Biotransformation of Liquiritigenin into Characteristic Metabolites by the Gut Microbiota
Abstract
1. Introduction
2. Results
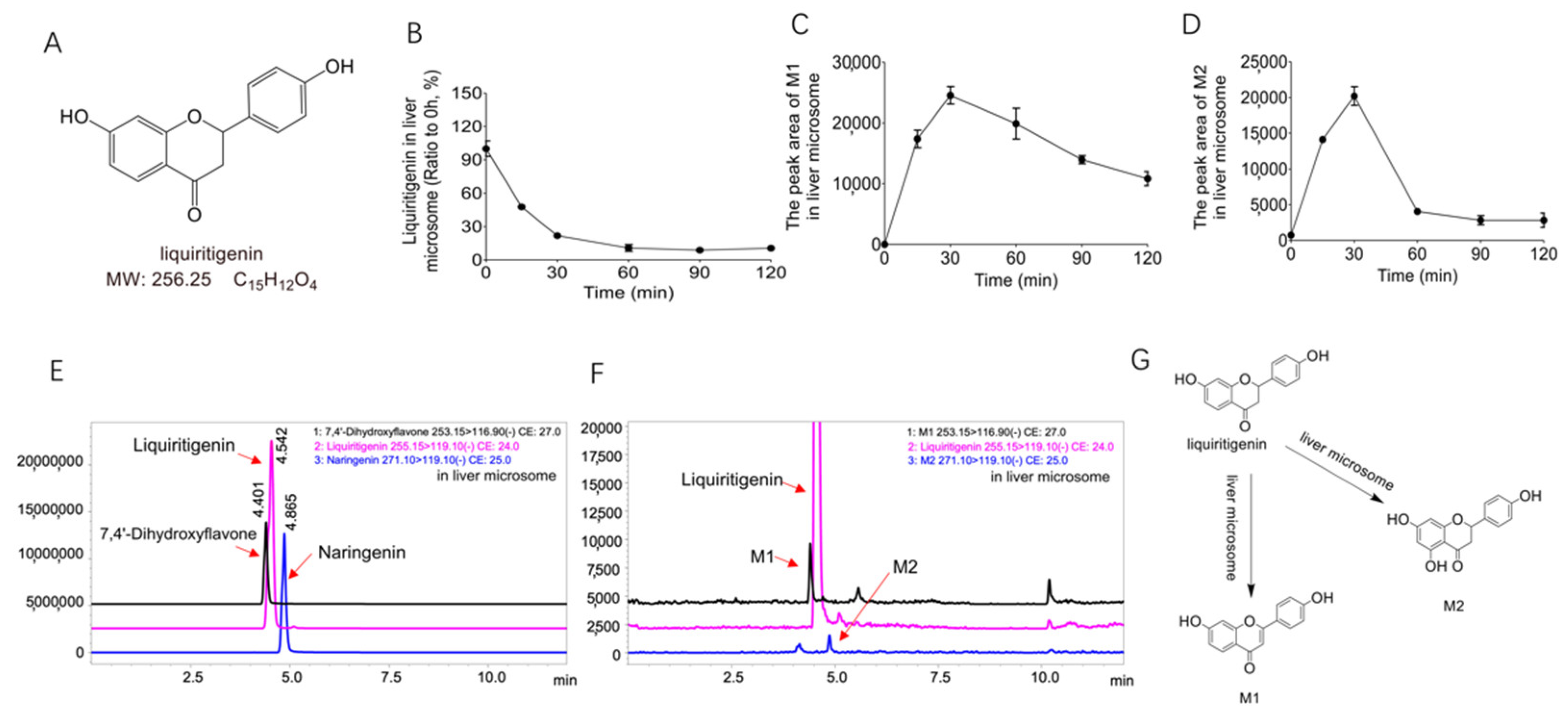
2.1. Biotransformation of Liquiritigenin in Liver Microsomes
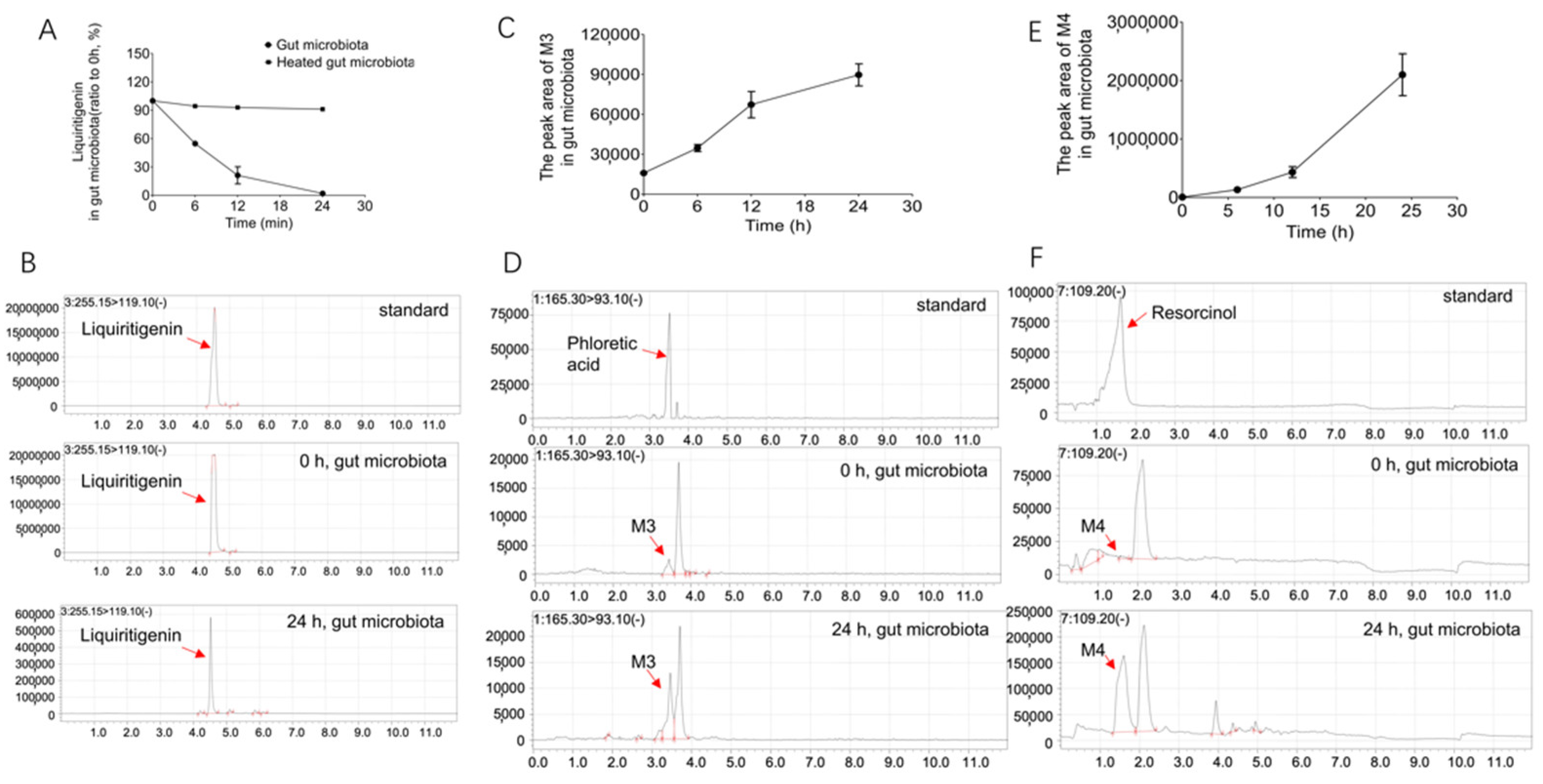
2.2. Biotransformation of Liquiritigenin in the Gut Microbiota
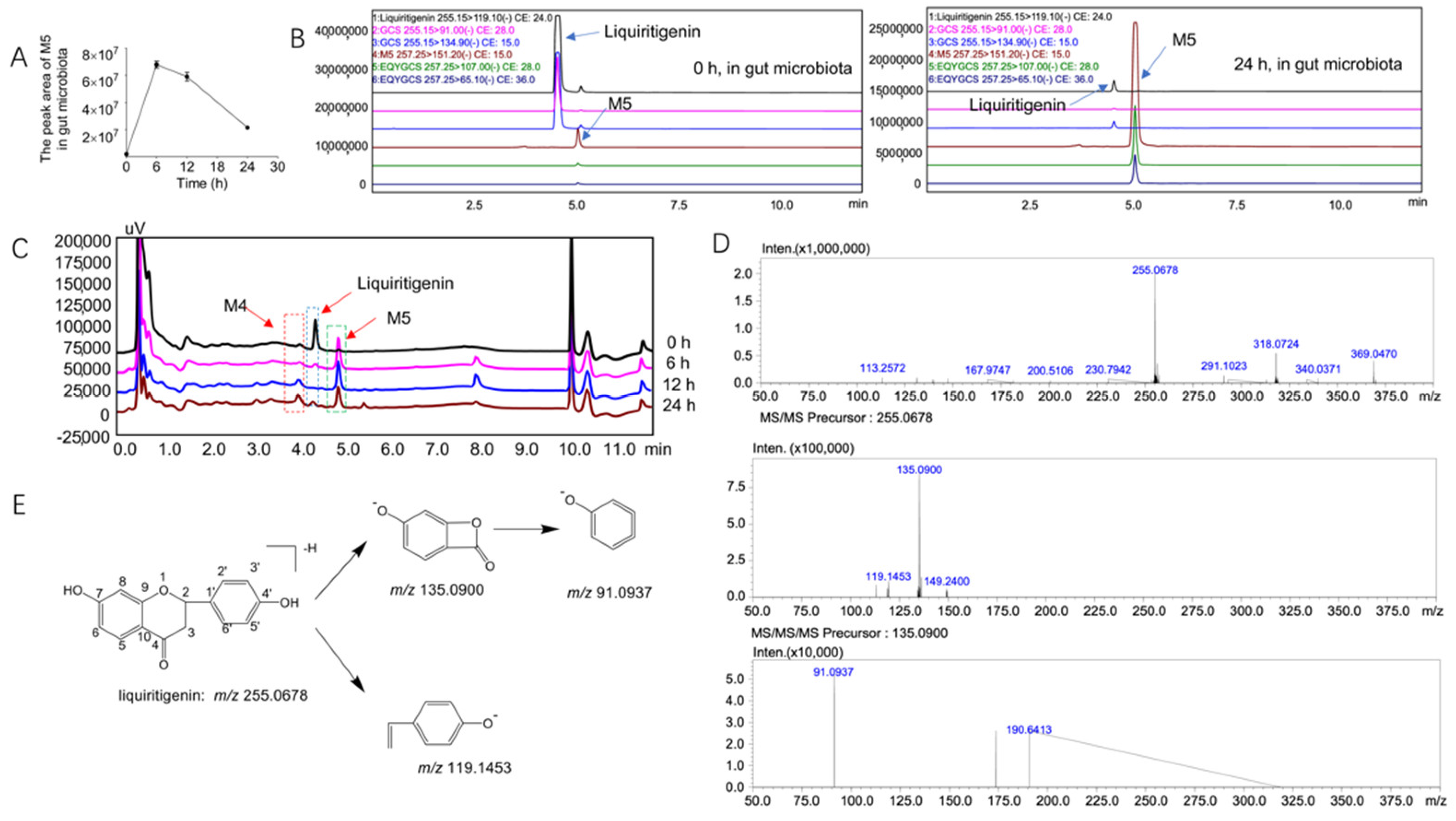
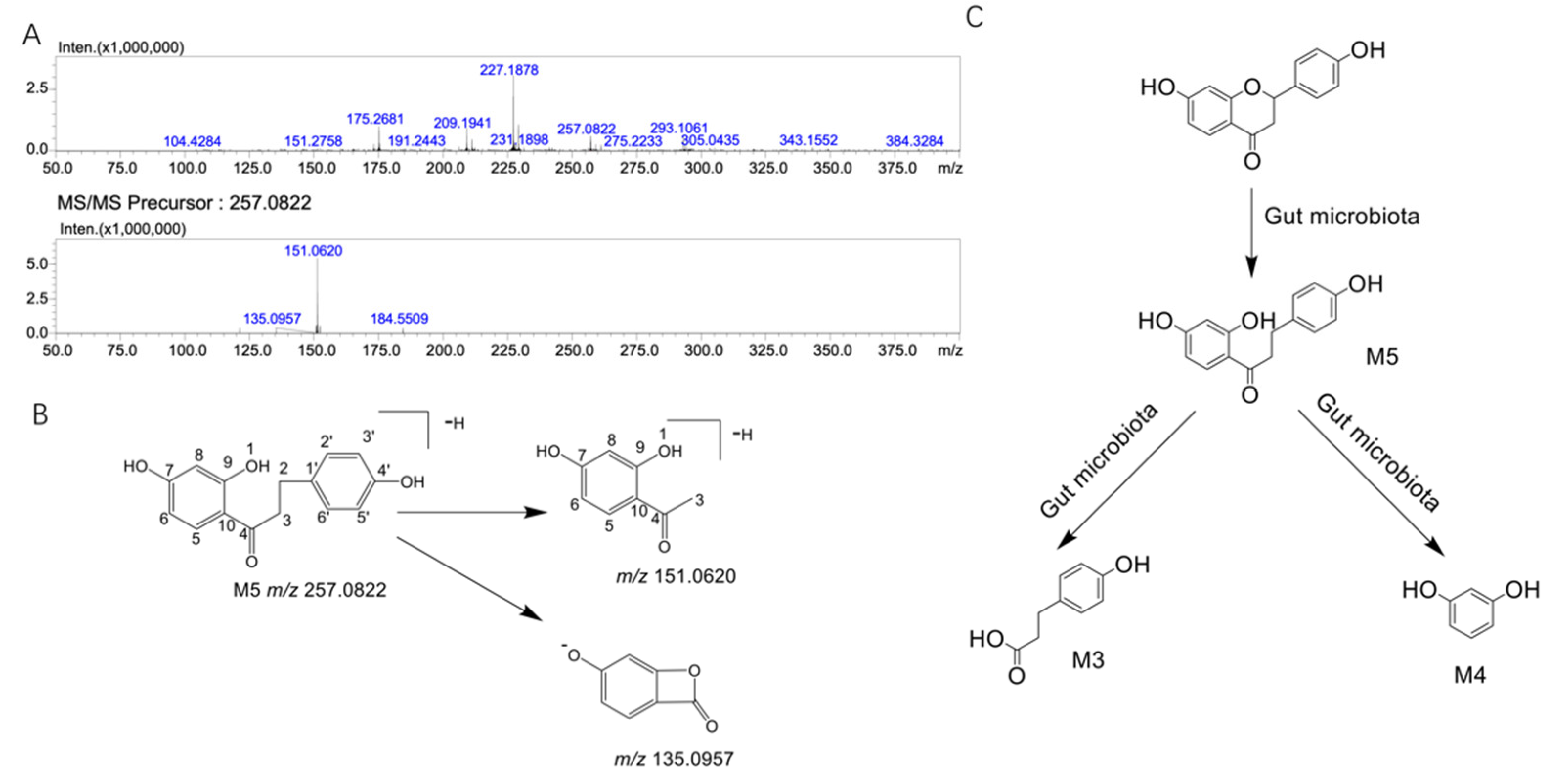
2.3. Structural Analysis of the Unknown Metabolite M5
3. Discussion
4. Materials and Methods
4.1. Instruments and Reagents
4.2. Animals
4.3. Determination of Liquiritigenin by LC/MSn-IT-TOF
4.4. Determination of Liquiritigenin by HPLC–MS/MS
4.5. In Vitro Incubation of Liquiritigenin with Liver Microsomes
4.6. In Vitro Incubation of Liquiritigenin with the Gut Microbiota
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.; Fang, X.; Yang, J.; Yao, Y.; Nandakumar, K.S.; Salem, M.L.; Cheng, K. Recent Research on Flavonoids and their Biomedical Applications. Curr. Med. Chem. 2021, 28, 1042–1066. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; Pinto, D.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.L.; Hodgson, J.M.; Croft, K.D.; Lewis, J.R.; Prince, R.L. Flavonoid intake and all-cause mortality. Am. J. Clin. Nutr. 2015, 101, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.A.; Lee, J.; Oh, J.H.; Chang, H.J.; Sohn, D.K.; Shin, A.; Kim, J. Dietary Flavonoids, CYP1A1 Genetic Variants, and the Risk of Colorectal Cancer in a Korean population. Sci. Rep. 2017, 7, 128. [Google Scholar] [CrossRef]
- Van Iersel, L.E.; Stevens, Y.R.; Conchillo, J.M.; Troost, F.J. The effect of citrus flavonoid extract supplementation on anaerobic capacity in moderately trained athletes: A randomized controlled trial. J. Int. Soc. Sports Nutr. 2021, 18, 2. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Herrera-Bravo, J.; Belén, L.H.; Kaur, R.; Kregiel, D.; Uprety, Y.; Beyatli, A.; Yeskaliyeva, B.; Kırkın, C.; et al. Glycyrrhiza Genus: Enlightening Phytochemical Components for Pharmacological and Health-Promoting Abilities. Oxid. Med. Cell. Longev. 2021, 2021, 7571132. [Google Scholar] [CrossRef]
- Ji, Y.; Hu, W.; Jin, Y.; Yu, H.; Fang, J. Liquiritigenin exerts the anti-cancer role in oral cancer via inducing autophagy-related apoptosis through PI3K/AKT/mTOR pathway inhibition in vitro and in vivo. Bioengineered 2021, 12, 6070–6082. [Google Scholar] [CrossRef]
- Wang, L.; Yang, R.; Yuan, B.; Liu, Y.; Liu, C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm. Sin. B 2015, 5, 310–315. [Google Scholar] [CrossRef]
- Zhang, M.; Xue, Y.; Zheng, B.; Li, L.; Chu, X.; Zhao, Y.; Wu, Y.; Zhang, J.; Han, X.; Wu, Z.; et al. Liquiritigenin protects against arsenic trioxide-induced liver injury by inhibiting oxidative stress and enhancing mTOR-mediated autophagy. Biomed. Pharm. 2021, 143, 112167. [Google Scholar] [CrossRef]
- Hu, K.; Yang, Z.H.; Pan, S.S.; Xu, H.J.; Ren, J. Synthesis and antitumor activity of liquiritigenin thiosemicarbazone derivatives. Eur. J. Med. Chem. 2010, 45, 3453–3458. [Google Scholar] [CrossRef] [PubMed]
- Houshyar, Y.; Massimino, L.; Lamparelli, L.A.; Danese, S.; Ungaro, F. Going Beyond Bacteria: Uncovering the Role of Archaeome and Mycobiome in Inflammatory Bowel Disease. Front. Physiol. 2021, 12, 783295. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Gul, L.; Modos, D.; Fonseca, S.; Madgwick, M.; Thomas, J.P.; Sudhakar, P.; Booth, C.; Stentz, R.; Carding, S.R.; Korcsmaros, T. Extracellular vesicles produced by the human commensal gut bacterium Bacteroides thetaiotaomicron affect host immune pathways in a cell-type specific manner that are altered in inflammatory bowel disease. J. Extracell. Vesicles 2022, 11, e12189. [Google Scholar] [CrossRef]
- Thibaut, M.M.; Bindels, L.B. Crosstalk between bile acid-activated receptors and microbiome in entero-hepatic inflammation. Trends Mol. Med. 2022, 28, 223–236. [Google Scholar] [CrossRef]
- Du, S.; Sun, X.; Zhang, J.; Lin, D.; Chen, R.; Cui, Y.; Xiang, S.; Wu, Z.; Ding, T. Metagenome-Assembled Genomes Reveal Mechanisms of Carbohydrate and Nitrogen Metabolism of Schistosomiasis-Transmitting Vector Biomphalaria Glabrata. Microbiol. Spectr. 2022, 10, e0184321. [Google Scholar] [CrossRef]
- Kim, I.S.; Yoo, D.H.; Jung, I.H.; Lim, S.; Jeong, J.J.; Kim, K.A.; Bae, O.N.; Yoo, H.H.; Kim, D.H. Reduced metabolic activity of gut microbiota by antibiotics can potentiate the antithrombotic effect of aspirin. Biochem. Pharmacol. 2016, 122, 72–79. [Google Scholar] [CrossRef]
- Kupfer, R.; Swanson, L.; Chow, S.; Staub, R.E.; Zhang, Y.L.; Cohen, I.; Christians, U. Oxidative in vitro metabolism of liquiritigenin, a bioactive compound isolated from the Chinese herbal selective estrogen beta-receptor agonist MF101. Drug Metab. Dispos. 2008, 36, 2261–2269. [Google Scholar] [CrossRef]
- Lee, Y.K.; Chin, Y.W.; Bae, J.K.; Seo, J.S.; Choi, Y.H. Pharmacokinetics of isoliquiritigenin and its metabolites in rats: Low bioavailability is primarily due to the hepatic and intestinal metabolism. Planta Med. 2013, 79, 1656–1665. [Google Scholar] [CrossRef]
- Yang, G.; Hong, S.; Yang, P.; Sun, Y.; Wang, Y.; Zhang, P.; Jiang, W.; Gu, Y. Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat. Commun. 2021, 12, 790. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shi, J.; Li, H. Liquiritigenin attenuates high glucose-induced mesangial matrix accumulation, oxidative stress, and inflammation by suppression of the NF-κB and NLRP3 inflammasome pathways. Biomed. Pharmacother. 2018, 106, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.C.; Lin, J.K. Liquiritigenin Inhibits Colorectal Cancer Proliferation, Invasion, and Epithelial-to-Mesenchymal Transition by Decreasing Expression of Runt-Related Transcription Factor 2. Oncol. Res. 2019, 27, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Besch-Williford, C.; Hyder, S.M. The estrogen receptor beta agonist liquiritigenin enhances the inhibitory effects of the cholesterol biosynthesis inhibitor RO 48-8071 on hormone-dependent breast-cancer growth. Breast Cancer Res. Treat. 2022, 192, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Sareddy, G.R.; Pratap, U.P.; Venkata, P.P.; Zhou, M.; Alejo, S.; Viswanadhapalli, S.; Tekmal, R.R.; Brenner, A.J.; Vadlamudi, R.K. Activation of estrogen receptor beta signaling reduces stemness of glioma stem cells. Stem Cells 2021, 39, 536–550. [Google Scholar] [CrossRef]
- Liu, J.; Viswanadhapalli, S.; Garcia, L.; Zhou, M.; Nair, B.C.; Kost, E.; Tekmal, R.R.; Li, R.; Rao, M.K.; Curiel, T.; et al. Therapeutic utility of natural estrogen receptor beta agonists on ovarian cancer. Oncotarget 2017, 8, 50002–50014. [Google Scholar] [CrossRef]
- Wang, D.; Lu, J.; Liu, Y.; Meng, Q.; Xie, J.; Wang, Z.; Teng, L. Liquiritigenin induces tumor cell death through mitogen-activated protein kinase- (MPAKs-) mediated pathway in hepatocellular carcinoma cells. Biomed Res. Int. 2014, 2014, 965316. [Google Scholar] [CrossRef]
- Shimamura, H.; Suzuki, H.; Hanano, M.; Suzuki, A.; Sugiyama, Y. Identification of tissues responsible for the conjugative metabolism of liquiritigenin in rats: An analysis based on metabolite kinetics. Biol. Pharm. Bull. 1993, 16, 899–907. [Google Scholar] [CrossRef][Green Version]
- Kang, H.E.; Jung, H.Y.; Cho, Y.K.; Kim, S.H.; Sohn, S.I.; Baek, S.R.; Lee, M.G. Pharmacokinetics of liquiritigenin in mice, rats, rabbits, and dogs, and animal scale-up. J. Pharm. Sci. 2009, 98, 4327–4342. [Google Scholar] [CrossRef]
- Guo, J.; Liu, A.; Cao, H.; Luo, Y.; Pezzuto, J.M.; van Breemen, R.B. Biotransformation of the chemopreventive agent 2’,4’,4-trihydroxychalcone (isoliquiritigenin) by UDP-glucuronosyltransferases. Drug Metab. Dispos. 2008, 36, 2104–2112. [Google Scholar] [CrossRef]
- Murota, K.; Nakamura, Y.; Uehara, M. Flavonoid metabolism: The interaction of metabolites and gut microbiota. Biosci. Biotechnol. Biochem. 2018, 82, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Biancatelli, R.M.L.C.; Berrill, M.; Catravas, J.D.; Marik, P.E. Quercetin and Vitamin C: An Experimental, Synergistic Therapy for the Prevention and Treatment of SARS-CoV-2 Related Disease (COVID-19). Front. Immunol. 2020, 11, 1451. [Google Scholar] [CrossRef] [PubMed]
- Harwood, M.; Danielewska-Nikiel, B.; Borzelleca, J.F.; Flamm, G.W.; Williams, G.M.; Lines, T.C. A critical review of the data related to the safety of quercetin and lack of evidence of in vivo toxicity, including lack of genotoxic/carcinogenic properties. Food Chem. Toxicol. 2007, 45, 2179–2205. [Google Scholar] [CrossRef] [PubMed]
- Logendra, S.; Ribnicky, D.M.; Yang, H.; Poulev, A.; Ma, J.; Kennelly, E.J.; Raskin, I. Bioassay-guided isolation of aldose reductase inhibitors from Artemisia dracunculus. Phytochemistry 2006, 67, 1539–1546. [Google Scholar] [CrossRef]
- Bohm, B.A.; Glennie, C.W. The isolation of 2′,4,4′-trihydroxydihydrochalcone from Viburnum davidi. Phytochemistry 1969, 8, 905–908. [Google Scholar] [CrossRef]
- Jensen, S.R.; Nielsen, B.J.; Norn, V. Dihydrochalcones from Viburnum davidii and V. lantanoides. Phytochemistry 1977, 16, 2036–2038. [Google Scholar] [CrossRef]
- Asano, T.; Ishihara, K.; Morota, T.; Takeda, S.; Aburada, M. Permeability of the flavonoids liquiritigenin and its glycosides in licorice roots and davidigenin, a hydrogenated metabolite of liquiritigenin, using human intestinal cell line Caco-2. J. Ethnopharmacol. 2003, 89, 285–289. [Google Scholar] [CrossRef]
- Kil, Y.S.; Park, J.; Jafari, M.; Woo, H.A.; Seo, E.K. Minor phenolics from Angelica keiskei and their proliferative effects on Hep3B cells. Bioorg. Med. Chem. Lett. 2017, 27, 3065–3070. [Google Scholar] [CrossRef]
- Yang, E.J.; Kim, M.; Woo, J.E.; Lee, T.; Jung, J.W.; Song, K.S. The comparison of neuroprotective effects of isoliquiritigenin and its Phase I metabolites against glutamate-induced HT22 cell death. Bioorg. Med. Chem. Lett. 2016, 26, 5639–5643. [Google Scholar] [CrossRef]
- Hong, L.; Ying, S.H. Ethanol extract and isolated constituents from Artemisia Dracunculus inhibit esophageal squamous cell carcinoma and induce apoptotic cell death. Drug Res. 2015, 65, 101–106. [Google Scholar] [CrossRef]
| Substance | Reaction | Theoretical Molecular Weight | Molecular Formula | Fragment Characteristics | |||
|---|---|---|---|---|---|---|---|
| MS1/[M-H]- | MS/MS | MS3 | |||||
| Gut microbiota system | Liquiritigenin | - | 256.0736 | C15H12O4 | 255.0678 | 135.0900 119.1453 | 91.0937 |
| M5 | +2H | 258.0892 | C15H14O4 | 257.0822 | 151.0620 135.0957 | ||
| Substance | Experimental Molecular Weight | Theoretical Molecular Weight | Predicted Molecular Formula | Ion Mode | Diff (ppm) | |
|---|---|---|---|---|---|---|
| Liquiritigenin | 255.0678 | 256.0736 | C15H12O4 | [M-H]- | 5.93 | |
| Liver microsomes system | M1 | 255.0670 | 254.0579 | C15H10O4 | [M+H]+ | 7.14 |
| M2 | 273.0736 | 272.0685 | C15H12O5 | [M+H]+ | 7.90 | |
| Gut microbiota system | M3 | 165.0577 | 166.0630 | C9H10O3 | [M-H]- | 0.11 |
| M4 | 111.0429 | 110.0368 | C6H6O2 | [M+H]+ | 10.51 | |
| M5 | 257.0822 | 258.0892 | C15H14O4 | [M-H]- | 1.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keranmu, A.; Pan, L.-B.; Fu, J.; Han, P.; Yu, H.; Zhang, Z.-W.; Xu, H.; Yang, X.-Y.; Hu, J.-C.; Zhang, H.-J.; et al. Biotransformation of Liquiritigenin into Characteristic Metabolites by the Gut Microbiota. Molecules 2022, 27, 3057. https://doi.org/10.3390/molecules27103057
Keranmu A, Pan L-B, Fu J, Han P, Yu H, Zhang Z-W, Xu H, Yang X-Y, Hu J-C, Zhang H-J, et al. Biotransformation of Liquiritigenin into Characteristic Metabolites by the Gut Microbiota. Molecules. 2022; 27(10):3057. https://doi.org/10.3390/molecules27103057
Chicago/Turabian StyleKeranmu, Adili, Li-Bin Pan, Jie Fu, Pei Han, Hang Yu, Zheng-Wei Zhang, Hui Xu, Xin-Yu Yang, Jia-Chun Hu, Hao-Jian Zhang, and et al. 2022. "Biotransformation of Liquiritigenin into Characteristic Metabolites by the Gut Microbiota" Molecules 27, no. 10: 3057. https://doi.org/10.3390/molecules27103057
APA StyleKeranmu, A., Pan, L.-B., Fu, J., Han, P., Yu, H., Zhang, Z.-W., Xu, H., Yang, X.-Y., Hu, J.-C., Zhang, H.-J., Bu, M.-M., Jiang, J.-D., Xing, N.-Z., & Wang, Y. (2022). Biotransformation of Liquiritigenin into Characteristic Metabolites by the Gut Microbiota. Molecules, 27(10), 3057. https://doi.org/10.3390/molecules27103057






