Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine
Abstract
1. Introduction
2. Results
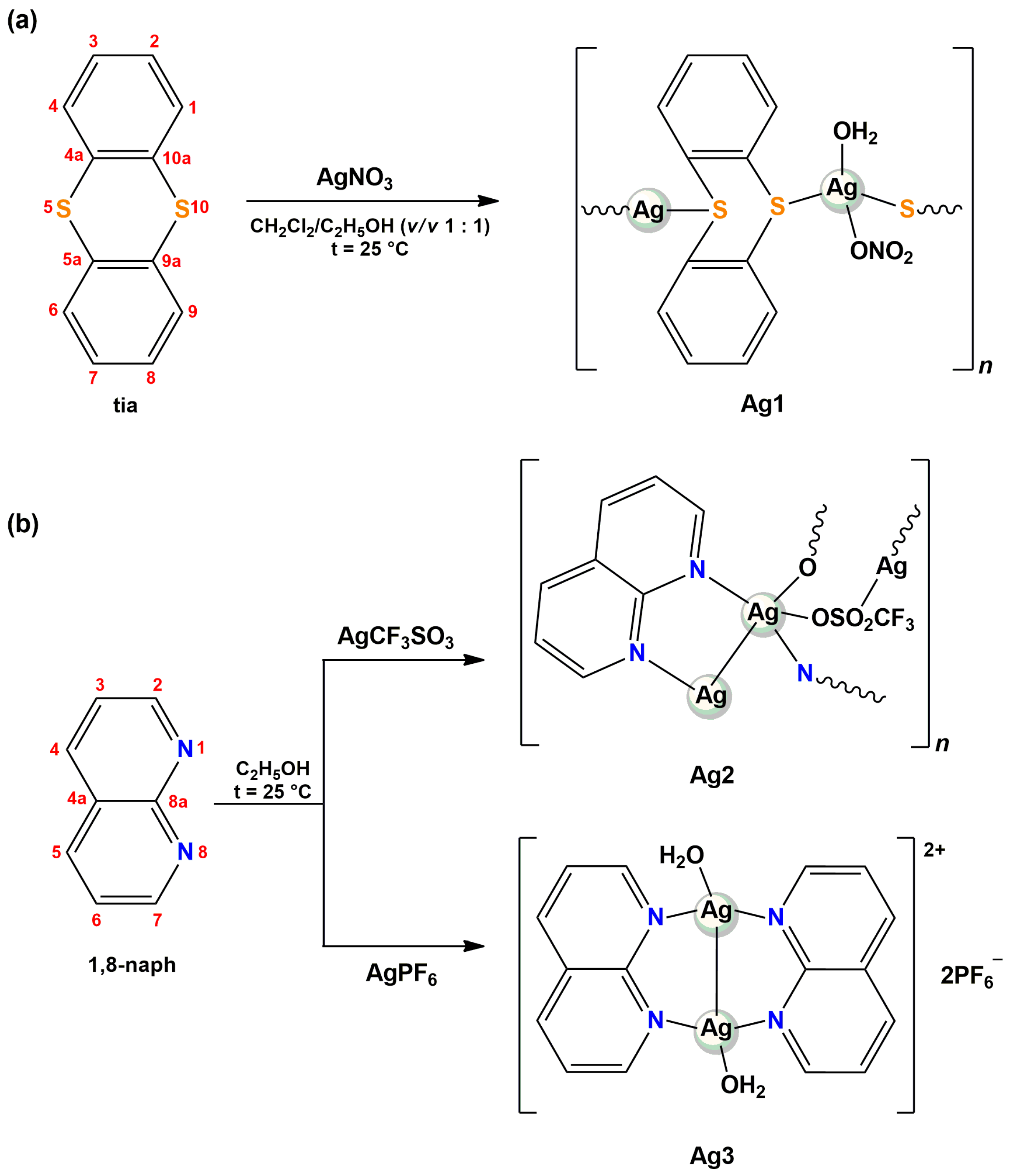
2.1. Synthesis and Characterization of Silver(I) Complexes Ag1–3
2.1.1. Solid-State Characterization
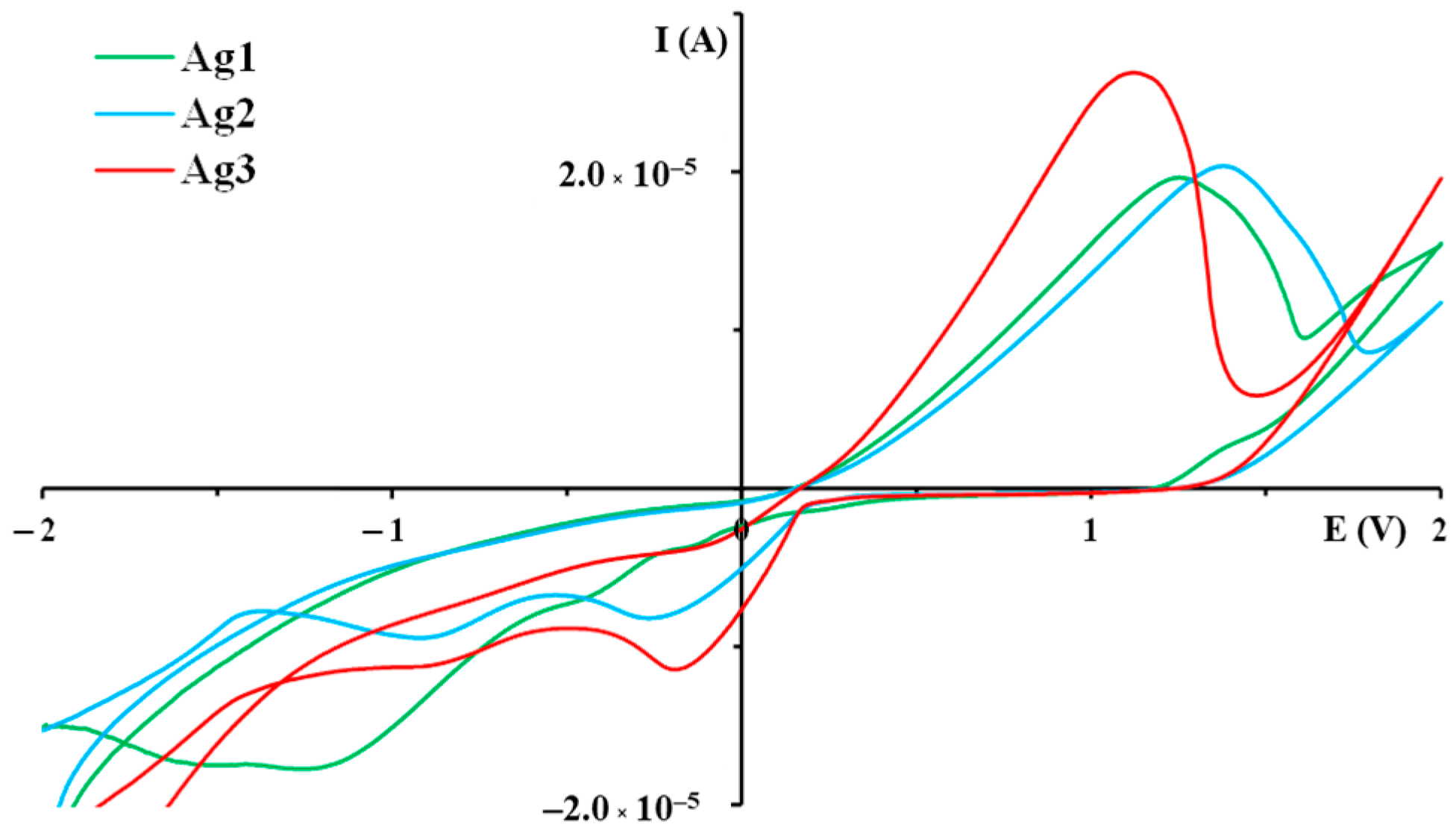
2.1.2. Solution Behavior
2.2. Biological Evaluation of the Silver(I) Complexes Ag1–3
2.2.1. Antimicrobial and Antiproliferative Effect of Silver(I) Complexes Ag1–3
2.2.2. C. elegans Toxicity
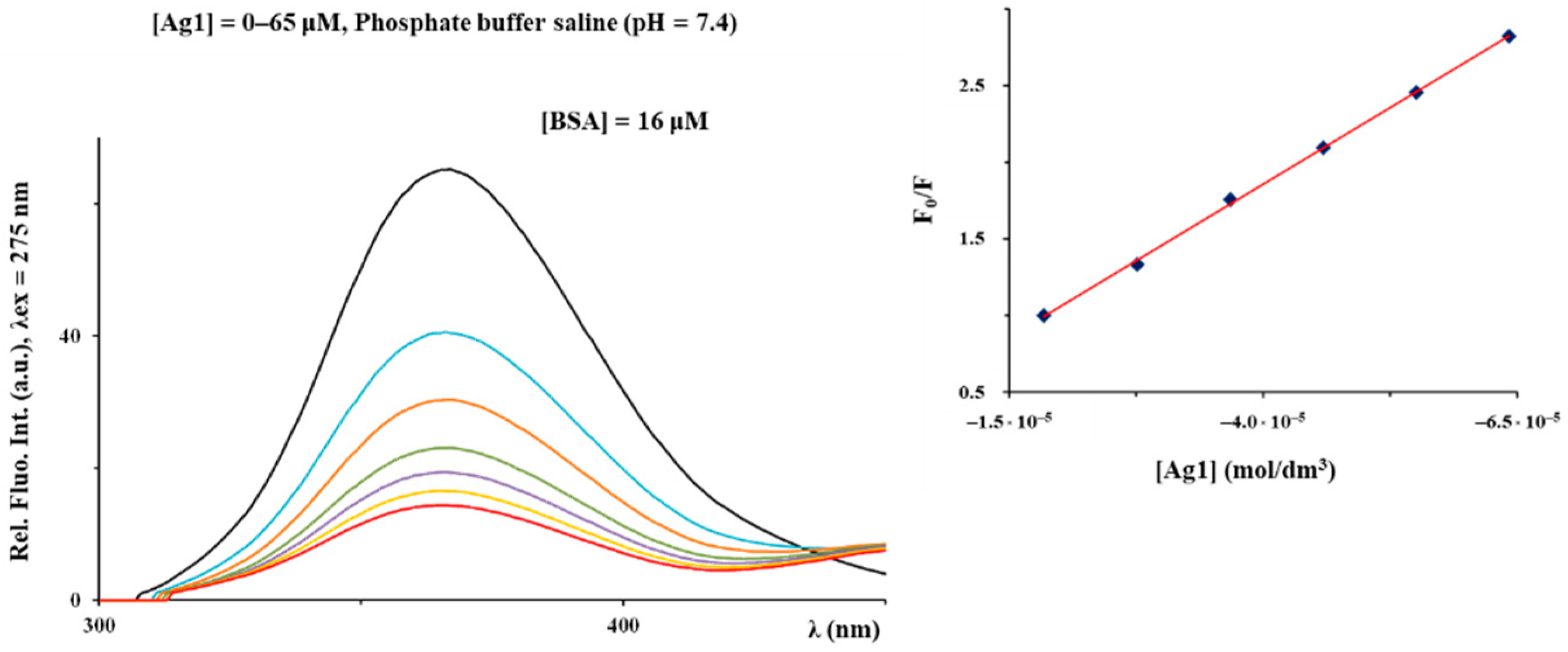
2.3. BSA Binding Study
2.4. Lipophilicity Assay
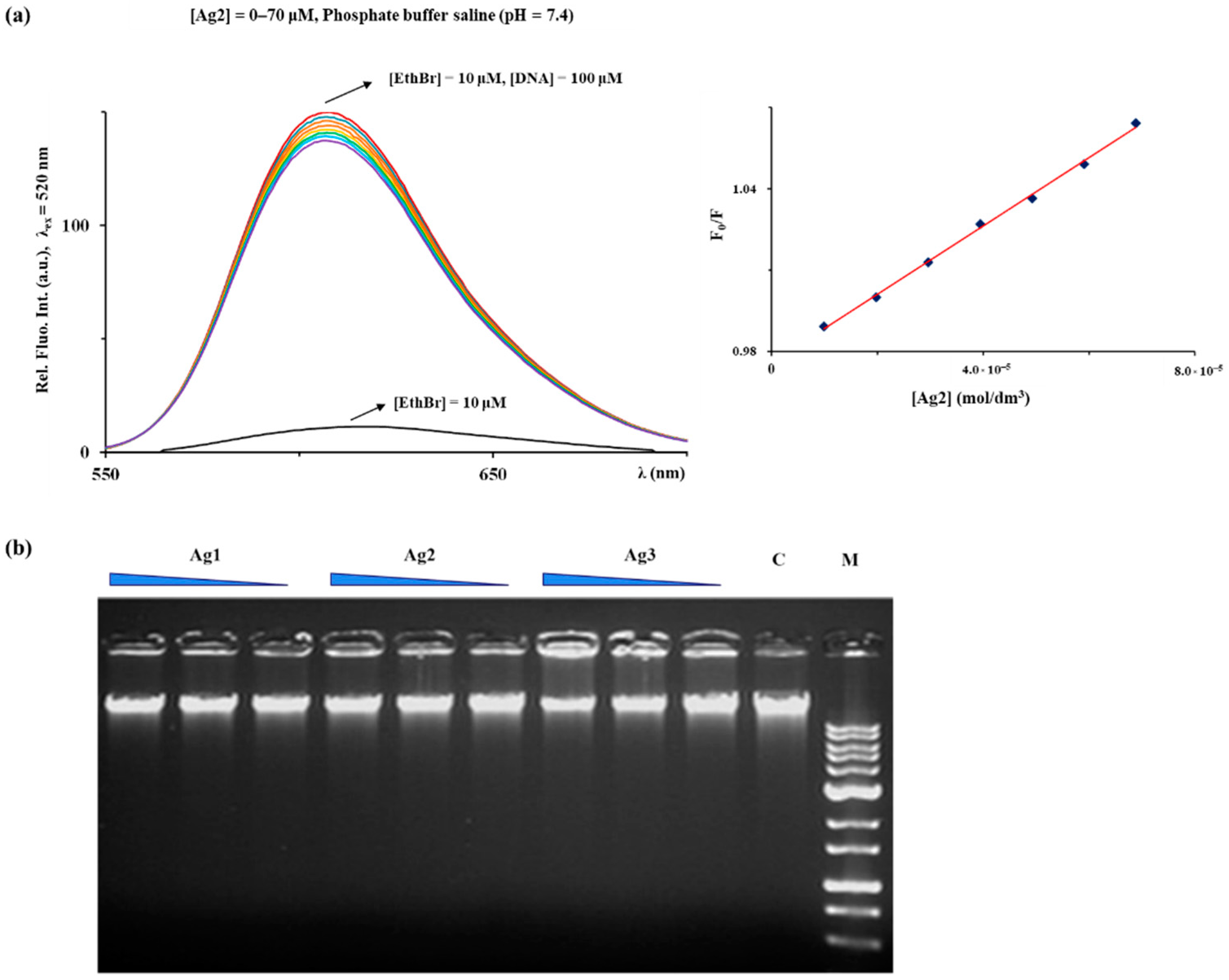
2.5. DNA Interaction
3. Materials and Methods
3.1. Materials
3.2. Measurements
3.3. Synthesis of Silver(I) Complexes Ag1–3
3.4. Air/Light Stability
3.5. Crystallographic Data Collection and Refinement of the Structures
3.6. Antimicrobial Studies
3.7. Toxicity Assessment
3.7.1. MTT Assay
3.7.2. C. elegans Survival Assay
3.8. BSA Binding Study
3.9. Lipophilicity Assay
3.10. DNA Interaction
3.10.1. Fluorescence Emission Spectroscopy
3.10.2. Gel Electrophoresis Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Tacconelli, E.; Pezzani, M.D. Public health burden of antimicrobial resistance in Europe. Lancet Infect. Dis. 2019, 19, 4–6. [Google Scholar] [CrossRef]
- O’Neill, J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Wellcome Trust: London, UK, 2014. [Google Scholar]
- Barillo, D.J.; Marx, D.E. Silver in medicine: A brief history BC 335 to present. Burns 2014, 40, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Huang, Y.-Y.; Sharma, S.K.; Hashmi, J.T.; Kurup, D.B.; Hamblin, M.R. Topical antimicrobials for burn wound infections. Recent Pat. Antiinfect. Drug Discov. 2010, 5, 124–151. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.H.B.; Bergamini, F.R.G.; Lustri, W.R.; de Paiva, P.P.; Ruiz, A.L.T.G.; de Carvalho, J.E.; Corbi, P.P. Synthesis, characterization and in vitro biological assays of a silver(I) complex with 5-fluorouracil: A strategy to overcome multidrug resistant tumor cells. J. Fluor. Chem. 2017, 195, 93–101. [Google Scholar] [CrossRef]
- Cavicchioli, M.; Massabni, A.C.; Heinrich, T.A.; Costa-Neto, C.M.; Abrão, E.P.; Fonseca, B.A.L.; Castellano, E.E.; Corbi, P.P.; Lustri, W.R.; Leite, C.Q.F. Pt(II) and Ag(I) complexes with acesulfame: Crystal structure and a study of their antitumoral, antimicrobial and antiviral activities. J. Inorg. Biochem. 2010, 104, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Zachariadis, P.C.; Hadjikakou, S.K.; Hadjiliadis, N.; Skoulika, S.; Michaelides, A.; Balzarini, J.; De Clercq, E. Synthesis, characterization and in vitro study of the cytostatic and antiviral activity of new polymeric silver(I) complexes with ribbon structures derived from the conjugated heterocyclic thioamide 2-mercapto-3,4,5,6-tetra- hydropyrimidine. Eur. J. Inorg. Chem. 2004, 2004, 1420–1426. [Google Scholar] [CrossRef]
- Lansdown, A.B.G. Silver in health care: Antimicrobial effects and safety in use. Curr. Probl. Dermatol. 2006, 33, 17–34. [Google Scholar] [PubMed]
- Rizzello, L.; Pompa, P.P. Nanosilver-based antibacterial drugs and devices: Mechanisms, methodological drawbacks, and guidelines. Chem. Soc. Rev. 2014, 43, 1501–1518. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Zoroddu, M.A. Medical uses of silver: History, myths, and scientific evidence. J. Med. Chem. 2019, 62, 5923–5943. [Google Scholar] [CrossRef]
- Eckhardt, S.; Brunetto, P.S.; Gagnon, J.; Priebe, M.; Giese, B.; Fromm, K.M. Nanobio silver: Its interactions with peptides and bacteria, and its uses in medicine. Chem. Rev. 2013, 113, 4708–4754. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.H.B.; de Paiva, R.E.F.; Cuin, A.; Lustri, W.R.; Corbi, P.P. Silver complexes with sulfathiazole and sulfamethoxazole: Synthesis, spectroscopic characterization, crystal structure and antibacterial assays. Polyhedron 2015, 85, 437–444. [Google Scholar] [CrossRef]
- Tomašić, T.; Mašič, L.P. Rhodanine as a privileged scaffold in drug discovery. Curr. Med. Chem. 2009, 16, 1596–1629. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J. Biological and pharmacological activities of 1,3,4-thiadiazole based compounds. Mini Rev. Med. Chem. 2015, 15, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Ganguly, S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: A mini-review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef]
- Pathania, S.; Narang, R.K.; Rawal, R.K. Role of sulphur-heterocycles in medicinal chemistry: An update. Eur. J. Med. Chem. 2019, 180, 486–508. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef]
- Kim, D.E.; Kim, Y.; Cho, D.-H.; Jeong, S.-Y.; Kim, S.-B.; Suh, N.; Lee, J.S.; Choi, E.K.; Koh, J.-Y.; Hwang, J.J. Raloxifene induces autophagy-dependent cell death in breast cancer cells via the activation of AMP-activated protein kinase. Mol. Cells 2015, 38, 138–144. [Google Scholar] [CrossRef]
- Herdeiro, M.T.; Soares, S.; Silva, T.; Roque, F.; Figueiras, A. Impact of rosiglitazone safety alerts on oral antidiabetic sales trends: A countrywide study in Portugal. Fundam. Clin. Pharmacol. 2016, 30, 440–449. [Google Scholar] [CrossRef]
- Séïde, M.; Marion, M.; Mateescu, M.A.; Averill-Bates, D.A. The fungicide thiabendazole causes apoptosis in rat hepatocytes. Toxicol. In Vitro 2016, 32, 232–239. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, X.-M.; Damu, G.L.V.; Geng, R.-X.; Zhou, C.-H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2013, 34, 340–437. [Google Scholar] [CrossRef]
- Jain, S.; Chandra, V.; Jain, P.K.; Pathak, K.; Pathak, D.; Vaidya, A. Comprehensive review on current developments of quinoline-based anticancer agents. Arabian J. Chem. 2019, 12, 4920–4946. [Google Scholar] [CrossRef]
- Savić, N.D.; Glišić, B.Đ.; Wadepohl, H.; Pavic, A.; Senerovic, L.; Nikodinovic-Runic, J.; Djuran, M.I. Silver(I) complexes with quinazoline and phthalazine: Synthesis, structural characterization and evaluation of biological activities. MedChemComm 2016, 7, 282–291. [Google Scholar] [CrossRef]
- Savić, N.D.; Milivojevic, D.R.; Glišić, B.Đ.; Ilic-Tomic, T.; Veselinovic, J.; Pavic, A.; Vasiljevic, B.; Nikodinovic-Runic, J.; Djuran, M.I. A comparative antimicrobial and toxicological study of gold(III) and silver(I) complexes with aromatic nitrogen-containing heterocycles: Synergistic activity and improved selectivity index of Au(III)/Ag(I) complexes mixture. RSC Adv. 2016, 6, 13193–13206. [Google Scholar] [CrossRef][Green Version]
- Glišić, B.Đ.; Senerovic, L.; Comba, P.; Wadepohl, H.; Veselinovic, A.; Milivojevic, D.R.; Djuran, M.I.; Nikodinovic-Runic, J. Silver(I) complexes with phthalazine and quinazoline as effective agents against pathogenic Pseudomonas aeruginosa strains. J. Inorg. Biochem. 2016, 155, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Coyle, B.; Kavanagh, K.; McCann, M.; Devereux, M.; Geraghty, M. Mode of anti-fungal activity of 1,10-phenanthroline and its Cu(II), Mn(II) and Ag(I) complexes. Biometals 2003, 16, 321–329. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.; Coyle, B.; McKay, S.; McCormack, P.; Kavanagh, K.; Devereux, M.; McKee, V.; Kinsella, P.; O’Connor, R.; Clynes, M. Synthesis and X-ray crystal structure of [Ag(phendio)2]ClO4 (phendio = 1,10-phenanthroline-5,6-dione) and its effects on fungal and mammalian cells. Biometals 2004, 17, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Rowan, R.; Moran, C.; McCann, M.; Kavanagh, K. Use of Galleria mellonella larvae to evaluate the in vivo anti-fungal activity of [Ag2(mal)(phen)3]. Biometals 2009, 22, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Savić, N.D.; Vojnovic, S.; Glišić, B.Đ.; Crochet, A.; Pavic, A.; Janjić, G.V.; Pekmezović, M.; Opsenica, I.M.; Fromm, K.M.; Nikodinovic-Runic, J.; et al. Mononuclear silver(I) complexes with 1,7-phenanthroline as potent inhibitors of Candida growth. Eur. J. Med. Chem. 2018, 156, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Pavic, A.; Savić, N.D.; Glišić, B.Đ.; Crochet, A.; Vojnovic, S.; Kurutos, A.; Stanković, D.M.; Fromm, K.M.; Nikodinovic-Runic, J.; Djuran, M.I. Silver(I) complexes with 4,7-phenanthroline efficient in rescuing the zebrafish embryos of lethal Candida albicans infection. J. Inorg. Biochem. 2019, 195, 149–163. [Google Scholar] [CrossRef]
- Đurić, S.; Vojnovic, S.; Pavic, A.; Mojicevic, M.; Wadepohl, H.; Savić, N.D.; Popsavin, M.; Nikodinovic-Runic, J.; Djuran, M.I.; Glišić, B.Đ. New polynuclear 1,5-naphthyridine-silver(I) complexes as potential antimicrobial agents: The key role of the nature of donor coordinated to the metal center. J. Inorg. Biochem. 2020, 203, 110872. [Google Scholar] [CrossRef]
- Aslanidis, P.; Hatzidimitriou, A.G.; Andreadou, E.G.; Pantazaki, A.A.; Voulgarakis, N. Silver(I) complexes of N-methylbenzothiazole-2-thione: Synthesis, structures and antibacterial activity. Mat. Sci. Eng. C 2015, 50, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Aulakh, J.K.; Lobana, T.S.; Sood, H.; Arora, D.S.; Kaur, R.; Singh, J.; Garcia-Santos, I.; Kaure, M.; Jasinski, J.P. Silver derivatives of multi-donor heterocyclic thioamides as antimicrobial/anticancer agents: Unusual bio-activity against methicillin resistant S. aureus, S. epidermidis, and E. faecalis and human bone cancer MG63 cell line. RSC Adv. 2019, 9, 15470–15487. [Google Scholar] [CrossRef]
- Leung, M.C.K.; Williams, P.L.; Benedetto, A.; Au, C.; Helmcke, K.J.; Aschner, M.; Meyer, J.N. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008, 106, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Potapov, A.S.; Nudnova, E.A.; Khlebnikov, A.I.; Ogorodnikov, V.D.; Petrenko, T.V. Synthesis, crystal structure and electrocatalytic activity of discrete and polymeric copper(II) complexes with bitopic bis(pyrazol-1-yl)methane ligands. Inorg. Chem. Commun. 2015, 53, 72–75. [Google Scholar] [CrossRef]
- Johnston, D.H.; Shriver, D.F. Vibrational study of the trifluoromethanesulfonate anion: Unambiguous assignment of the asymmetric stretching modes. Inorg. Chem. 1993, 32, 1045–1047. [Google Scholar] [CrossRef]
- Van Albada, G.A.; Smeets, W.J.J.; Spek, A.L.; Reedijk, J. Synthesis, spectroscopic properties and X-ray crystal structures of two dinuclear alkoxo-bridged copper(II) compounds with the ligand bis(1-methyl-2-benzimidazolyl) propane. A unique alkoxo-bridged Cu(II) dinuclear compound with an additional bidentate bridging triflate anion. Inorg. Chim. Acta 1997, 260, 151–161. [Google Scholar]
- El Hamdani, H.; El Amane, M.; Duhayon, C. Studies on the syntheses, structural characterization, antimicrobial of the co-crystal 1,10-phenanthrolin-1-ium(1,10-phenH+)-caffeine(caf)-hexafluorophosphate. J. Mol. Struct. 2018, 1155, 789–796. [Google Scholar] [CrossRef]
- Nakajima, Y.; Shiraishi, Y.; Tsuchimoto, T.; Ozawa, F. Synthesis and coordination behavior of CuI bis(phosphaethenyl)pyridine complexes. Chem. Commun. 2011, 47, 6332–6334. [Google Scholar] [CrossRef][Green Version]
- Munakata, M.; Yan, S.G.; Ino, I.; Kuroda-Sowa, T.; Maekawa, M.; Suenaga, Y. Synthesis and structure of a novel bis(μ-η2-thianthrene )disilver(I) bis(perchlorate). Inorg. Chim. Acta 1998, 271, 145–150. [Google Scholar] [CrossRef]
- Glišić, B.Đ.; Warżajtis, B.; Hoffmann, M.; Rychlewska, U.; Djuran, M.I. Mononuclear gold(III) complexes with diazanaphthalenes: The influence of the position of nitrogen atoms in the aromatic rings on the complex crystalline properties. RSC Adv. 2020, 10, 44481–44493. [Google Scholar] [CrossRef]
- Zhang, J.-A.; Pan, M.; Zhang, J.-Y.; Zhang, H.-K.; Fan, Z.-J.; Kang, B.-S.; Su, C.-Y. Syntheses, structures and bioactivities of silver(I) complexes with a tridentate heterocyclic N- and S-ligand. Polyhedron 2009, 28, 145–149. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, C.-F.; Zheng, Z.; He, J.-B.; Wang, Y. Synthesis, characterization and antibacterial activity of a biocompatible silver complex based on 2,2′-bipyridine and 5-sulfoisophthalate. Inorg. Chim. Acta 2016, 451, 143–147. [Google Scholar] [CrossRef]
- Kalinowska-Lis, U.; Felczak, A.; Chęcińska, L.; Zawadzka, K.; Patyna, E.; Lisowska, K.; Ochocki, J. Synthesis, characterization and antimicrobial activity of water-soluble silver(I) complexes of metronidazole drug and selected counter-ions. Dalton Trans. 2015, 44, 8178–8189. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.M.S.; Correia, I.; Galvão, A.M.; Marques, F.; Carvalho, M.F.N.N. Synthesis of Ag(I) camphor sulphonylimine complexes and assessment of their cytotoxic properties against cisplatin-resistant A2780cisR and A2780 cell lines. J. Inorg. Biochem. 2017, 166, 55–63. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Wu, J.-Y.; Pan, Y.-L.; Zhang, X.-J.; Sun, T.; Tian, Y.-P.; Yang, J.-X.; Chen, Z.-N. Synthesis, photoluminescence and electrochemical properties of a series of carbazole-functionalized ligands and their silver(I) complexes. Inorg. Chim. Acta 2007, 360, 2083–2091. [Google Scholar] [CrossRef]
- Połczyński, P.; Jurczakowski, R.; Grochala, W. Strong and long-lived free-radical oxidizer based on silver(II). Mechanism of Ag(I) electrooxidation in concentrated H2SO4. J. Phys. Chem. C 2013, 117, 20689–20696. [Google Scholar] [CrossRef]
- Andrejević, T.P.; Milivojevic, D.; Glišić, B.Đ.; Kljun, J.; Stevanović, N.L.; Vojnovic, S.; Medic, S.; Nikodinovic-Runic, J.; Turel, I.; Djuran, M.I. Silver(I) complexes with different pyridine-4,5-dicarboxylate ligands as efficient agents for the control of cow mastitis associated pathogens. Dalton Trans. 2020, 49, 6084–6096. [Google Scholar] [CrossRef] [PubMed]
- Milivojevic, D.; Šumonja, N.; Medić, S.; Pavic, A.; Moric, I.; Vasiljevic, B.; Senerovic, L.; Nikodinovic-Runic, J. Biofilm-forming ability and infection potential of Pseudomonas aeruginosa strains isolated from animals and humans. Pathog. Dis. 2018, 76, fty041. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, S.K. Anti-proliferative and anti-tumor activity of silver(I) compounds. Metallomics 2013, 5, 569–596. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed]
- Ortego, L.; Gonzalo-Asensio, J.; Laguna, A.; Villacampa, M.D.; Gimeno, M.C. (Aminophosphane)gold(I) and silver(I) complexes as antibacterial agents. J. Inorg. Biochem. 2015, 146, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Francius, G.; Domenech, O.; Mingeot-Leclercq, M.P.; Dufrêne, Y.F. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin. J. Bacteriol. 2008, 190, 7904–7909. [Google Scholar] [CrossRef]
- Xiong, H.; Pears, C.; Woollard, A. An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 2017, 7, 9839. [Google Scholar] [CrossRef]
- Hunt, P.R. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017, 37, 50–59. [Google Scholar] [CrossRef]
- Shi, J.-H.; Pan, D.-Q.; Jiang, M.; Liu, T.-T.; Wang, Q. In vitro study on binding interaction of quinapril with bovine serum albumin (BSA) using multi-spectroscopic and molecular docking methods. J. Biomol. Struct. Dyn. 2017, 35, 2211–2223. [Google Scholar] [CrossRef]
- Shi, J.-H.; Zhu, Y.-Y.; Wang, J.; Chen, J.; Shen, Y.-J. Intermolecular interaction of prednisolone with bovine serum albumin: Spectroscopic and molecular docking methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 103, 287–294. [Google Scholar] [CrossRef]
- Anbazhagan, V.; Renganathan, R. Study on the binding of 2,3-diazabicyclo [2.2.2]oct-2-ene with bovine serum albumin by fluorescence spectroscopy. J. Lumin. 2008, 128, 1454–1458. [Google Scholar] [CrossRef]
- Li, Z.; Niu, M.; Chang, G.; Zhao, G.C. Chiral manganese(IV) complexes derived from Schiff base ligands: Synthesis, characterization, in vitro cytotoxicity and DNA/BSA interaction. J. Photochem. Photobiol. B Biol. 2015, 153, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Kumbhar, A.A.; Heptullah, H.; Khan, A.A.; Gobre, V.V.; Gejji, S.P.; Puranik, V.G. Synthesis, electronic structure, DNA and protein binding, DNA cleavage, and anticancer activity of fluorophore-labeled copper(II) complexes. Inorg. Chem. 2011, 50, 545–558. [Google Scholar] [CrossRef]
- Đurić, S.Ž.; Vojnovic, S.; Andrejević, T.P.; Stevanović, N.L.; Savić, N.D.; Nikodinovic-Runic, J.; Glišić, B.Đ.; Djuran, M.I. Antimicrobial activity and DNA/BSA binding affinity of polynuclear silver(I) complexes with 1,2-bis(4-pyridyl)ethane/ethene as bridging ligands. Bioinorg. Chem. Appl. 2020, 2020, 3812050. [Google Scholar] [CrossRef]
- Smolénski, P.; Pettinari, C.; Marchetti, F.; Guedes da Silva, M.F.C.; Lupidi, G.; Patzmay, G.V.B.; Petrelli, D.; Vitali, L.A.; Pomberio, A.J.L. Syntheses, structures, and antimicrobial activity of new remarkably light-stable and water-soluble tris(pyrazolyl)methanesulfonate silver(I) derivatives of N-methyl-1,3,5-triaza-7-phosphaadamantane salt—[mPTA]BF4. Inorg. Chem. 2015, 54, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Timerbaev, A.R.; Hartinger, C.G.; Aleksenko, S.S.; Keppler, B.K. Interactions of antitumor metallodrugs with serum proteins: advances in characterization using modern analytical methodology. Chem. Rev. 2006, 106, 2224–2248. [Google Scholar] [CrossRef]
- Shi, Y.; Guo, C.; Sun, Y.; Liu, Z.; Xu, F.; Zhang, Y.; Wen, Z.; Li, Z. Interaction between DNA and microcystin-LR studied by spectra analysis and atomic force microscopy. Biomacromolecules 2011, 12, 797–803. [Google Scholar] [CrossRef]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-ligand copper(II)-phenolate complexes: effect of coligand on enhanced DNA and protein binding, DNA cleavage, and anticancer activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef] [PubMed]
- Fetzer, L.; Boff, B.; Ali, M.; Xiangjun, M.; Collin, J.-P.; Sirlin, C.; Gaiddon, C.; Pfeffer, M. Library of second-generation cycloruthenated compounds and evaluation of their biological properties as potential anticancer drugs: Passing the nanomolar barrier. Dalton Trans. 2011, 40, 8869–8878. [Google Scholar] [CrossRef]
- Li, J.J.; Tian, M.; Tian, Z.; Zhang, S.; Yan, C.; Shao, C.; Liu, Z. Half-sandwich iridium(III) and ruthenium(II) complexes containing P^P-chelating ligands: A new class of potent anticancer agents with unusual redox features. Inorg. Chem. 2018, 57, 1705–1716. [Google Scholar] [CrossRef]
- Puckett, C.A.; Barton, J.K. Methods to explore cellular uptake of ruthenium complexes. J. Am. Chem. Soc. 2007, 129, 46–47. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Turel, I.; Kljun, J. Interactions of metal ions with DNA, its constituents and derivatives, which may be relevant for anticancer research. Curr. Top. Med. Chem. 2011, 11, 2661–2687. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-L.; Li, W.-Y.; He, X.-W.; Miao, K.; Liang, H. Spectral studies of the binding of lucigenin, a bisacridinium derivative, with double-helix DNA. Anal. Bioanal. Chem. 2002, 373, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Movahedi, E.; Rezvani, A.R.; Razmazma, H. Binding interaction of a heteroleptic silver(I) complex with DNA: A joint experimental and computational study. Int. J. Biol. Macromol. 2019, 126, 1244–1254. [Google Scholar] [CrossRef]
- Ašanin, D.P.; Andrejević, T.P.; Skaro-Bogojevic, S.; Stevanović, N.L.; Aleksic, I.; Milivojevic, D.; Perdih, F.; Turel, I.; Djuran, M.I.; Glišić, B.Đ. Polynuclear silver(I) complex with thianthrene: Structural characterization, antimicrobial activity and interaction with biomolecules. Proceedings 2020, 67, 4. [Google Scholar] [CrossRef]
- Ašanin, D.P.; Andrejević, T.P.; Skaro-Bogojevic, S.; Perdih, F.; Turel, I.; Nikodinovic-Runic, J.; Djuran, M.I.; Glišić, B.Đ. Antimicrobial activity and DNA/BSA binding study of new silver(I) complexes with 1,8-naphthyridine. In Proceedings of the 6th International Electronic Conference on Medicinal Chemistry, Session General: Presentations, 1–30 November 2020. [Google Scholar] [CrossRef]
- Agilent Technologies Ltd. CrysAlisPro, Version 1.171.39.46; Agilent Technologies: Yarnton, UK, 2013. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Cuenca-Estrella, M.; Lass-Flörl, C.; Hope, W. EUCAST-AFST. EUCAST technical note on the EUCAST definitive document EDef 7.2: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 2012, 18, E246–E247. [Google Scholar] [CrossRef]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef]
- Stiernagle, T. Maintenance of C. elegans. In WormBook; Oxford University Press: Oxford, UK, 2006; pp. 1–11. [Google Scholar]
- Scoffone, V.C.; Chiarelli, L.R.; Makarov, V.; Brackman, G.; Israyilova, A.; Azzalin, A.; Forneris, F.; Riabova, O.; Savina, S.; Coenye, T.; et al. Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo. Sci. Rep. 2016, 6, 32487. [Google Scholar] [CrossRef]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H., Jr.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Bera, R.; Sahoo, B.K.; Ghosh, K.S.; Dasgupta, S. Studies on the interaction of isoxazolcurcumin with calf thymus DNA. Int. J. Biol. Macromol. 2008, 42, 14–21. [Google Scholar] [CrossRef] [PubMed]
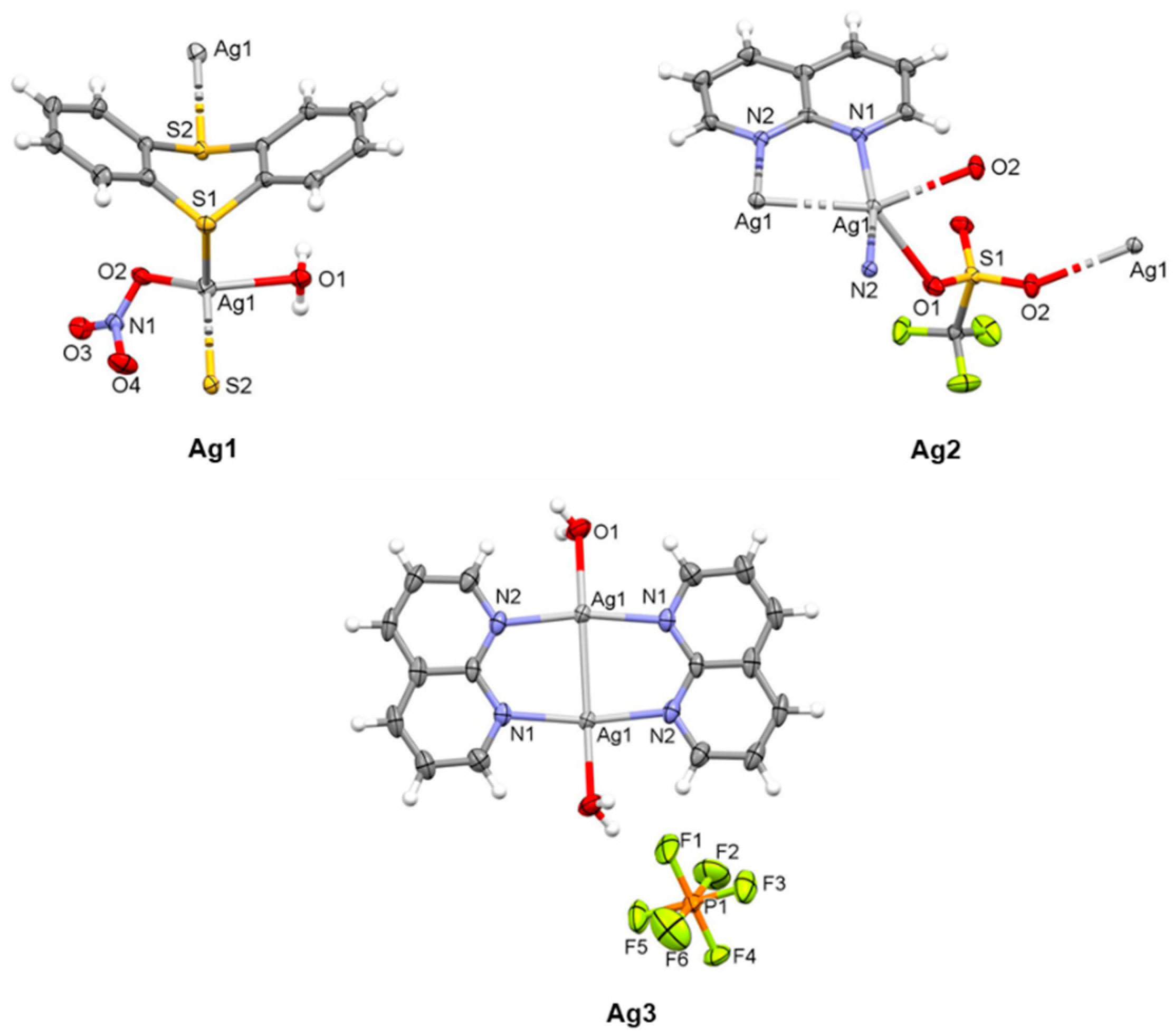
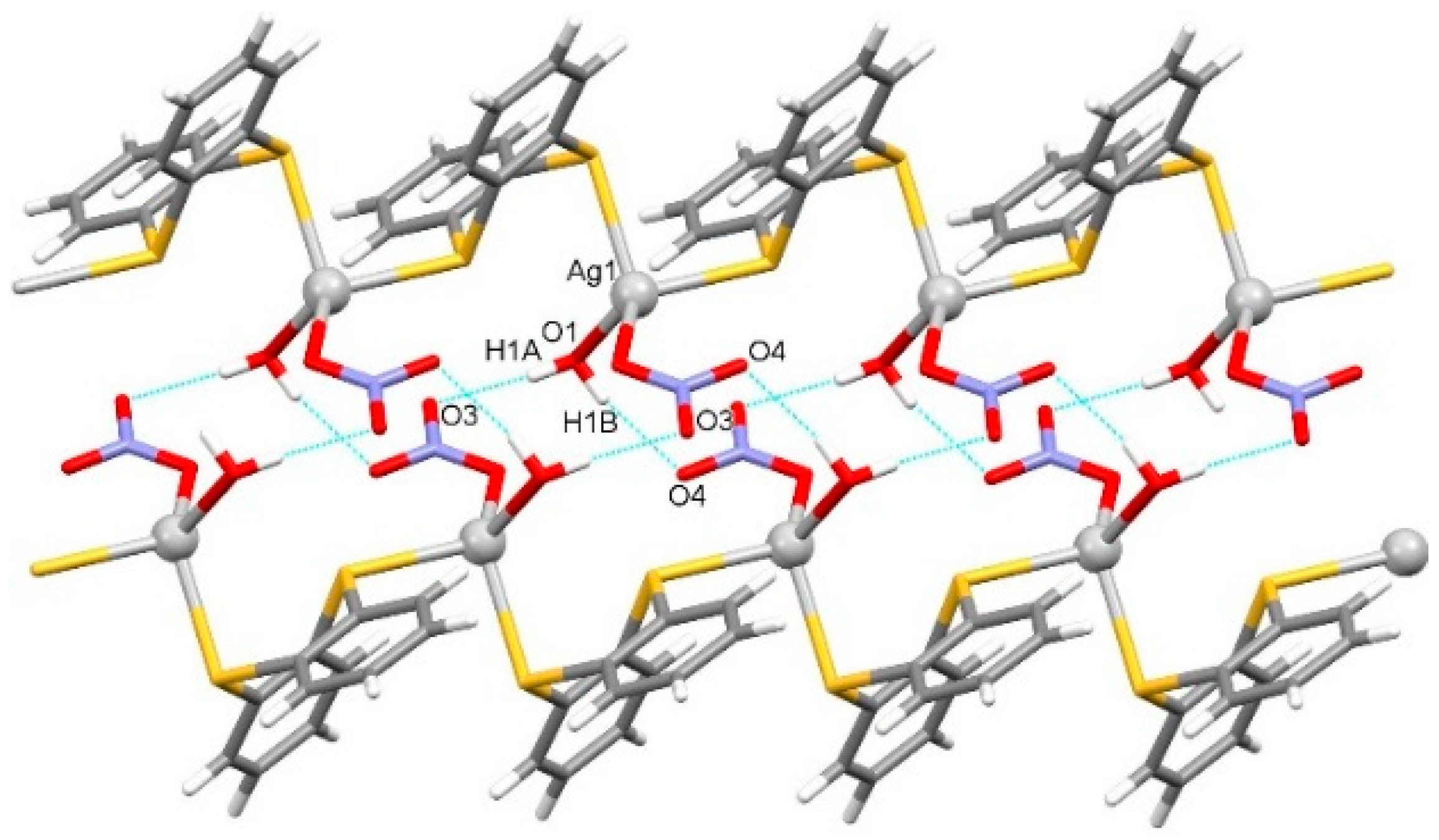
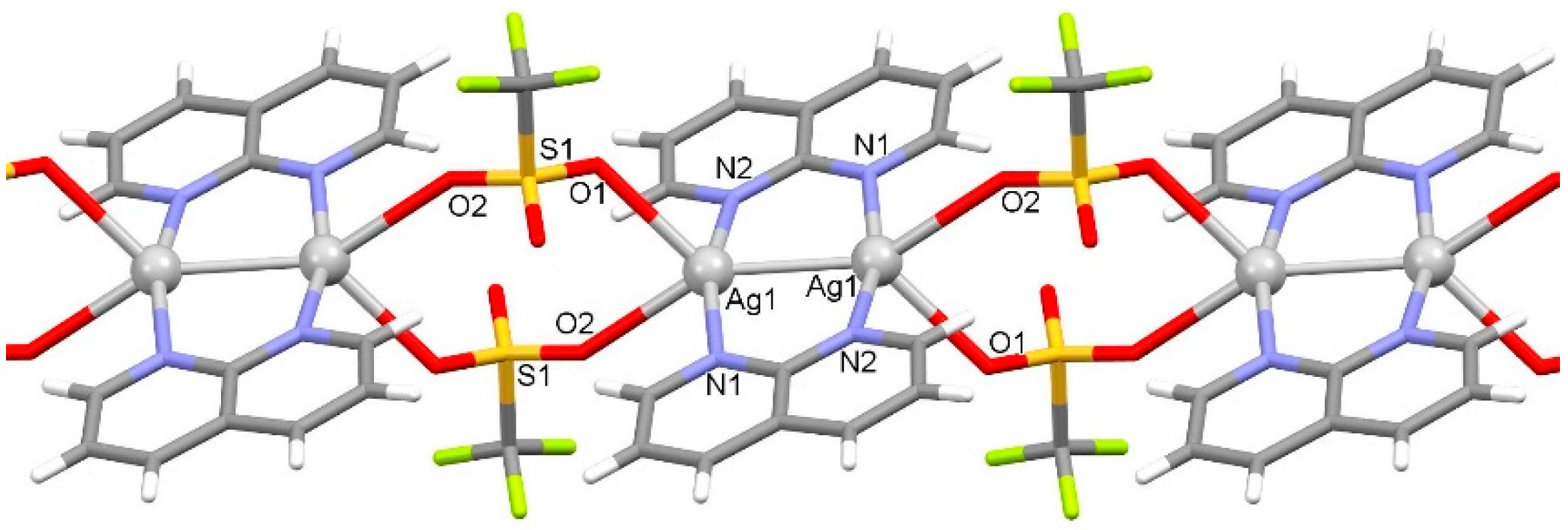

| Distance (Å) | Ag1 | Distance (Å) | Ag2 | Ag3 | |
|---|---|---|---|---|---|
| Ag1–O1 | 2.360(2) | Ag1–O1 | 2.555(2) | 2.449(4) | |
| Ag1–O2 | 2.328(2) | Ag1···O2 ii | 2.645(2) | – | |
| Ag1–S1 | 2.6198(7) | Ag1–N1 | 2.200(2) | 2.186(3) | |
| Ag1–S2 i | 2.6747(8) | Ag1–N2 i | 2.218(2) | 2.184(3) | |
| Ag1···Ag1 i | 2.7871(5) | 2.7235(6) | |||
| Angle (°) | Angle (°) | ||||
| O1–Ag1–O2 | 108.07(7) | O1–Ag1–N1 | 112.75(8) | 91.94(14) | |
| O1–Ag1–S1 | 105.18(5) | O1–Ag1–N2 i | 82.66(8) | 96.78(14) | |
| O2–Ag1–S1 | 124.24(5) | N1–Ag1–N2 i | 164.29(10) | 169.76(12) | |
| O1–Ag1–S2 i | 103.56(5) | ||||
| O2–Ag1–S2 i | 121.54(5) | ||||
| S1–Ag1–S2 i | 91.44(2) |
| D–H···A | D–H (Å) | H···A (Å) | D···A (Å) | D–H···A (o) |
|---|---|---|---|---|
| Ag1 | ||||
| O1–H1A···O3 i | 0.820(18) | 2.007(18) | 2.825(3) | 175(3) |
| O1–H1B···O4 ii | 0.82(3) | 2.06(3) | 2.849(3) | 160(3) |
| C3–H3···O1 iii | 0.95 | 2.50 | 3.428(3) | 166.6 |
| C5–H5···O2 iv | 0.95 | 2.55 | 3.202(3) | 125.6 |
| C5–H5···O4 v | 0.95 | 2.51 | 3.279(4) | 137.8 |
| Ag2 | ||||
| C1–H1···O2 ii | 0.95 | 2.43 | 3.169(4) | 134 |
| C7–H7···F2 iii | 0.95 | 2.46 | 3.301(4) | 148 |
| C8–H8···O3 iv | 0.95 | 2.40 | 3.299(4) | 157 |
| Ag3 | ||||
| O1–H1A···F1 | 0.91 | 2.12 | 2.787(7) | 129.0 |
| O1–H1A···F1 ii | 0.91 | 2.11 | 2.810(7) | 133.0 |
| O1–H1B···F5 | 0.92 | 2.49 | 3.121(6) | 126.6 |
| O1–H1B···F5 iii | 0.92 | 2.51 | 3.084(6) | 121.2 |
| C1–H1···O1 | 0.95 | 2.45 | 3.152(6) | 130.3 |
| C8–H8···F5 iv | 0.95 | 2.52 | 3.141(5) | 123.0 |
| Silver(I) Complex | Oxidation Process (E, V) | Reduction Processes (E, V) | |
|---|---|---|---|
| Ag(I)→Ag(II) | Ag(II)→Ag(I) | Ag(I)→Ag(0) | |
| Ag1 | +1.25 | −0.43 | −1.26 |
| Ag2 | +1.38 | −0.27 | −0.92 |
| Ag3 | +1.12 | −0.19 | -0.82 |
| Compound Test | tia | Ag1 | 1,8-naph | Ag2 | Ag3 | 1,5-naph | [Ag(NO3) (1,5-naph)]n | [Ag(CF3COO) (1,5-naph)]n | [Ag(CF3SO3) (1,5-naph)]n | AgSD | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. albicans | µg/mL | >200 | 7.81 | >200 | 3.91 | 3.91 | >250 | 3.1 | 3.1 | 1.25 | 3.6 |
| µM | >925 | 19.3 | >1537 | 10.1 | 5.0 | >1921 | 10.3 | 8.8 | 3.2 | 10 | |
| C. krusei | µg/mL | >200 | 1.56 | >200 | 1.56 | 1.56 | >250 | 0.78 | 1.56 | 1.25 | 0.89 |
| µM | >925 | 3.6 | >1537 | 4.0 | 1.0 | >1921 | 2.6 | 4.4 | 3.2 | 2.5 | |
| C. parapsilosis | µg/mL | >200 | 3.91 | >200 | 3.91 | 7.81 | >250 | 6.25 | 3.1 | 2.5 | 0.89 |
| µM | >925 | 9.7 | >1537 | 10.1 | 9.9 | >1921 | 20.8 | 8.8 | 6.5 | 2.5 | |
| S. aureus | µg/mL | >250 | 3.91 | >250 | 7.81 | 7.81 | >250 | 50 | 50 | 25 | 27 |
| µM | >1157 | 9.7 | >1921 | 20.2 | 9.9 | >1921 | 167 | 142 | 65 | 75 | |
| L. monocytogenes | µg/mL | >250 | 15.62 | >250 | 15.62 | 125 | >250 | NT | NT | NT | NT |
| µM | >1157 | 38.6 | >1921 | 40.4 | 160 | >1921 | NT | NT | NT | NT | |
| E. coli | µg/mL | >250 | 15.62 | >250 | 31.25 | 15.62 | >250 | 25 | 12.5 | 12.5 | 7.14 |
| µM | >1157 | 38.6 | >1921 | 80.7 | 19.8 | >1921 | 83 | 36 | 32 | 20 | |
| P. aeruginosa ATCC 10332 | µg/mL | >200 | 6.25 | >200 | 3.13 | 3.13 | >250 | 25 | 25 | 25 | 8.93 |
| µM | >925 | 15.5 | >1537 | 8.1 | 4.0 | >1921 | 83 | 71 | 65 | 25 | |
| P. aeruginosa BK25H | µg/mL | >250 | 3.91 | >250 | 3.91 | 3.91 | >250 | NT | NT | NT | NT |
| µM | >1157 | 9.7 | >1921 | 10.1 | 5.0 | >1921 | NT | NT | NT | NT | |
| MRC-5 cells | µg/mL | >100 | 4.25 | >100 | 3.75 | 3.65 | >250 | NT | NT | NT | 3.6 |
| µM | >462 | 10.5 | >768 | 9.7 | 4.6 | >1921 | NT | NT | NT | 10 |
| Complex | Ksv (M−1) | Hypochromism (%) | Kq (M−1·s−1) | KA (M−1) | n |
|---|---|---|---|---|---|
| Ag1 | (1.56 ± 0.01) × 105 | 78.0 | 1.56 × 1013 | 3.05 × 106 | 1.41 |
| Ag2 | (1.34 ± 0.01) × 104 | 70.2 | 1.34 × 1012 | 2.88 × 104 | 1.12 |
| Ag3 | (2.70 ± 0.05) × 104 | 68.7 | 2.70 × 1012 | 6.08 × 105 | 1.37 |
| Complex | Ksv (M−1) | Hypochromism (%) | Kq (M−1·s−1) | KA (M−1) | n |
|---|---|---|---|---|---|
| Ag1 | (7.33 ± 0.33) × 102 | 12.7 | 7.33 × 1010 | 1.05 × 102 | 0.76 |
| Ag2 | (1.29 ± 0.03) × 103 | 15.2 | 1.29 × 1011 | 4.83 × 102 | 0.90 |
| Ag3 | (1.15 ± 0.07) × 103 | 15.1 | 1.15 × 1011 | 4.33 × 102 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ašanin, D.P.; Skaro Bogojevic, S.; Perdih, F.; Andrejević, T.P.; Milivojevic, D.; Aleksic, I.; Nikodinovic-Runic, J.; Glišić, B.Đ.; Turel, I.; Djuran, M.I. Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine. Molecules 2021, 26, 1871. https://doi.org/10.3390/molecules26071871
Ašanin DP, Skaro Bogojevic S, Perdih F, Andrejević TP, Milivojevic D, Aleksic I, Nikodinovic-Runic J, Glišić BĐ, Turel I, Djuran MI. Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine. Molecules. 2021; 26(7):1871. https://doi.org/10.3390/molecules26071871
Chicago/Turabian StyleAšanin, Darko P., Sanja Skaro Bogojevic, Franc Perdih, Tina P. Andrejević, Dusan Milivojevic, Ivana Aleksic, Jasmina Nikodinovic-Runic, Biljana Đ. Glišić, Iztok Turel, and Miloš I. Djuran. 2021. "Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine" Molecules 26, no. 7: 1871. https://doi.org/10.3390/molecules26071871
APA StyleAšanin, D. P., Skaro Bogojevic, S., Perdih, F., Andrejević, T. P., Milivojevic, D., Aleksic, I., Nikodinovic-Runic, J., Glišić, B. Đ., Turel, I., & Djuran, M. I. (2021). Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine. Molecules, 26(7), 1871. https://doi.org/10.3390/molecules26071871












