Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice
Abstract
1. Introduction
2. Results
2.1. Fragmentation of LW6
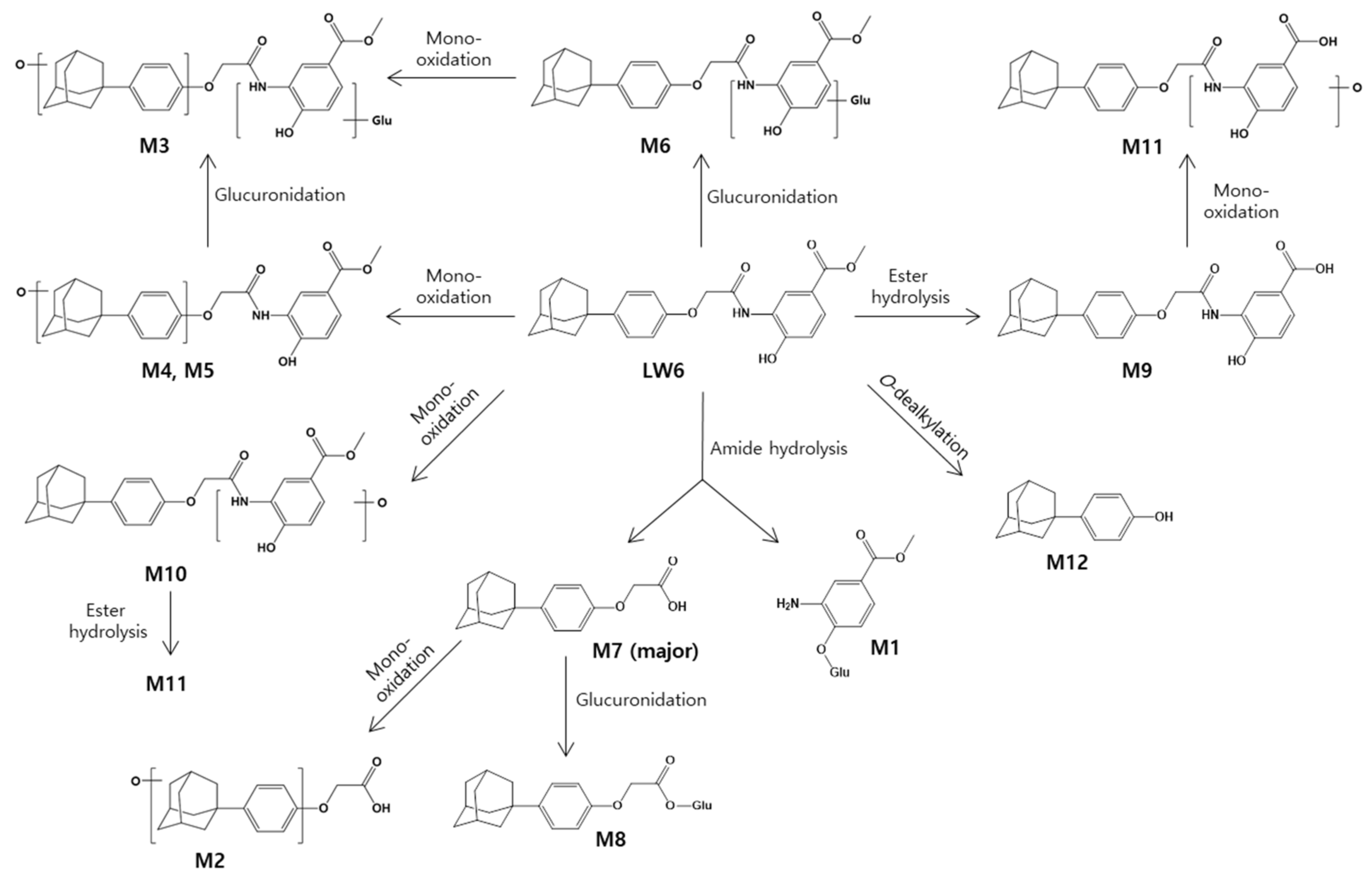
2.2. Metabolite Profiling of LW6 in Serial Mouse Plasma
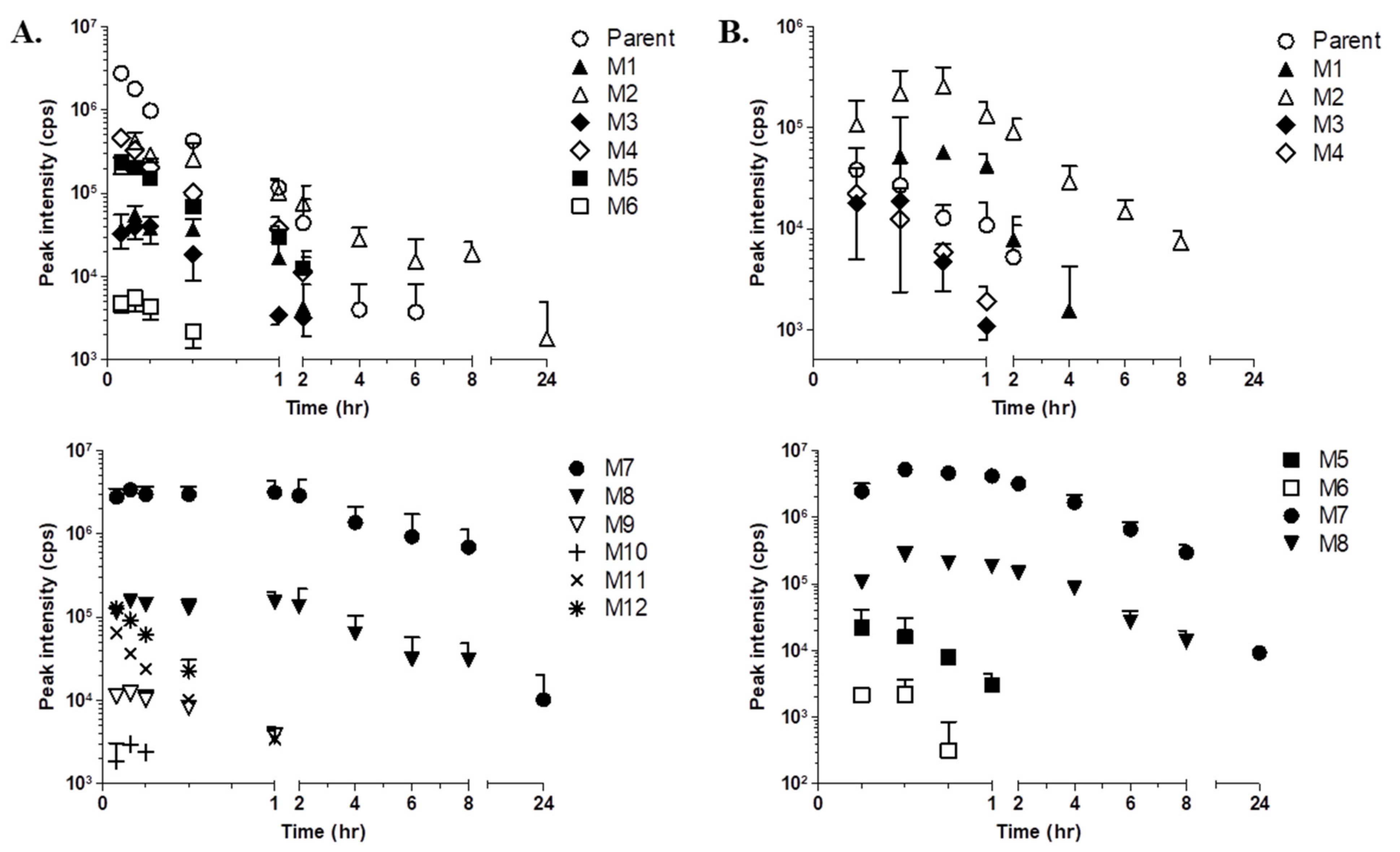
2.3. Time Profiling and Relative Quantification of LW6 and its Metabolites
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animal Study
4.3. Sample Preparation
4.4. Chromatographic and Mass Spectrometric Methods
4.5. Predicted MRM-Enhanced Product Ion Scan (pMRM-EPI) Method
4.6. Data Processing Method and Set Criteria for Metabolite Hits
4.7. Semi-Quantitation of Targeted Metabolites
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Carrero, P.; Okamoto, K.; Coumailleau, P.; O’Brien, S.; Tanaka, H.; Poellinger, L. Redox-regulated recruitment of the tran-scriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol. Cell Biol. 2000, 20, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Lando, D.; Pongratz, I.; Poellinger, L.; Whitelaw, M.L. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 2000, 275, 4618–4627. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia–A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Semenza, G.L. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol. Med. 2012, 9, 534–543. [Google Scholar] [CrossRef]
- Lee, K.; Kang, J.E.; Park, S.K.; Jin, Y.; Kim, S.; Lee, K.; Lee, M.R.; Yang, E.G.; Lee, J.J.; Won, M. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1 alpha via up regulation of VHL in a colon cancer cell line. Biochem. Pharmacol. 2010, 80, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Baranczewski, P.; Kautiainen, A.; Sandin, P.O. Introduction to early in vitro identification of me-tabolites of new chemical entities in drug discovery and development. Pharmacol. Rep. 2006, 58, 341–352. [Google Scholar]
- Pandey, S.; Tiwari, G. Bioanalysis in drug discovery and development. Pharm. Methods 2010, 1, 14–24. [Google Scholar] [CrossRef]
- Zhu, M.; Ma, L.; Zhang, D.; Ray, K.; Zhao, W.; Humphreys, W.G.; Skiles, G.; Sanders, M.; Zhang, H. Detection and charac-terization of metabolites in biological matrices using mass defect filtering of liquid chromatography/high resolution mass spectrometry data. Drug Metab. Dispos. 2006, 34, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Prakash, C.; Shaffer, C.L.; Nedderman, A. Analytical strategies for identifying drug metabolites. Mass Spectrom. Rev. 2007, 26, 340–369. [Google Scholar] [CrossRef]
- Baillie, T.A. Metabolism and toxicity of drugs. Two decades of progress in industrial drug metabolism. Chem. Res. Toxicol. 2008, 21, 129–137. [Google Scholar] [CrossRef]
- Gao, H.; Materne, O.L.; Howe, D.L.; Brummel, C.L. Method for rapid metabolite profiling of drug candidates in fresh hepatocytes using liquid chromatography coupled with a hybrid quadrupole linear ion trap. Rapid Commun. Mass. Spectrom. 2007, 21, 3683–3693. [Google Scholar] [CrossRef]
- Xia, Y.Q.; Miller, J.D.; Bakhtiar, R.; Franklin, R.B.; Liu, D.Q. Use of a quadrupole linear ion trap mass spectrometer in metabolite identification and bioanalysis. Rapid Commun. Mass. Spectrom. 2003, 17, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Li, A.C.; Alton, D.; Bryant, M.S.; Shou, W.Z. Simultaneously quantifying parent drugs and screening for metabolites in plasma pharmacokinetic samples using selected reaction monitoring information-dependent acquisition on a QTrap instrument. Rapid Commun Mass Spectrom. 2005, 19, 1943–1950. [Google Scholar] [CrossRef]
- Song, Y.L.; Jing, W.H.; Yan, R.; Wang, Y.T. Metabolic characterization of (±)-praeruptorin a in vitro and in vivo by high performance liquid chromatography coupled with hybrid triple quadrupole-linear ion trap mass spectrometry and time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2014, 90, 98–110. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, S.Y.; Lee, K.; Oh, S.J.; Kim, S.K. Determination of species-difference in microsomal metabolism of amitriptyline using a predictive MRM-IDA-EPI method. Chem. Biol. Interact. 2015, 229, 109–118. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Pace, N.; Kerns, E.H.; Kleintop, T.; Kagan, N.; Sakuma, T. Hybrid triple quadrupole-linear ion trap mass spec-trometry in fragmentation mechanism studies: Application to structure elucidation of buspirone and one of its metabolites. J Mass Spectrom. 2005, 40, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Rousu, T.; Herttuainen, J.; Tolonen, A. Comparison of triple quadrupole, hybrid linear ion trap triple quadrupole, time-of-flight and LTQ-Orbitrap mass spectrometers in drug discovery phase metabolite screening and identification in vitro--amitriptyline and verapamil as model compounds. Rapid Commun. Mass Spectrom. 2010, 24, 939–957. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Ma, L.; Humphreys, W.G.; Zhu, M. Rapid screening and characterization of drug metabolites using a multiple ion monitoring-dependent MS/MS acquisition method on a hybrid triple quadrupole-linear ion trap mass spectrometer. J. Mass Spectrom. 2008, 43, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Deng, S.; Obach, R.S. A simple liquid chromatography-tandem mass spectrometry method to determine relative plasma exposures of drug metabolites across species for metabolite safety assessments. Drug Metab. Dispos. 2010, 38, 2147–2156. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration Guidance for Industry: Safety Testing of Drug Metabolites. 2008. Available online: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079266.pdf (accessed on 13 January 2015).
- ICH Harmonised Tripartite Guideline, Guidance on Nonclinical Safety Studies for the Conduct of Human Clinical Trials and Marketing Authorization for Pharmaceuticals M3 (R2). 2009. Available online: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Multidisciplinary/M3_R2/Step4/M3_R2__Guideline.pdf (accessed on 13 January 2015).
- Shou, W.Z.; Magis, L.; Li, A.C.; Naidong, W.; Bryant, M.S. A novel approach to perform metabolite screening during the quantitative LC-MS/MS analyses of in vitro metabolic stability samples using a hybrid triple-quadrupole linear ion trap mass spectrometer. J. Mass Spectrom. 2005, 40, 1347–1356. [Google Scholar] [CrossRef]
- Huang, J.; Si, L.; Fan, Z.; Hu, L.; Qiu, J.; Li, G. In vitro metabolic stability and metabolite profiling of TJ0711 hydrochloride, a newly developed vasodilatory β-blocker, using a liquid chromatography-tandem mass spectrometry method. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011, 879, 3386–3392. [Google Scholar] [CrossRef]
- Lutz, J.D.; Fujioka, Y.; Isoherranen, N. Rationalization and prediction of in vivo metabolite exposures: The role of metabolite kinetics, clearance predictions and in vitro parameters. Expert Opin. Drug Metab. Toxicol. 2010, 6, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.X.; Rockafellow, B.A.; Skedzielewski, T.M.; Huebert, N.D.; Hafeman, W. Improved pharmacokinetic and bioavailability support of drug discovery using serial blood sampling in mice. J. Pharm. Sci. 2009, 98, 1877–1884. [Google Scholar] [CrossRef] [PubMed]
- Polson, C.; Sarkar, P.; Incledon, B.; Raguvaran, V.; Grant, R. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. J. Chromatogr. 2003, 785, 263–275. [Google Scholar] [CrossRef]
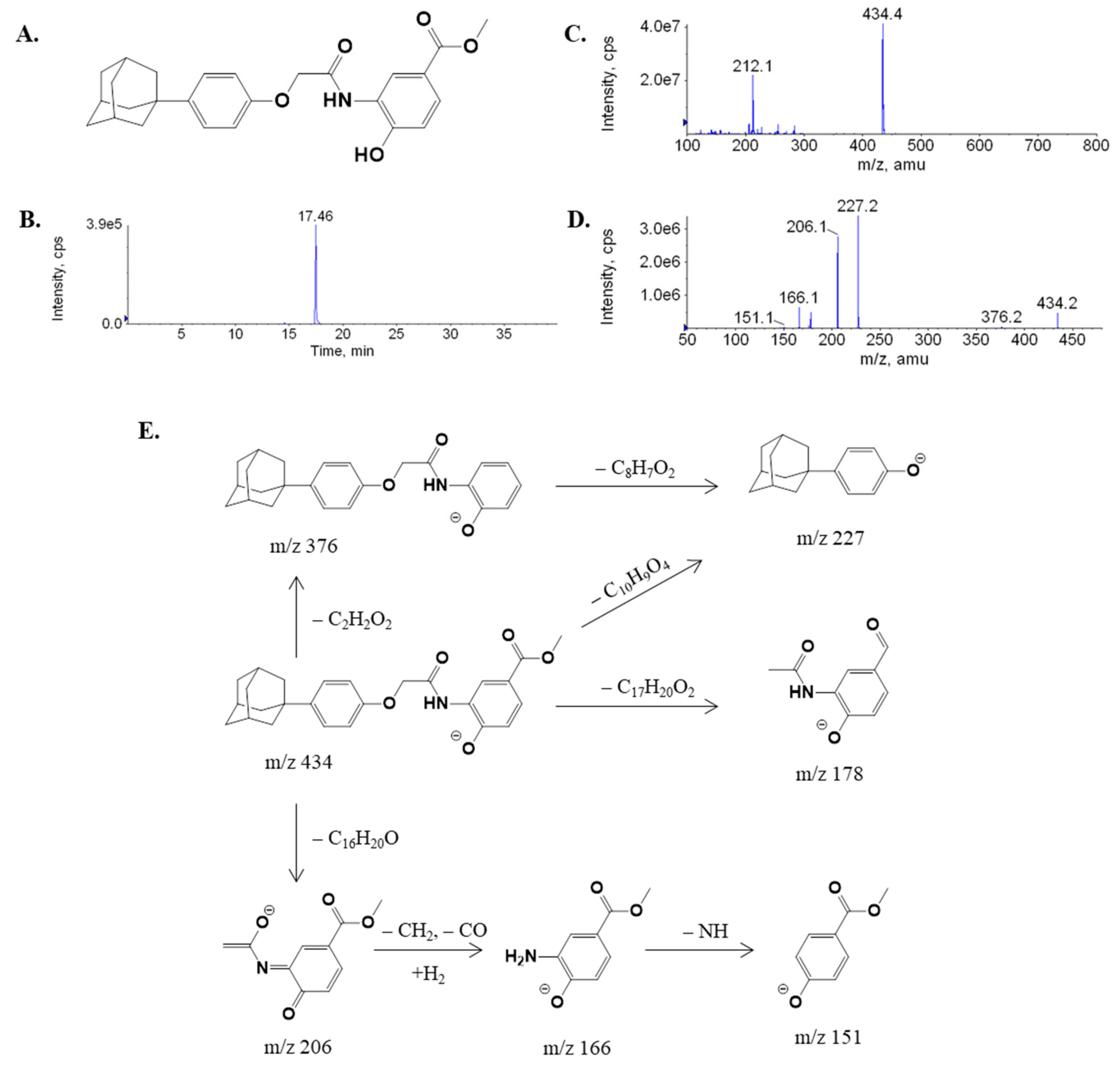

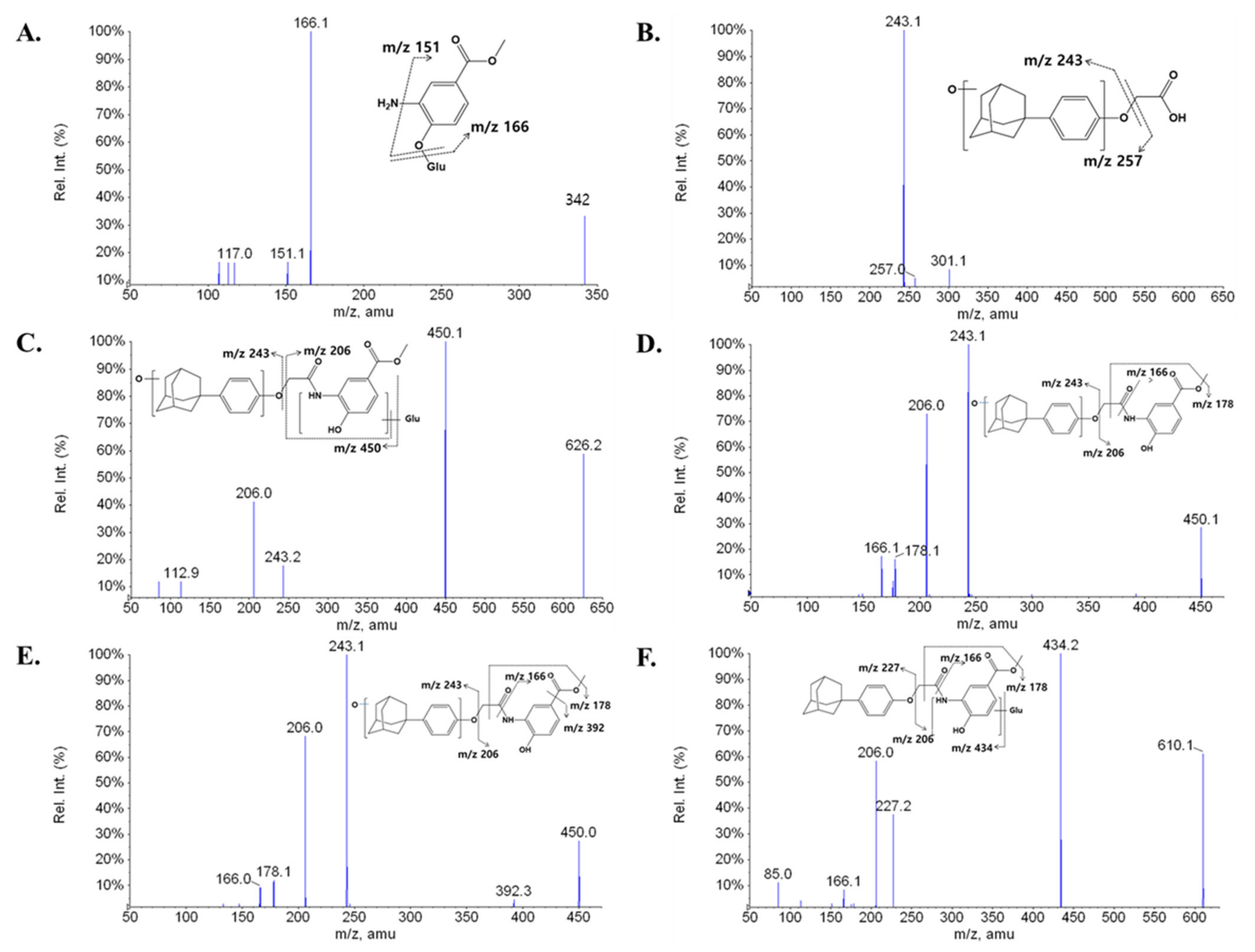
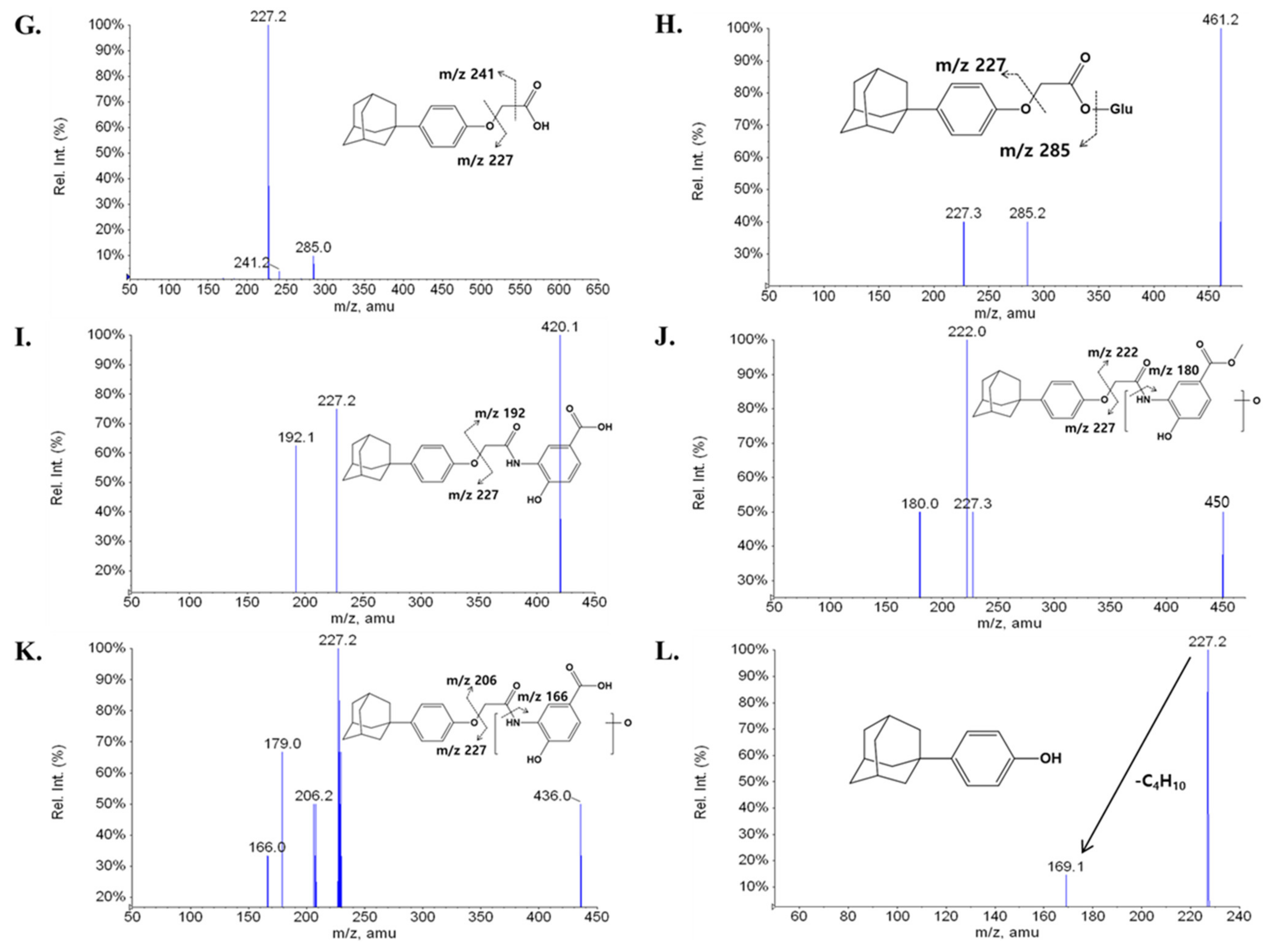
| Metabolite | Retention Time (min) | [M-H]− | Major Fragments |
|---|---|---|---|
| LW6 (parent) | 17.46 | 434 | 227, 206,166 |
| APA (M7) | 15.73 | 285 | 241, 227 |
| M1 | 2.84 | 342 | 166, 151, 117 |
| M2 | 10.62 | 301 | 257, 243 |
| M3 | 10.96 | 626 | 450, 243, 206, 113, 85 |
| M4 | 13.22 | 450 | 243, 206, 178, 166 |
| M5 | 13.37 | 450 | 392, 243, 206, 178, 166 |
| M6 | 14.59 | 610 | 434, 227, 206, 178, 166, 85 |
| M8 | 15.76 | 461 | 285, 227 |
| M9 | 16.09 | 420 | 227, 192 |
| M10 | 16.81 | 450 | 227, 222, 180 |
| M11 | 17.49 | 436 | 227, 206, 179, 166 |
| M12 | 17.48 | 227 | 227, 169 |
| Metabolite | i.v. | p.o. | ||||
|---|---|---|---|---|---|---|
| Average AUC0→t (Peak Intensity, Cps) | Relative % of Parent AUC0→t (i.v.) 1 | Relative % of Total Metabolite AUC0→t 2 | Average AUC0→t (Peak inTensity, cps) | Relative % of Parent AUC0→t (p.o.) 1 | Relative % of Total Metabolite AUC0→t 2 | |
| LW6 (Parent) | 1,048,883 | 100.00 | - | 28,735 | 2.74 | - |
| APA (M7) | 19,697,150 | 1877.92 | 92.09 | 17,688,080 | 1686.37 | 93.32 |
| M1 | 42,982 | 4.10 | 0.20 | 66,545 | 6.34 | 0.35 |
| M2 | 653,800 | 62.33 | 3.06 | 457,710 | 43.64 | 2.41 |
| M3 | 23,904 | 2.28 | 0.11 | 10,404 | 0.99 | 0.05 |
| M4 | 170,950 | 16.30 | 0.80 | 10,340 | 0.99 | 0.05 |
| M5 | 116,583 | 11.11 | 0.55 | 11,845 | 1.13 | 0.06 |
| M6 | 2445 | 0.23 | 0.01 | 1127 | 0.11 | 0.01 |
| M8 | 622,040 | 59.30 | 2.91 | 708,375 | 67.54 | 3.74 |
| M9 | 7672 | 0.73 | 0.04 | - | - | - |
| M10 | 511 | 0.05 | 0.00 | - | - | - |
| M11 | 17,015 | 1.62 | 0.08 | - | - | - |
| M12 | 31,345 | 2.99 | 0.15 | - | - | - |
| Type of Biotransformation | [M-H]− m/z | pMRM |
|---|---|---|
| Parent (LW6) | 434 | 434 > 227 |
| Mono-oxidation | 450 | 450 > 227, 450 > 243 |
| Di-oxidation | 466 | 466 > 227, 466 > 243 |
| Glucuronidation | 610 | 610 > 227, 610 > 243 |
| Mono-oxidation + glucuronidation | 626 | 626 > 450, 626 > 243 |
| Ester hydrolysis | 420 | 420 > 227 |
| Ester hydrolysis + mono-oxidation | 436 | 436 > 227, 436 > 243 |
| Ester hydrolysis + mono-oxidation + glucuronidation | 612 | 612 > 227, 612 > 243 |
| Ester hydrolysis + glucuronidation | 596 | 596 > 420, 596 > 227 |
| Amide hydrolysis (amine part) | 166 | 166 > 166 |
| Amide hydrolysis + ester hydrolysis | 152 | 152 > 152 |
| Amide hydrolysis + glucuronidation | 342 | 342 > 166 |
| Amide hydrolysis + ester hydrolysis + glucuronidation | 328 | 328 > 152 |
| Amide hydrolysis (acid part) | 285 | 285 > 227 |
| Amide hydrolysis + glucuronidation | 461 | 461 > 227 |
| Amide hydrolysis + mono-oxidation | 301 | 301 > 243 |
| Amide hydrolysis + mono-oxidation + glucuronidation | 477 | 477 > 301, 477 > 243 |
| O-dealkylation (alcohol part) | 227 | 227 > 227 |
| O-dealkylation + glucuronidation | 403 | 403 > 227 |
| O-dealkylation + mono-oxidation | 243 | 243 > 243 |
| O-dealkylation + mono-oxidation + glucuronidation | 419 | 419 > 243 |
| O-dealkylation (aldehyde part) | 222 | 222 > 222 |
| O-dealkylation + glucuronidation | 398 | 398 > 222 |
| O-dealkylation + aldehyde oxidation | 238 | 238 > 238 |
| O-dealkylation + aldehyde oxidation + glucuronidation | 414 | 414 > 238 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Lee, J.-Y.; Lee, K.; Jung, C.-R.; Kim, M.J.; Kim, J.A.; Yoo, D.G.; Shin, E.J.; Oh, S.J. Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice. Molecules 2021, 26, 1951. https://doi.org/10.3390/molecules26071951
Lee K, Lee J-Y, Lee K, Jung C-R, Kim MJ, Kim JA, Yoo DG, Shin EJ, Oh SJ. Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice. Molecules. 2021; 26(7):1951. https://doi.org/10.3390/molecules26071951
Chicago/Turabian StyleLee, Kiho, Ji-Yoon Lee, Kyeong Lee, Cho-Rock Jung, Min Ju Kim, Jung Ah Kim, Dong Gu Yoo, Eun Jin Shin, and Soo Jin Oh. 2021. "Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice" Molecules 26, no. 7: 1951. https://doi.org/10.3390/molecules26071951
APA StyleLee, K., Lee, J.-Y., Lee, K., Jung, C.-R., Kim, M. J., Kim, J. A., Yoo, D. G., Shin, E. J., & Oh, S. J. (2021). Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice. Molecules, 26(7), 1951. https://doi.org/10.3390/molecules26071951






