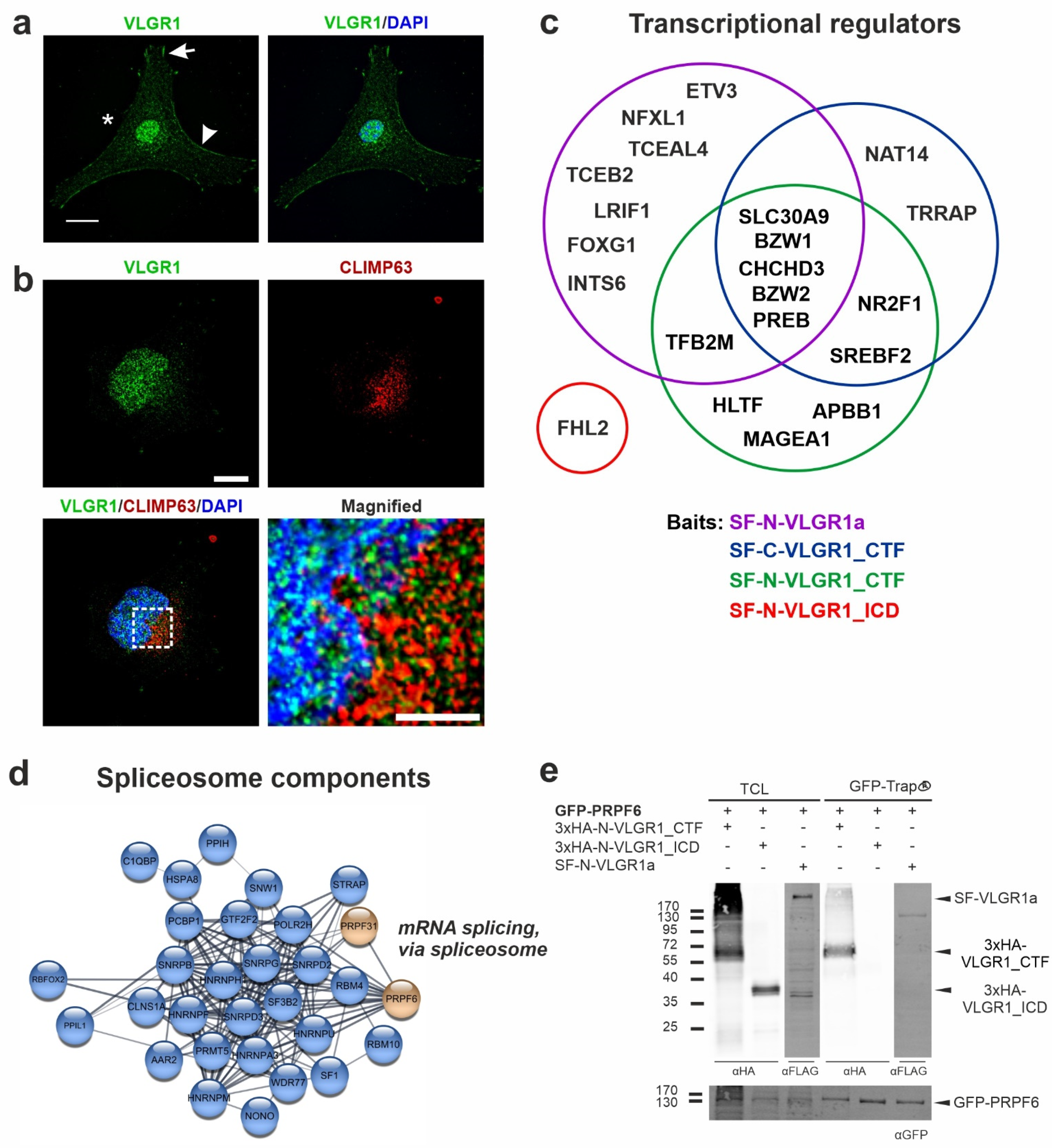
2.6. VLGR1 Is Part of Protein Networks in the ER and Nucleus
2.7. VLGR1 Is Associated with the γ-Secretase Complex
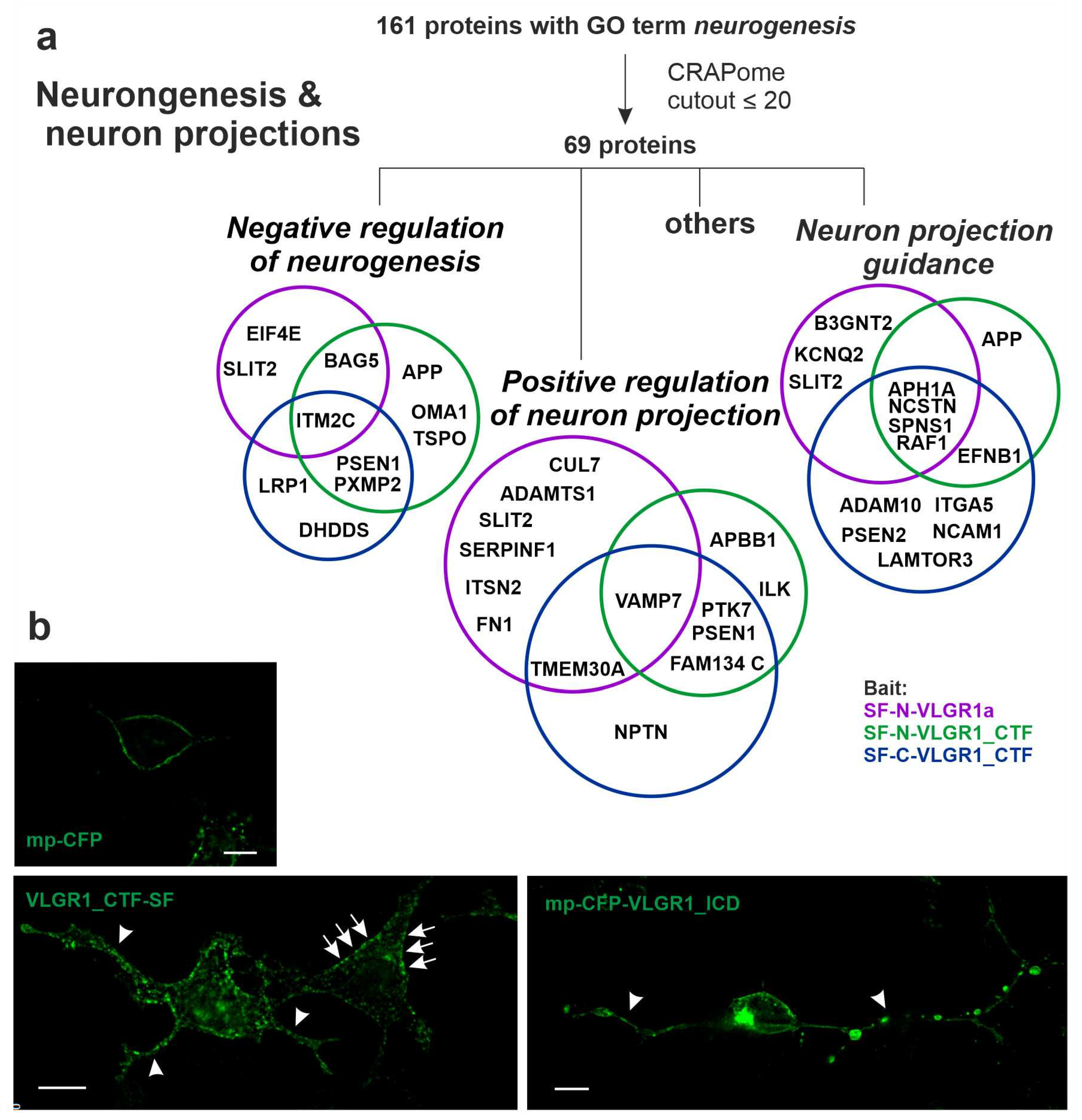
2.8. VLGR1 Interacts with Proteins Involved in Neurogenesis
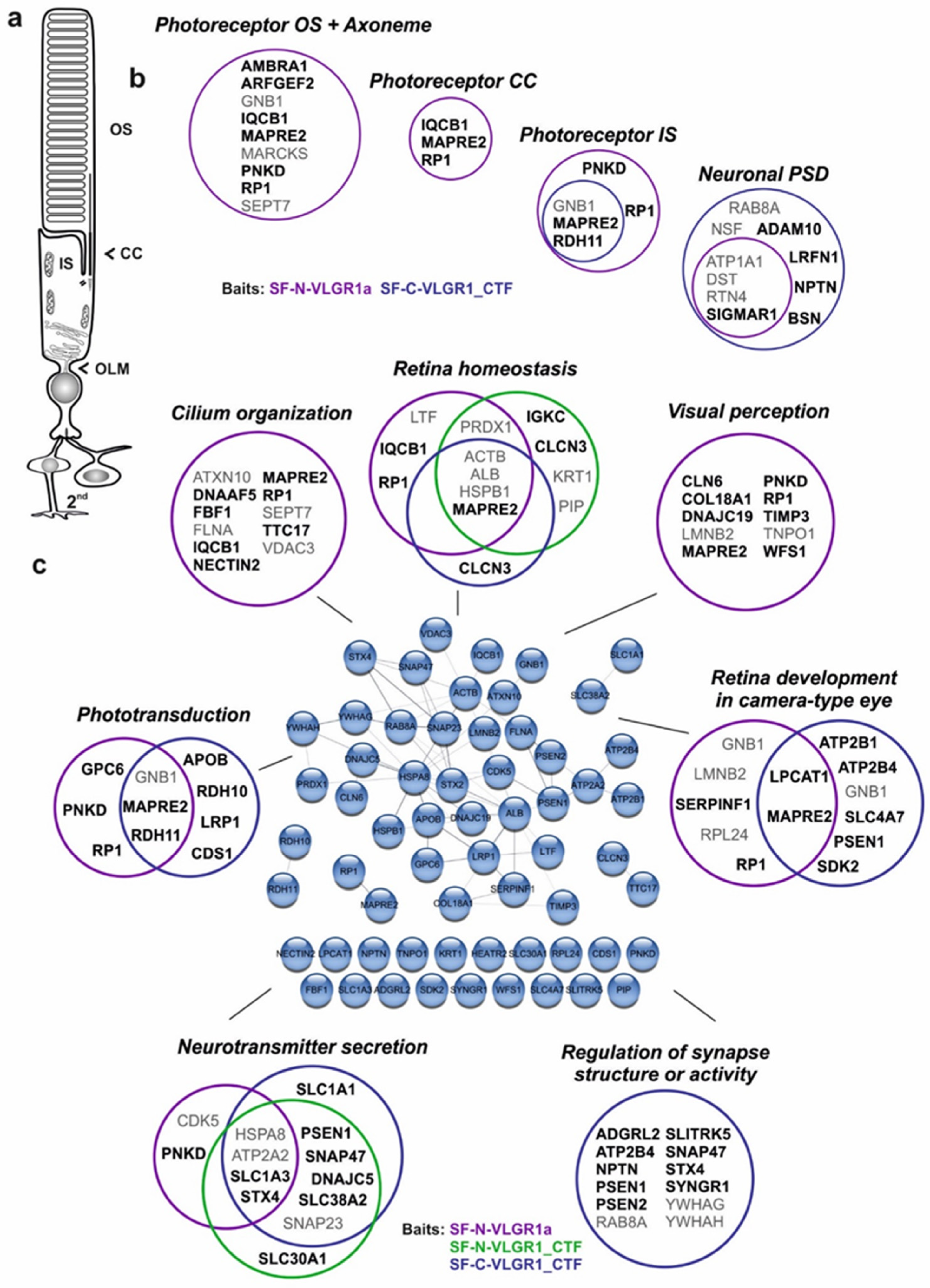
2.9. VLGR1 TAPs Confirm and Specify Roles in the Function of the Mature Retina and Photoreceptor Cells
2.10. TAP Data Support a Role of VLGR1 Associated with Epilepsy
2.11. VLGR1 Protein Networks Are Linked to Neuron Death and Degenerative Diseases of the Central Nervous System
2.12. VLGR1 Is Found in Alzheimer’s Disease-Related Protein Complexes
3. Materials and Methods
3.1. Plasmids
3.2. Cell Culture
3.3. Tandem Affinity Purification (TAP)
3.4. Mass Spectrometry
3.5. Data Processing
3.6. Antibodies
3.7. Immunocytochemistry
3.8. siRNA Knock Down
3.9. GFP-Trap®
3.10. Immunoprecipitation
3.11. Second Messenger Assays
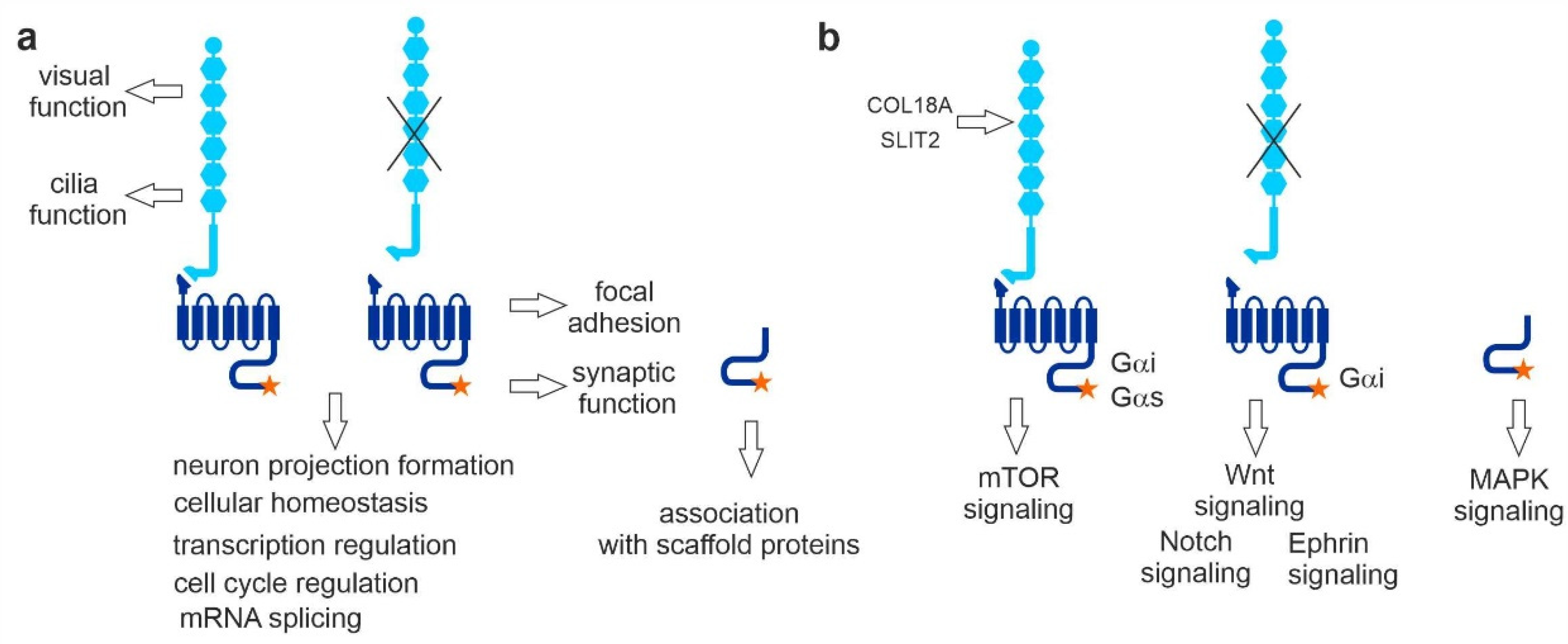
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McMillan, D.R.; Kayes-Wandover, K.M.; Richardson, J.A.; White, P.C. Very Large G Protein-coupled Receptor-1, the Largest Known Cell Surface Protein, Is Highly Expressed in the Developing Central Nervous System. J. Biol. Chem. 2002, 277, 785–792. [Google Scholar] [CrossRef] [PubMed]
- McMillan, D.R.; White, P.C. Studies on the Very Large G Protein-Coupled Receptor: From Initial Discovery to Determining its Role in Sensorineural Deafness in Higher Animals. Adhes.-Gpcrs Struct. Funct. 2010, 706, 76–86. [Google Scholar] [CrossRef]
- Hamann, J.; Aust, G.; Araç, D.; Engel, F.; Formstone, C.; Fredriksson, R.; Hall, R.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G Protein–Coupled Receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef] [PubMed]
- Stemerdink, M.; García-Bohórquez, B.; Schellens, R.; Garcia-Garcia, G.; Van Wijk, E.; Millan, J.M. Genetics, pathogenesis and therapeutic developments for Usher syndrome type 2. Hum. Genet. 2021, 141, 737–758. [Google Scholar] [CrossRef]
- Reiners, J.; Nagel-Wolfrum, K.; Jürgens, K.; Märker, T.; Wolfrum, U. Molecular basis of human Usher syndrome: Deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 2006, 83, 97–119. [Google Scholar] [CrossRef]
- Wolfrum, U. Protein networks related to the Usher syndrome gain insights in the molecular basis of the disease. In Usher Syndrome: Pathogenesis, Diagnosis and Therapy; Satpal, A., Ed.; Nova Science Publishers: New York, NY, USA, 2011; pp. 51–73. [Google Scholar]
- Nakayama, J.; Fu, Y.-H.; Clark, A.M.; Nakahara, S.; Hamano, K.; Iwasaki, N.; Matsui, A.; Arinami, T.; Ptacek, L.J. A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann. Neurol. 2002, 52, 654–657. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, X.; Zhang, W.; Zhang, C.; Wang, J.; Jiang, T.; Wang, L. Deficiency of very large G-protein-coupled receptor-1 is a risk factor of tumor-related epilepsy: A whole transcriptome sequencing analysis. J. Neuro-Oncol. 2015, 121, 609–616. [Google Scholar] [CrossRef]
- Zhou, X.-W.; Wang, X.; Yang, Y.; Luo, J.-W.; Dong, H.; Liu, Y.-H.; Mao, Q. Biomarkers related with seizure risk in glioma patients: A systematic review. Clin. Neurol. Neurosurg. 2016, 151, 113–119. [Google Scholar] [CrossRef]
- Myers, K.A.; Nasioulas, S.; Boys, A.; McMahon, J.M.; Slater, H.; Lockhart, P.; du Sart, D.; Scheffer, I.E. ADGRV1 is implicated in myoclonic epilepsy. Epilepsia 2018, 59, 381–388. [Google Scholar] [CrossRef]
- Skradski, S.L.; Clark, A.M.; Jiang, H.; White, H.S.; Fu, Y.-H.; Ptáček, L.J. A Novel Gene Causing a Mendelian Audiogenic Mouse Epilepsy. Neuron 2001, 31, 537–544. [Google Scholar] [CrossRef]
- McMillan, D.R.; White, P.C. Loss of the transmembrane and cytoplasmic domains of the very large G-protein-coupled receptor-1 (VLGR1 or Mass1) causes audiogenic seizures in mice. Mol. Cell. Neurosci. 2004, 26, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Takamura, Y.; Yoneda, T.; Konno, D.; Akagi, Y.; Yoshida, K.; Sato, M. Vlgr1 knockout mice show audiogenic seizure susceptibility. J. Neurochem. 2005, 92, 191–202. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.; Goodyear, R.J.; McMillan, D.R.; Stauffer, E.A.; Holt, J.R.; Locke, K.G.; Birch, D.G.; Legan, P.K.; White, P.C.; Walsh, E.J.; et al. The Very Large G-Protein-Coupled Receptor VLGR1: A Component of the Ankle Link Complex Required for the Normal Development of Auditory Hair Bundles. J. Neurosci. 2006, 26, 6543–6553. [Google Scholar] [CrossRef] [PubMed]
- van Wijk, E.; van der Zwaag, B.; Peters, T.; Zimmermann, U.; Brinke, H.T.; Kersten, F.F.; Märker, T.; Aller, E.; Hoefsloot, L.H.; Cremers, C.W.; et al. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum. Mol. Genet. 2006, 15, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Michalski, N.; Michel, V.; Bahloul, A.; Lefèvre, G.; Barral, J.; Yagi, H.; Chardenoux, S.; Weil, D.; Martin, P.; Hardelin, J.-P.; et al. Molecular Characterization of the Ankle-Link Complex in Cochlear Hair Cells and Its Role in the Hair Bundle Functioning. J. Neurosci. 2007, 27, 6478–6488. [Google Scholar] [CrossRef]
- Reiners, J.; Van Wijk, E.; Märker, T.; Zimmermann, U.; Jürgens, K.; Brinke, H.T.; Overlack, N.; Roepman, R.; Knipper, M.; Kremer, H.; et al. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum. Mol. Genet. 2005, 14, 3933–3943. [Google Scholar] [CrossRef]
- Specht, D.; Wu, S.-B.; Turner, P.; Dearden, P.; Koentgen, F.; Wolfrum, U.; Maw, M.; Brandstätter, J.H.; Dieck, S.T. Effects of Presynaptic Mutations on a Postsynaptic Cacna1s Calcium Channel Colocalized with mGluR6 at Mouse Photoreceptor Ribbon Synapses. Investig. Opthalmology Vis. Sci. 2009, 50, 505–515. [Google Scholar] [CrossRef]
- Yagi, H.; Tokano, H.; Maeda, M.; Takabayashi, T.; Nagano, T.; Kiyama, H.; Fujieda, S.; Kitamura, K.; Sato, M. Vlgr1 is required for proper stereocilia maturation of cochlear hair cells. Genes Cells 2007, 12, 235–250. [Google Scholar] [CrossRef]
- Maerker, T.; Van Wijk, E.; Overlack, N.; Kersten, F.F.; McGee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008, 17, 71–86. [Google Scholar] [CrossRef]
- Lin, H.-H.; Chang, G.-W.; Davies, J.Q.; Stacey, M.; Harris, J.; Gordon, S. Autocatalytic Cleavage of the EMR2 Receptor Occurs at a Conserved G Protein-coupled Receptor Proteolytic Site Motif. J. Biol. Chem. 2004, 279, 31823–31832. [Google Scholar] [CrossRef]
- Araç, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Südhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef] [PubMed]
- Prömel, S.; Frickenhaus, M.; Hughes, S.; Mestek, L.; Staunton, D.; Woollard, A.; Vakonakis, I.; Schöneberg, T.; Schnabel, R.; Russ, A.P.; et al. The GPS Motif Is a Molecular Switch for Bimodal Activities of Adhesion Class G Protein-Coupled Receptors. Cell Rep. 2012, 2, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Patra, C.; van Amerongen, M.J.; Ghosh, S.; Ricciardi, F.; Sajjad, A.; Novoyatleva, T.; Mogha, A.; Monk, K.R.; Mühlfeld, C.; Engel, F.B. Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent. Proc. Natl. Acad. Sci. USA 2013, 110, 16898–16903. [Google Scholar] [CrossRef] [PubMed]
- Okajima, D.; Kudo, G.; Yokota, H. Brain-specific angiogenesis inhibitor 2 (BAI2) may be activated by proteolytic processing. J. Recept. Signal Transduct. 2010, 30, 143–153. [Google Scholar] [CrossRef]
- Paavola, K.J.; Stephenson, J.; Ritter, S.L.; Alter, S.P.; Hall, R.A. The N Terminus of the Adhesion G Protein-coupled Receptor GPR56 Controls Receptor Signaling Activity. J. Biol. Chem. 2011, 286, 28914–28921. [Google Scholar] [CrossRef]
- Ward, Y.; Lake, R.; Yin, J.J.; Heger, C.D.; Raffeld, M.; Goldsmith, P.K.; Merino, M.; Kelly, K. LPA Receptor Heterodimerizes with CD97 to Amplify LPA-Initiated RHO-Dependent Signaling and Invasion in Prostate Cancer Cells. Cancer Res. 2011, 71, 7301–7311. [Google Scholar] [CrossRef]
- Stephenson, J.; Paavola, K.J.; Schaefer, S.A.; Kaur, B.; Van Meir, E.G.; Hall, R.A. Brain-specific Angiogenesis Inhibitor-1 Signaling, Regulation, and Enrichment in the Postsynaptic Density. J. Biol. Chem. 2013, 288, 22248–22256. [Google Scholar] [CrossRef]
- Liebscher, I.; Schöneberg, T. Tethered Agonism: A Common Activation Mechanism of Adhesion GPCRs. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2016; Volume 234, pp. 111–125. [Google Scholar] [CrossRef]
- Purcell, R.H.; Hall, R.A. Adhesion G Protein–Coupled Receptors as Drug Targets. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 429–449. [Google Scholar] [CrossRef]
- Braun, D.A.; Hildebrandt, F. Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028191. [Google Scholar] [CrossRef]
- Bujakowska, K.; Liu, Q.; Pierce, E.A. Photoreceptor Cilia and Retinal Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028274. [Google Scholar] [CrossRef]
- May-Simera, H.; Nagel-Wolfrum, K.; Wolfrum, U. Cilia—The sensory antennae in the eye. Prog. Retin. Eye Res. 2017, 60, 144–180. [Google Scholar] [CrossRef] [PubMed]
- Satir, P. CILIA: Before and after. Cilia 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Wolfrum, U. Adhesion GPCR-Related Protein Networks. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2016; Volume 234, pp. 147–178. [Google Scholar] [CrossRef]
- Shin, D.; Lin, S.-T.; Fu, Y.-H.; Ptáček, L.J. Very large G protein-coupled receptor 1 regulates myelin-associated glycoprotein via Gαs/Gαq-mediated protein kinases A/C. Proc. Natl. Acad. Sci. USA 2013, 110, 19101–19106. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.-X.; Dong, J.-H.; Du, H.-B.; Zhang, D.-L.; Ren, H.-Z.; Ma, M.-L.; Cai, Y.; Zhao, T.-C.; Yin, X.-L.; Yu, X.; et al. Constitutive Gαi Coupling Activity of Very Large G Protein-coupled Receptor 1 (VLGR1) and Its Regulation by PDZD7 Protein. J. Biol. Chem. 2014, 289, 24215–24225. [Google Scholar] [CrossRef]
- Boldt, K.; Van Reeuwijk, J.; Lu, Q.; Koutroumpas, K.; Nguyen, T.-M.T.; Texier, Y.; Van Beersum, S.E.C.; Horn, N.; Willer, J.R.; Mans, D.A.; et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 2016, 7, 11491. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.-P.; St-Denis, N.A.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A contaminant repository for affinity purification—Mass spectrometry data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Hsiao, C.-C.; Chu, T.-Y.; Wu, C.-J.; van den Biggelaar, M.; Pabst, C.; Hébert, J.; Kuijpers, T.W.; Scicluna, B.P.; I, K.-Y.; Chen, T.-C.; et al. The Adhesion G Protein-Coupled Receptor GPR97/ADGRG3 Is Expressed in Human Granulocytes and Triggers Antimicrobial Effector Functions. Front. Immunol. 2018, 9, 2830. [Google Scholar] [CrossRef]
- Appert-Collin, A.; Baisamy, L.; Diviani, D. Regulation of G Protein-Coupled Receptor Signaling by A-Kinase Anchoring Proteins. J. Recept. Signal Transduct. 2006, 26, 631–646. [Google Scholar] [CrossRef]
- Kennedy, B.; Lamming, D.W. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016, 23, 990–1003. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-Inducible Factor 1 (HIF-1) Pathway. Sci. STKE 2007, 2007, cm8. [Google Scholar] [CrossRef] [PubMed]
- Liebscher, I.; Schön, J.; Petersen, S.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Cöster, M.; Simon, K.-U.; Rothemund, S.; et al. A Tethered Agonist within the Ectodomain Activates the Adhesion G Protein-Coupled Receptors GPR126 and GPR133. Cell Rep. 2014, 9, 2018–2026. [Google Scholar] [CrossRef] [PubMed]
- Pisoni, G.B.; Molinari, M. Five Questions (with their Answers) on ER-Associated Degradation. Traffic 2016, 17, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Kusuluri, D.K.; Güler, B.E.; Knapp, B.; Horn, N.; Boldt, K.; Ueffing, M.; Aust, G.; Wolfrum, U. Adhesion G protein-coupled receptor VLGR1/ADGRV1 regulates cell spreading and migration by mechanosensing at focal adhesions. iScience 2021, 24, 102283. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Guo, S.S.; Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016, 215, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Owens, J.C.; Detweiler, C.S.; Li, J.J. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc. Natl. Acad. Sci. USA 1997, 94, 12521–12526. [Google Scholar] [CrossRef]
- Kumagai, H.; Sato, N.; Yamada, M.; Mahony, D.; Seghezzi, W.; Lees, E.; Arai, K.; Masai, H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol. 1999, 19, 5083–5095. [Google Scholar] [CrossRef]
- Lunn, C.L.; Chrivia, J.C.; Baldassare, J.J. Activation of Cdk2/Cyclin E complexes is dependent on the origin of replication licensing factor Cdc6 in mammalian cells. Cell Cycle 2010, 9, 4533–4541. [Google Scholar] [CrossRef][Green Version]
- Adams, M.; Simms, R.J.; Abdelhamed, Z.; Dawe, H.R.; Szymanska, K.; Logan, C.; Wheway, G.; Pitt, E.; Gull, K.; Knowles, M.A.; et al. A meckelin–filamin A interaction mediates ciliogenesis. Hum. Mol. Genet. 2012, 21, 1272–1286. [Google Scholar] [CrossRef]
- Wei, Q.; Xu, Q.; Zhang, Y.; Li, Y.; Zhang, Q.; Hu, Z.; Harris, P.C.; Torres, V.E.; Ling, K.; Hu, J. Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat. Commun. 2013, 4, 2750. [Google Scholar] [CrossRef] [PubMed]
- Kypri, E.; Christodoulou, A.; Maimaris, G.; Lethan, M.; Markaki, M.; Lysandrou, C.; Lederer, C.W.; Tavernarakis, N.; Geimer, S.; Pedersen, L.B.; et al. The nucleotide-binding proteins Nubp1 and Nubp2 are negative regulators of ciliogenesis. Cell. Mol. Life Sci. 2014, 71, 517–538. [Google Scholar] [CrossRef] [PubMed]
- Ronquillo, C.C.; Hanke-Gogokhia, C.; Revelo, M.P.; Frederick, J.M.; Jiang, L.; Baehr, W. Ciliopathy-associated IQCB1/NPHP5 protein is required for mouse photoreceptor outer segment formation. FASEB J. 2016, 30, 3400–3412. [Google Scholar] [CrossRef] [PubMed]
- Tanos, B.E.; Yang, H.-J.; Soni, R.; Wang, W.-J.; Macaluso, F.P.; Asara, J.M.; Tsou, M.-F.B. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013, 27, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zuo, J.; Pierce, E. The Retinitis Pigmentosa 1 Protein Is a Photoreceptor Microtubule-Associated Protein. J. Neurosci. 2004, 24, 6427–6436. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, P.; Simpson, M.; Mendola, A.; Vasudevan, P.; Connell, F.C.; van Impel, A.; Moore, A.T.; Loeys, B.; Ghalamkarpour, A.; Onoufriadis, A.; et al. Mutations in KIF11 Cause Autosomal-Dominant Microcephaly Variably Associated with Congenital Lymphedema and Chorioretinopathy. Am. J. Hum. Genet. 2012, 90, 356–362. [Google Scholar] [CrossRef]
- Blangy, A.; Lane, H.A.; D’Hérin, P.; Harper, M.; Kress, M.; Nigg, E. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Et Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef]
- Knapp, B.; Rödig, J.; Boldt, K.; Krzysko, J.; Horn, N.; Ueffing, M.; Wolfrum, U. Affinity proteomics identifies novel functional modules related to adhesion GPCRs. Ann. N. Y. Acad. Sci. 2019, 1456, 144–167. [Google Scholar] [CrossRef]
- Li, H.J.; Haque, Z.K.; Chen, A.; Mendelsohn, M. RIF-1, a novel nuclear receptor corepressor that associates with the nuclear matrix. J. Cell. Biochem. 2007, 102, 1021–1035. [Google Scholar] [CrossRef]
- Xuan, S.; Baptista, C.A.; Balas, G.; Tao, W.; Soares, V.C.; Lai, E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 1995, 14, 1141–1152. [Google Scholar] [CrossRef]
- Pauley, S.; Lai, E.; Fritzsch, B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 2006, 235, 2470–2482. [Google Scholar] [CrossRef]
- Inoue, M.; Iida, A.; Satoh, S.; Kodama, T.; Watanabe, S. COUP-TFI and -TFII nuclear receptors are expressed in amacrine cells and play roles in regulating the differentiation of retinal progenitor cells. Exp. Eye Res. 2010, 90, 49–56. [Google Scholar] [CrossRef]
- Lau, K.-F.; Chan, W.-M.; Perkinton, M.S.; Tudor, E.L.; Chang, R.C.C.; Chan, H.-Y.E.; McLoughlin, D.M.; Miller, C.C.J. Dexras1 Interacts with FE65 to Regulate FE65-Amyloid Precursor Protein-dependent Transcription. J. Biol. Chem. 2008, 283, 34728–34737. [Google Scholar] [CrossRef]
- Ba, A.N.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef]
- Papasaikas, P.; Valcárcel, J. The Spliceosome: The Ultimate RNA Chaperone and Sculptor. Trends Biochem. Sci. 2016, 41, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.G.; Boutz, P.L.; Dougherty, J.D.; Stoilov, P.; Black, D.L. Homologues of the Caenorhabditis elegans Fox-1 Protein Are Neuronal Splicing Regulators in Mammals. Mol. Cell. Biol. 2005, 25, 10005–10016. [Google Scholar] [CrossRef]
- Folk, P.; Půta, F.; Skružný, M. Transcriptional coregulator SNW/SKIP: The concealed tie of dissimilar pathways. Cell. Mol. Life Sci. 2004, 61, 629–640. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Liu, W.-T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef]
- Wolfe, M.S. Structure and Function of the γ-Secretase Complex. Biochemistry 2019, 58, 2953–2966. [Google Scholar] [CrossRef]
- Zhou, S.; Zhou, H.; Walian, P.J.; Jap, B.K. CD147 is a regulatory subunit of the γ-secretase complex in Alzheimer’s disease amyloid β-peptide production. Proc. Natl. Acad. Sci. USA 2005, 102, 7499–7504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Xu, H.; Zhang, Y.-W. The Î3-secretase complex: From structure to function. Front. Cell. Neurosci. 2014, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Chauvet, V.; Tian, X.; Husson, H.; Grimm, D.H.; Wang, T.; Hieseberger, T.; Igarashi, P.; Bennett, A.M.; Ibraghimov-Beskrovnaya, O.; Somlo, S.; et al. Mechanical stimuli induce cleavage and nuclear translocation of the polycystin-1 C terminus. J. Clin. Investig. 2004, 114, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Low, S.H.; Vasanth, S.; Larson, C.H.; Mukherjee, S.; Sharma, N.; Kinter, M.; Kane, M.E.; Obara, T.; Weimbs, T. Polycystin-1, STAT6, and P100 Function in a Pathway that Transduces Ciliary Mechanosensation and Is Activated in Polycystic Kidney Disease. Dev. Cell 2006, 10, 57–69. [Google Scholar] [CrossRef]
- Lal, M.; Song, X.; Pluznick, J.L.; Di Giovanni, V.; Merrick, D.M.; Rosenblum, N.D.; Chauvet, V.; Gottardi, C.J.; Pei, Y.; Caplan, M.J. Polycystin-1 C-terminal tail associates with β-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet. 2008, 17, 3105–3117. [Google Scholar] [CrossRef]
- Merrick, D.; Chapin, H.; Baggs, J.E.; Yu, Z.; Somlo, S.; Sun, Z.; Hogenesch, J.B.; Caplan, M.J. The γ-Secretase Cleavage Product of Polycystin-1 Regulates TCF and CHOP-Mediated Transcriptional Activation through a p300-Dependent Mechanism. Dev. Cell 2012, 22, 197–210. [Google Scholar] [CrossRef]
- Chalasani, S.H.; Sabol, A.; Xu, H.A.; Gyda, M.A.; Rasband, K.; Granato, M.; Chien, C.-B.; Raper, J.A. Stromal Cell-Derived Factor-1 Antagonizes Slit/Robo Signaling In Vivo. J. Neurosci. 2007, 27, 973–980. [Google Scholar] [CrossRef]
- Bellon, A.; Iyer, A.; Bridi, S.; Lee, F.; Ovando-Vázquez, C.; Corradi, E.; Longhi, S.; Roccuzzo, M.; Strohbuecker, S.; Naik, S.; et al. miR-182 Regulates Slit2-Mediated Axon Guidance by Modulating the Local Translation of a Specific mRNA. Cell Rep. 2017, 18, 1171–1186. [Google Scholar] [CrossRef]
- Leyva-Díaz, E.; del Toro, D.; Menal, M.J.; Cambray, S.; Susín, R.; Tessier-Lavigne, M.; Klein, R.; Egea, J.; López-Bendito, G. FLRT3 Is a Robo1-Interacting Protein that Determines Netrin-1 Attraction in Developing Axons. Curr. Biol. 2014, 24, 494–508. [Google Scholar] [CrossRef]
- O’Sullivan, M.L.; de Wit, J.; Savas, J.N.; Comoletti, D.; Otto-Hitt, S.; Yates, J.R., 3rd; Ghosh, A. FLRT Proteins Are Endogenous Latrophilin Ligands and Regulate Excitatory Synapse Development. Neuron 2012, 73, 903–910. [Google Scholar] [CrossRef]
- Puppo, F.; Thomé, V.; Lhoumeau, A.; Cibois, M.; Gangar, A.; Lembo, F.; Belotti, E.; Marchetto, S.; Lécine, P.; Prébet, T.; et al. Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 2011, 12, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Matsuda, Y.; D’Adamio, L. BRI3 Inhibits Amyloid Precursor Protein Processing in a Mechanistically Distinct Manner from Its Homologue Dementia Gene BRI2. J. Biol. Chem. 2009, 284, 15815–15825. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, M.; Weiner, J.; Sanes, J.R. Sidekicks: Synaptic Adhesion Molecules that Promote Lamina-Specific Connectivity in the Retina. Cell 2002, 110, 649–660. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. Synaptic Localization and Function of Sidekick Recognition Molecules Require MAGI Scaffolding Proteins. J. Neurosci. 2010, 30, 3579–3588. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stevens, C.B.; Cameron, D.A.; Stenkamp, D.L. Plasticity of photoreceptor-generating retinal progenitors revealed by prolonged retinoic acid exposure. BMC Dev. Biol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Zhang, C.; Baudino, T.A.; Dowd, D.R.; Tokumaru, H.; Wang, W.; MacDonald, P.N. Ternary Complexes and Cooperative Interplay between NCoA-62/Ski-interacting Protein and Steroid Receptor Coactivators in Vitamin D Receptor-mediated Transcription. J. Biol. Chem. 2001, 276, 40614–40620. [Google Scholar] [CrossRef]
- Carpio, R.V.-D.; Kaplan, F.M.; Weaver, K.L.; VanWye, J.D.; Alves-Guerra, M.-C.; Robbins, D.J.; Capobianco, A.J. Assembly of a Notch Transcriptional Activation Complex Requires Multimerization. Mol. Cell. Biol. 2011, 31, 1396–1408. [Google Scholar] [CrossRef]
- Birtel, J.; Gliem, M.; Mangold, E.; Tebbe, L.; Spier, I.; Müller, P.L.; Holz, F.G.; Neuhaus, C.; Wolfrum, U.; Bolz, H.J.; et al. Novel Insights into the Phenotypical Spectrum of KIF11-Associated Retinopathy, Including a New Form of Retinal Ciliopathy. Investig. Opthalmology Vis. Sci. 2017, 58, 3950–3959. [Google Scholar] [CrossRef]
- Ronquillo, C.C.; Bernstein, P.S.; Baehr, W. Senior–Løken syndrome: A syndromic form of retinal dystrophy associated with nephronophthisis. Vis. Res. 2012, 75, 88–97. [Google Scholar] [CrossRef]
- Gao, J.; Cheon, K.; Nusinowitz, S.; Liu, Q.; Bei, D.; Atkins, K.; Azimi, A.; Daiger, S.P.; Farber, D.B.; Heckenlively, J.R.; et al. Progressive photoreceptor degeneration, outer segment dysplasia, and rhodopsin mislocalization in mice with targeted disruption of the retinitis pigmentosa-1 (Rp1) gene. Proc. Natl. Acad. Sci. USA 2002, 99, 5698–5703. [Google Scholar] [CrossRef]
- Liu, Q.; Lyubarsky, A.; Skalet, J.H.; Pugh, E.N., Jr.; Pierce, E.A. RP1 Is Required for the Correct Stacking of Outer Segment Discs. Investig. Opthalmology Vis. Sci. 2003, 44, 4171–4183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmidt-Kastner, R.; Kreczmanski, P.; Preising, M.; Diederen, R.; Schmitz, C.; Reis, D.; Blanks, J.; Dorey, C.K. Expression of the diabetes risk gene wolframin (WFS1) in the human retina. Exp. Eye Res. 2009, 89, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tan, G.; Levenkova, N.; Li, T.; Pugh, E.N., Jr.; Rux, J.J.; Speicher, D.W.; Pierce, E.A. The Proteome of the Mouse Photoreceptor Sensory Cilium Complex. Mol. Cell. Proteom. 2007, 6, 1299–1317. [Google Scholar] [CrossRef] [PubMed]
- Tranebjaerg, L.; Barrett, T.; Rendtorff, N.D. WFS1-Related Disorders. In GeneReviews (R); Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Ledbetter, N., Mefford, H.C., Smith, R.J.H., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Wang, J.; Zhong, J.; Chen, G.; Li, M.; Wu, F.-X.; Pan, Y. ClusterViz: A Cytoscape APP for Cluster Analysis of Biological Network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 12, 815–822. [Google Scholar] [CrossRef]
- Raggenbass, M.; Bertrand, D. Nicotinic receptors in circuit excitability and epilepsy. J. Neurobiol. 2002, 53, 580–589. [Google Scholar] [CrossRef]
- Chen, T.T.; Klassen, T.L.; Goldman, A.M.; Marini, C.; Guerrini, R.; Noebels, J.L. Novel brain expression of ClC-1 chloride channels and enrichment of CLCN1 variants in epilepsy. Neurology 2013, 80, 1078–1085. [Google Scholar] [CrossRef]
- Xie, H.; Zhang, Y.; Zhang, P.; Wang, J.; Wu, Y.; Wu, X.; Netoff, T.; Jiang, Y. Functional Study of NIPA2 Mutations Identified from the Patients with Childhood Absence Epilepsy. PLoS ONE 2014, 9, e109749. [Google Scholar] [CrossRef]
- Greeve, I.; Hermans-Borgmeyer, I.; Brellinger, C.; Kasper, D.; Gomez-Isla, T.; Behl, C.; Levkau, B.; Nitsch, R.M. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer’s disease-associated neurodegeneration and oxidative stress. J. Neurosci. 2000, 20, 7345–7352. [Google Scholar] [CrossRef]
- Khanim, F.; Kirk, J.; Latif, F.; Barrett, T. WFS1/wolframin mutations, Wolfram syndrome, and associated diseases. Hum. Mutat. 2001, 17, 357–367. [Google Scholar] [CrossRef]
- Al-Saif, A.; Al-Mohanna, F.; Bohlega, S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann. Neurol. 2011, 70, 913–919. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid Precursor Protein Processing and Alzheimer’s Disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Kojro, E.; Fahrenholz, F. The Non-Amyloidogenic Pathway: Structure and Function of α-Secretases. Sub-Cell. Biochem. 2005, 38, 105–127. [Google Scholar] [CrossRef]
- Chow, V.W.; Mattson, M.P.; Wong, P.C.; Gleichmann, M. An Overview of APP Processing Enzymes and Products. Neuromol. Med. 2010, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kounnas, M.Z.; Moir, R.D.; Rebeck, G.W.; Bush, A.I.; Argraves, W.S.; Tanzi, R.E.; Hyman, B.T.; Strickland, D.K. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell 1995, 82, 331–340. [Google Scholar] [CrossRef]
- Cao, X.; Südhof, T.C. A Transcriptively Active Complex of APP with Fe65 and Histone Acetyltransferase Tip60. Science 2001, 293, 115–120. [Google Scholar] [CrossRef]
- Sumioka, A.; Nagaishi, S.; Yoshida, T.; Lin, A.; Miura, M.; Suzuki, T. Role of 14-3-3γ in FE65-dependent Gene Transactivation Mediated by the Amyloid β-Protein Precursor Cytoplasmic Fragment. J. Biol. Chem. 2005, 280, 42364–42374. [Google Scholar] [CrossRef]
- Sabo, S.L.; Lanier, L.M.; Ikin, A.F.; Khorkova, O.; Sahasrabudhe, S.; Greengard, P.; Buxbaum, J.D. Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J. Biol. Chem. 1999, 274, 7952–7957. [Google Scholar] [CrossRef]
- Klug, W.; Dietl, A.; Simon, B.; Sinning, I.; Wild, K. Phosphorylation of LRP1 regulates the interaction with Fe65. FEBS Lett. 2011, 585, 3229–3235. [Google Scholar] [CrossRef][Green Version]
- Malicki, J.J.; Johnson, C.A. The cilium: Cellular antenna and central processing unit. Trends Cell Biol. 2017, 27, 126–140. [Google Scholar] [CrossRef]
- Waterham, H.R.; Koster, J.; Romeijn, G.J.; Hennekam, R.C.; Vreken, P.; Andersson, H.C.; FitzPatrick, D.R.; Kelley, R.I.; Wanders, R.J. Mutations in the 3β-Hydroxysterol Δ24-Reductase Gene Cause Desmosterolosis, an Autosomal Recessive Disorder of Cholesterol Biosynthesis. Am. J. Hum. Genet. 2001, 69, 685–694. [Google Scholar] [CrossRef]
- Canault, M.; Tellier, E.; Bonardo, B.; Mas, E.; Aumailley, M.; Juhan-Vague, I.; Nalbone, G.; Peiretti, F. FHL2 interacts with both ADAM-17 and the cytoskeleton and regulates ADAM-17 localization and activity. J. Cell. Physiol. 2006, 208, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, H.; Tabira, T. Alzheimer’s disease-associated presenilin 2 interacts with DRAL, an LIM-domain protein. Hum. Mol. Genet. 2000, 9, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Güler, B.E.; Krzysko, J.; Wolfrum, U. Isolation and culturing of primary mouse astrocytes for the analysis of focal adhesion dynamics. STAR Protoc. 2021, 2, 100954. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, C.J.; Boldt, K.; Schumacher, A.; Roepman, R.; Ueffing, M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 2007, 7, 4228–4234. [Google Scholar] [CrossRef] [PubMed]
- Bohnekamp, J.; Schöneberg, T. Cell Adhesion Receptor GPR133 Couples to Gs Protein. J. Biol. Chem. 2011, 286, 41912–41916. [Google Scholar] [CrossRef]
- Berridge, M.J. Rapid accumulation of inositol trisphosphate reveals that agonists hydrolyse polyphosphoinositides instead of phosphatidylinositol. Biochem. J. 1983, 212, 849–858. [Google Scholar] [CrossRef]
- Schoneberg, T.; Schulz, A.; Biebermann, H.; Gruters, A.; Grimm, T.; Hubschmann, K.; Filler, G.; Gudermann, T.; Schultz, G. V2 vasopressin receptor dysfunction in nephrogenic diabetes insipidus caused by different molecular mechanisms. Hum. Mutat. 1998, 12, 196–205. [Google Scholar] [CrossRef]
- Wilde, C.; Fischer, L.; Lede, V.; Kirchberger, J.; Rothemund, S.; Schöneberg, T.; Liebscher, I. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 2015, 30, 666–673. [Google Scholar] [CrossRef]

| Pathways | Prey Proteins | VLGR1a | Baits N-CTF | C-CTF | Prey Proteins | VLGR1a | Baits N-CTF | C-CTF |
|---|---|---|---|---|---|---|---|---|
| AMPK pathway: coordinates cell growth, autophagy and metabolism | CPT1A G6PC3 HMGCR IGF1R PCK2 | + + + | + + + + | + + + + | PIK3R2 PRKAG1 SCD SCD5 | + + + | + + | + + |
| Ephrin signaling: development | ADAM10 APH1A EFNB1 | + | + + | + + + | NCSTN PSEN2 | + | + | + + |
| FoxO pathway: regulation of cell cycle, apoptosis and metabolism | FOXG1 G6PC3 IGF1R PIK3R2 | + + + | + + | PCK2 PRKAG1 RAF1 TGFBR1 | + + + | + + + | ||
| HIF 1 pathway: regulates hypoxia inducible genes under hypoxic conditions | CUL2 EIF4E HK2 HMOX1 | + + + | + + | + | IGF1R PIK3R2 TCEB1 TCEB2 | + + + + | + | + |
| Insulin receptor pathway: controls critical energy functions such as glucose and lipid metabolism, connected to FoxO signaling pathway | ATP6AP1 ATP6V0A1 ATP6V0A2 ATP6V0D1 ATP6V1F ATP6V1H EIF4E IGF1R LAMTOR1 | + + + | + + + + + + + + | + + + + + + + + | LAMTOR3 NCAM1 PIGU PIK3R2 PIK3R4 PRKAG1 RAF1 SPNS1 | + + + | + + + + + | + + + + + + |
| Integrin signaling:development, tissue maintenance and repair | ADAM10 ADAMTS CD47 | + | + + | ITGA5 ITGA6 SPNS1 | + | + | + + | |
| MAPK pathway: involved in proliferation, differentiation, motility, stress response, apoptosis and survival | ARL6IP5 ATP6AP1 ATP6AP2 CD81 CDK5RAP3 FN1 FZD7 GDF15 HMGCR IGF1R ILK INHBE IRAK1 LAMTOR1 LAMTOR3 | + + + + + + | + + + + + + + + + | + + + + + + + + + | LEMD2 MAP2K7 MTCH1 NCAM1 NPTN PIGU PPM1L PSEN1 RAF1 RHBDD3 SPNS1 SYNJ2BP TAB2 TGFBR1 VRK2 | + + + + + + + | + + + + + + + + + + + | + + + + + + + + + + |
| mTOR pathway: senses and integrates nutritional and environmental cues | EIF4E LAMTOR1 LAMTOR3P | + | + | + + | K3R2 RICTOR | + + | ||
| Neurotrophin pathway: regulates survival, development and function of neurons | ADAM17 APH1A IRAK1 LAMTOR3 NCAM1 NCSTN PIGU | + + + | + + + + + | + + + + + + | PIK3R2 PSEN2 RAF1 RICTOR SMPD2 SORT1 SPNS1 | + + + + + | + + + + | + + + + |
| Notch pathway: regulates cell fate determination during development and maintains adult tissue homeostasis | ADAM10 ADAM17 APH1A APP GALNT11 GOLT1B MAGEA1 | + | + + + + + | + + + + | NCSTN PSEN1 PSEN2 SEL1L SYNJ2BP TSPAN15 | + | + + + + + | + + + + + + |
| SMAD pathway: regulates cell growth, differentiation & development | GDF15 INHBE LNPEP | + + | + | + | NCEH1 SLC33A1 STOML1 | + | + + | + + |
| Sphingolipid pathway: regulate cellular responses to stress | ABCC1 BAX CERS1 CERS2 CERS4 CERS5 CERS6 DEGS1 PIK3R2 | + + + | + + + + + + + | + + + + + + + | PLCB4 PLD2 RAF1 SGPL1 SGPP1 SMPD2 SPTLC1 SPTLC2 | + + + + | + + + + + + + | + + + + + + |
| Wnt pathway: regulates cell migration, cell polarity, neural patterning and organogenesis | ATP6AP2 FZD7 GPC6 ILK LRP1 MESDC2 | + + | + + + | + + | PSEN1 PTK7 SMO SPNS1 UBR5 WLS | + + | + + + + + | + + + + + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knapp, B.; Roedig, J.; Roedig, H.; Krzysko, J.; Horn, N.; Güler, B.E.; Kusuluri, D.K.; Yildirim, A.; Boldt, K.; Ueffing, M.; et al. Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1. Molecules 2022, 27, 3108. https://doi.org/10.3390/molecules27103108
Knapp B, Roedig J, Roedig H, Krzysko J, Horn N, Güler BE, Kusuluri DK, Yildirim A, Boldt K, Ueffing M, et al. Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1. Molecules. 2022; 27(10):3108. https://doi.org/10.3390/molecules27103108
Chicago/Turabian StyleKnapp, Barbara, Jens Roedig, Heiko Roedig, Jacek Krzysko, Nicola Horn, Baran E. Güler, Deva Krupakar Kusuluri, Adem Yildirim, Karsten Boldt, Marius Ueffing, and et al. 2022. "Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1" Molecules 27, no. 10: 3108. https://doi.org/10.3390/molecules27103108
APA StyleKnapp, B., Roedig, J., Roedig, H., Krzysko, J., Horn, N., Güler, B. E., Kusuluri, D. K., Yildirim, A., Boldt, K., Ueffing, M., Liebscher, I., & Wolfrum, U. (2022). Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1. Molecules, 27(10), 3108. https://doi.org/10.3390/molecules27103108







