Expressed Soybean Leghemoglobin: Effect on Escherichia coli at Oxidative and Nitrosative Stress
Abstract
1. Introduction
2. Results
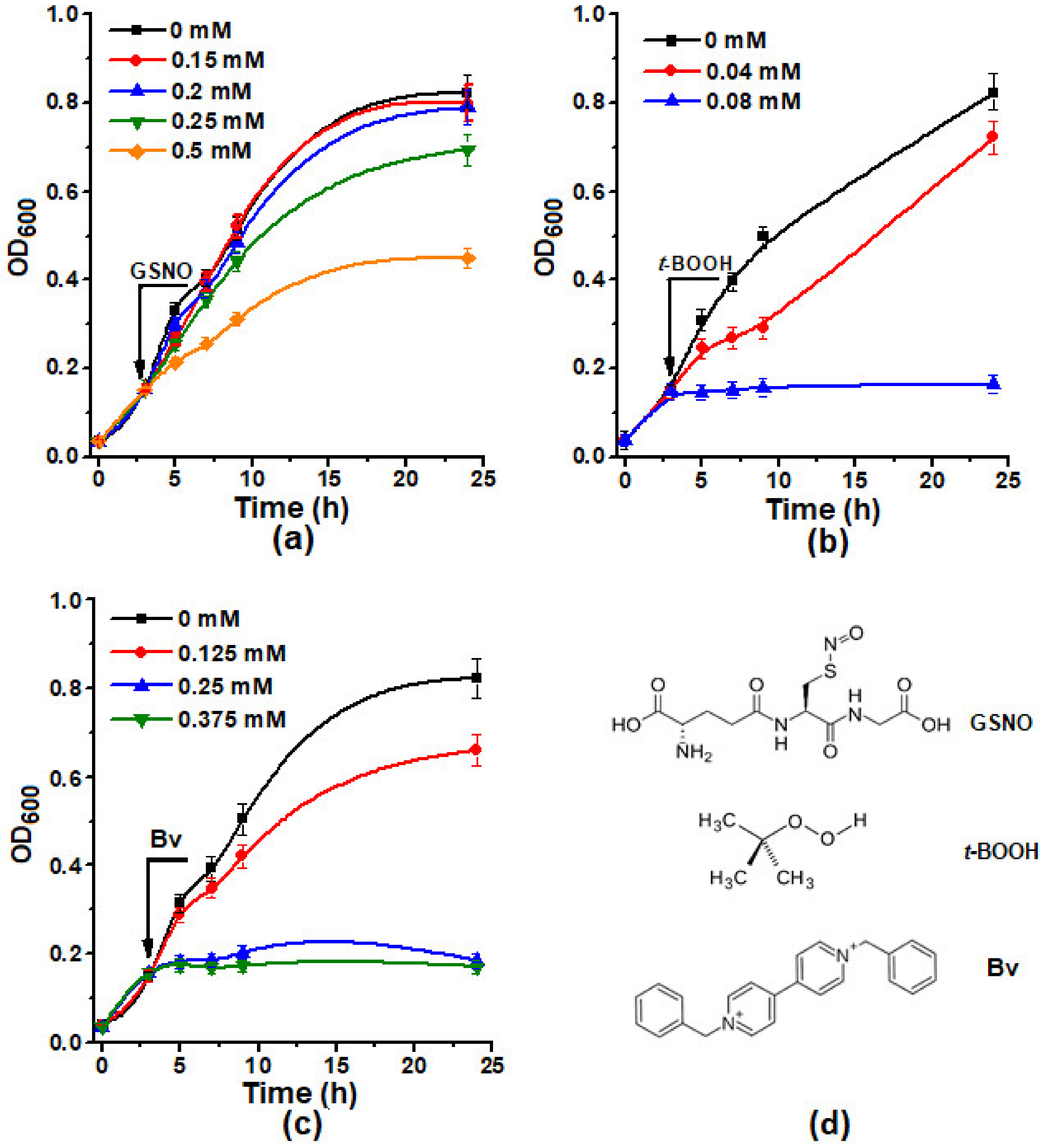
2.1. The Effect of Different Concentrations of Inducers of Oxidative and Nitrosative Stress Inductors on the Growth of E. coli Cells
2.2. The Effect of Lb Expression on the Growth of E. coli Cells
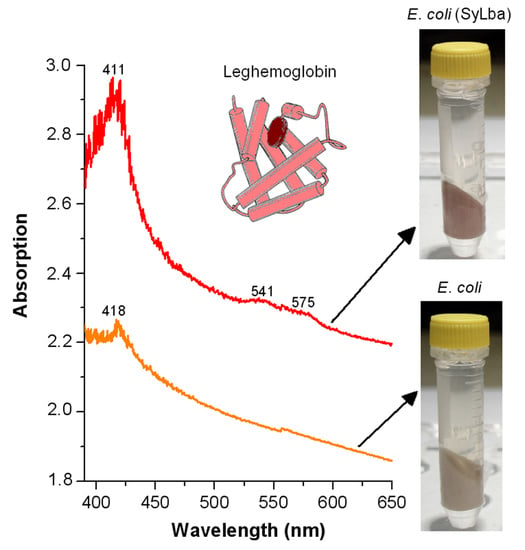
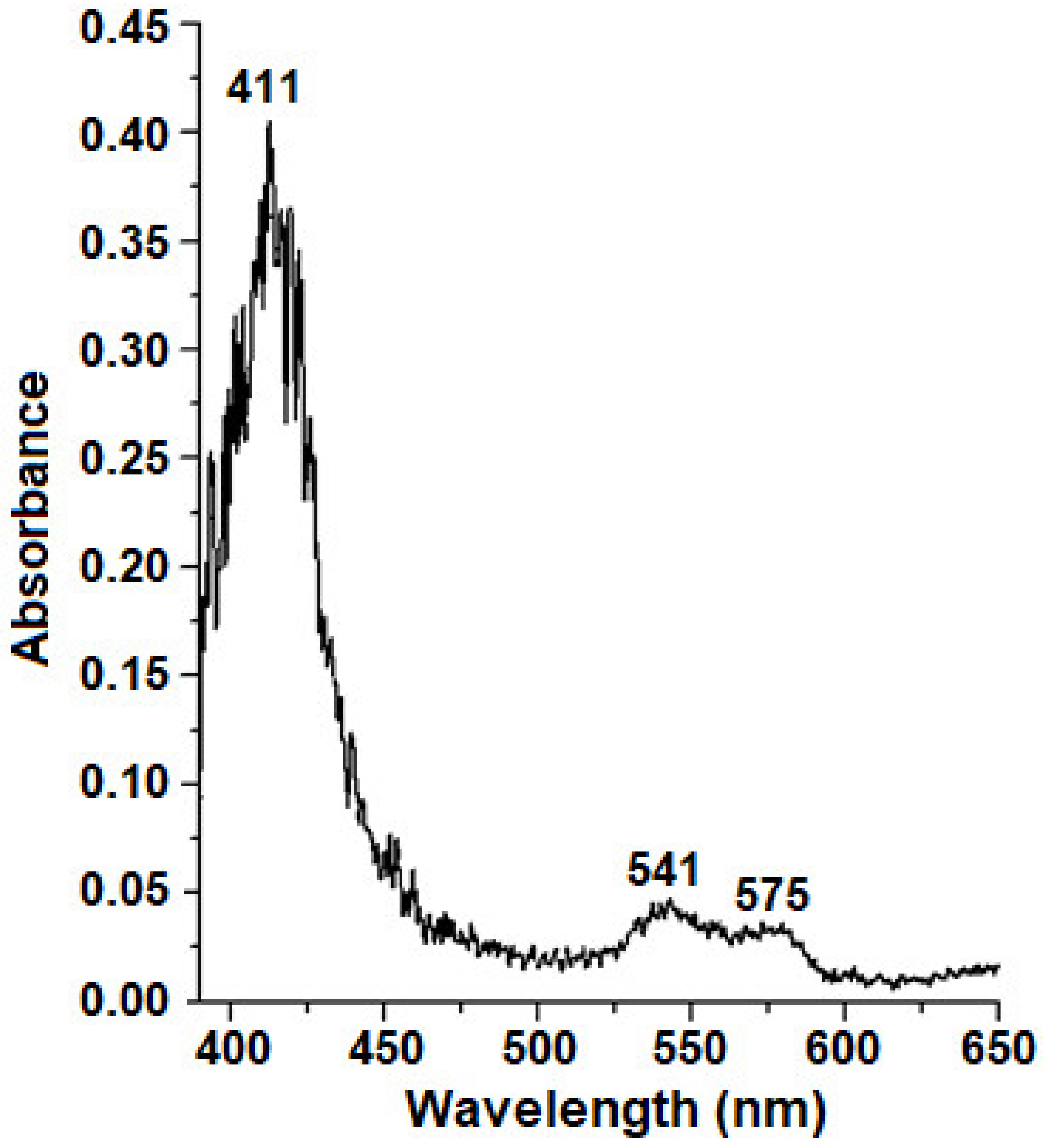
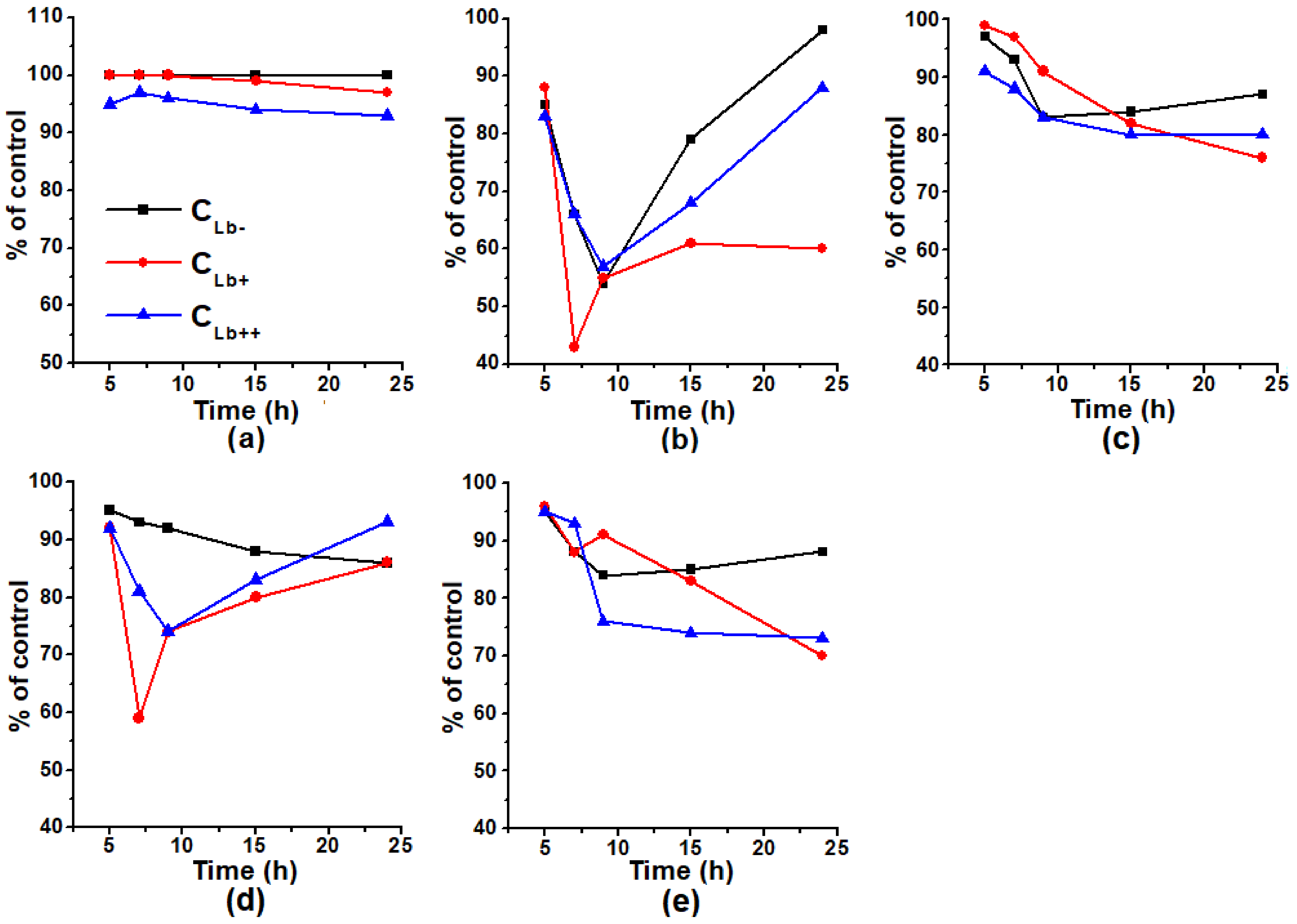
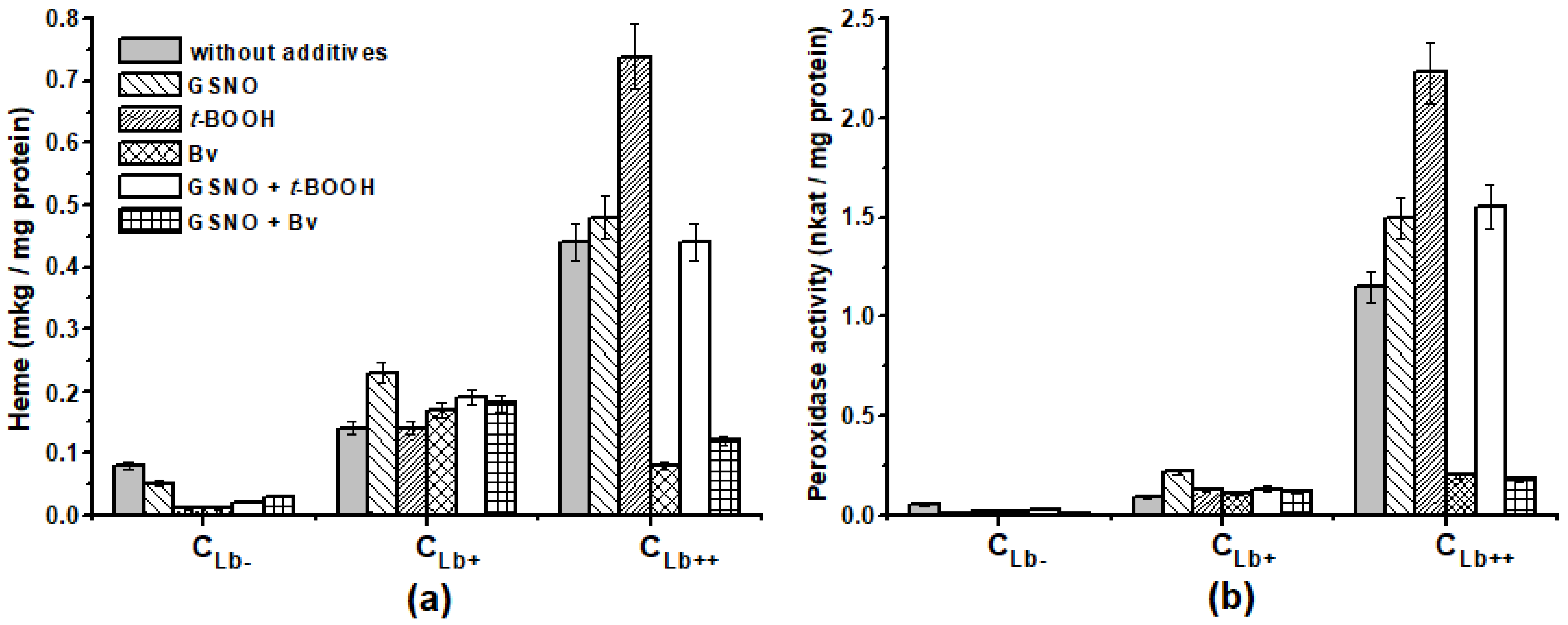
2.3. Effect of Lb Expression on Heme Concentration and Peroxidase Activity in Cellular Protein Extracts
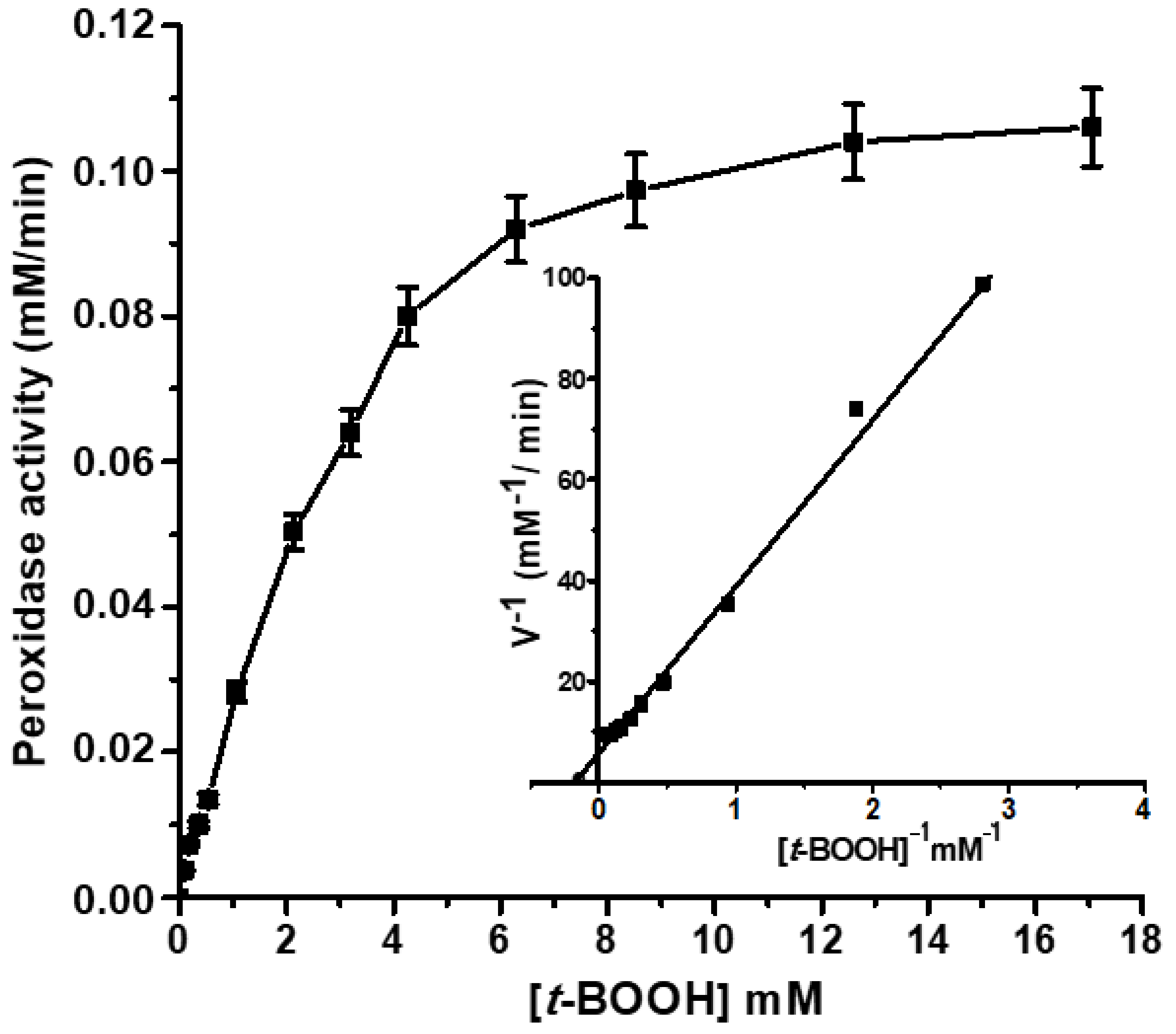
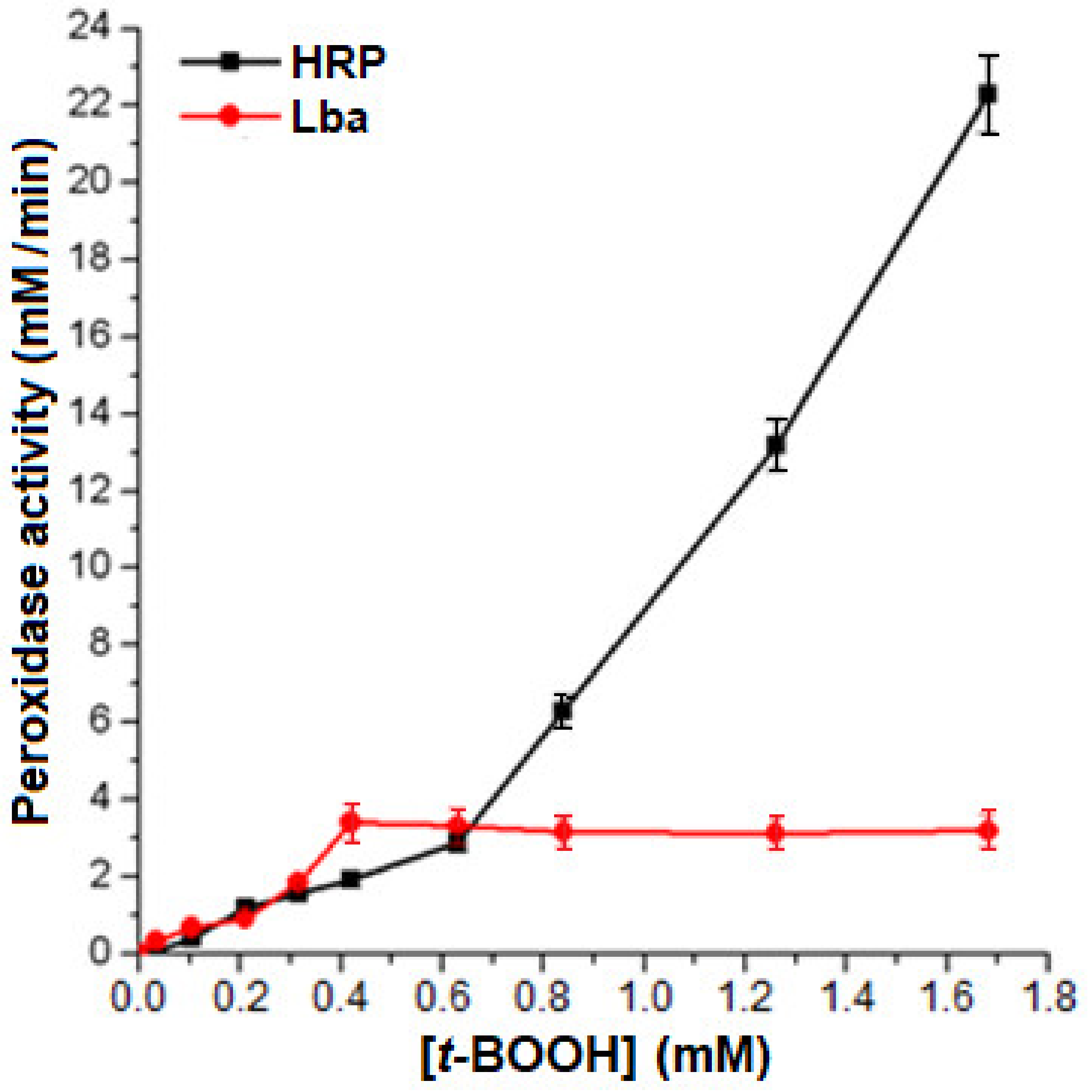
2.4. Peroxidase Activity of the Isolated Lb
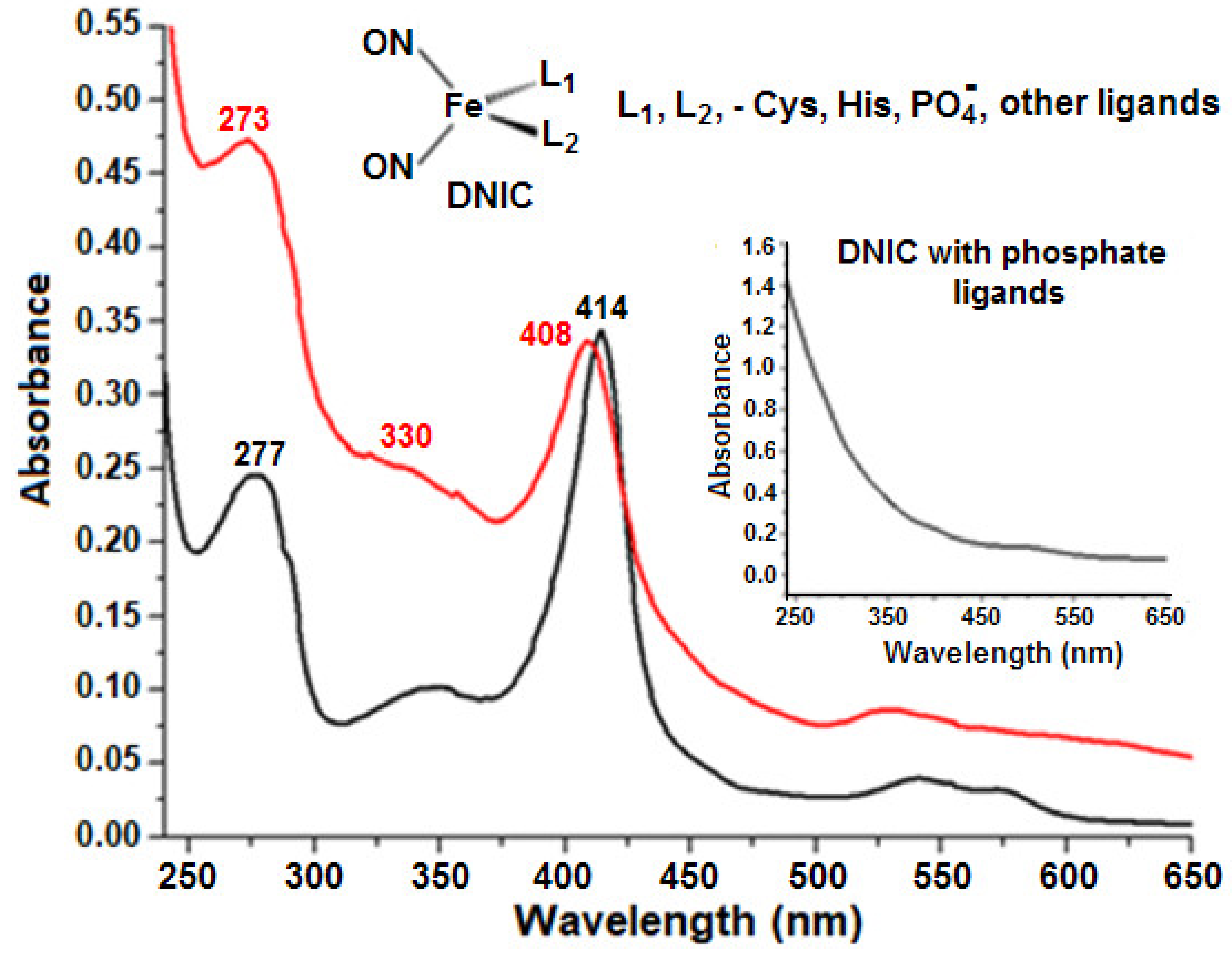
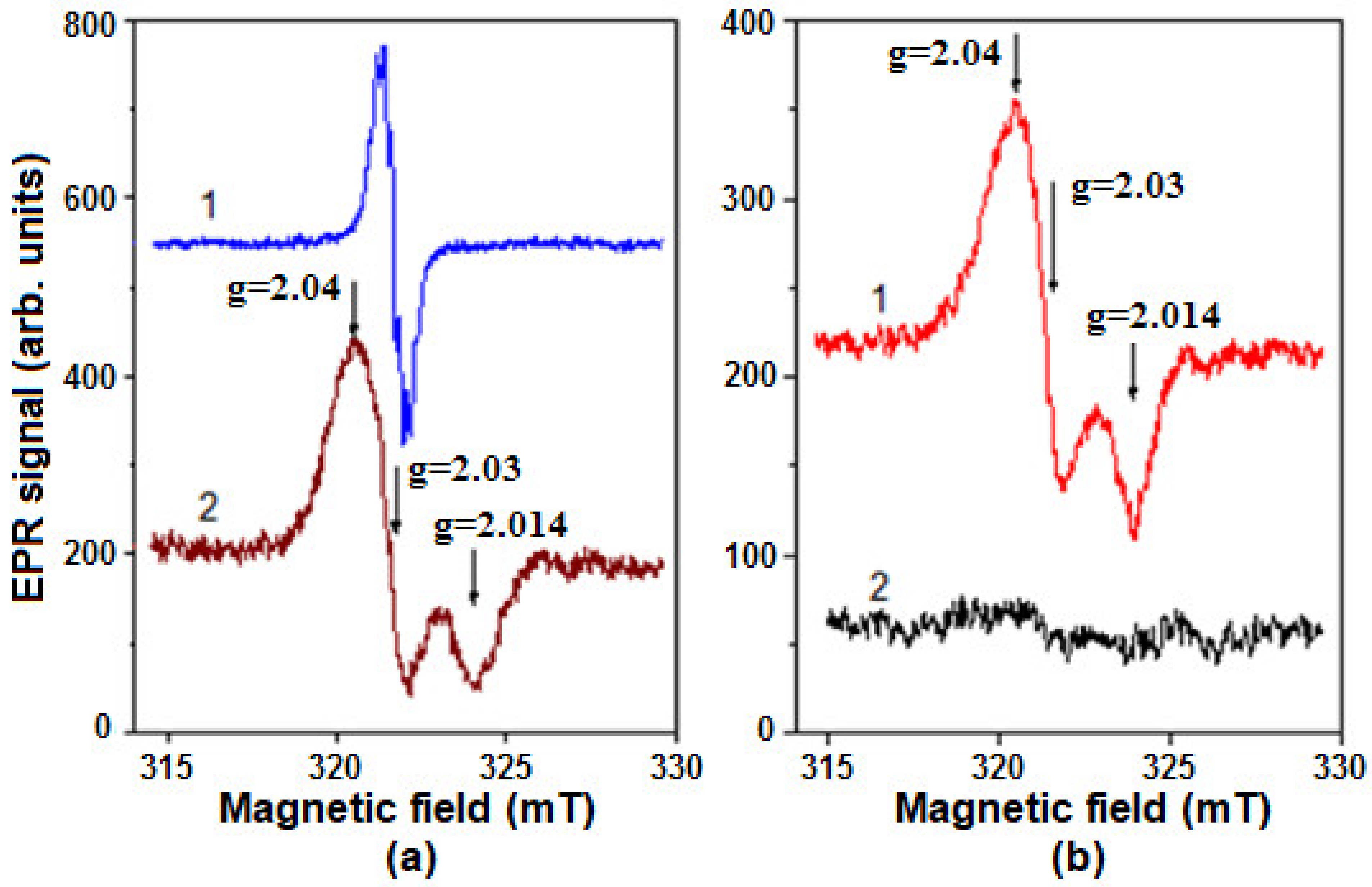
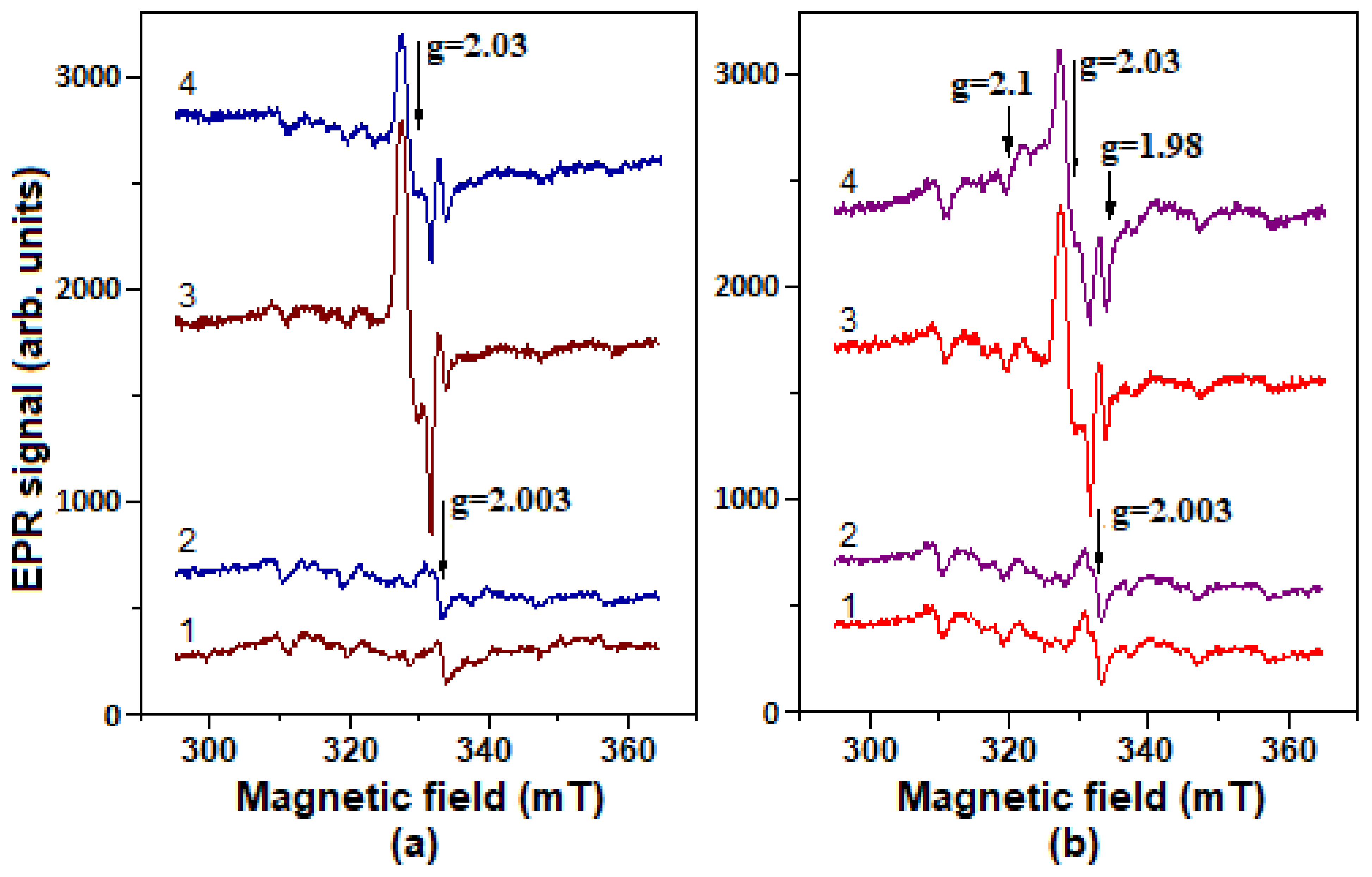
2.5. Formation of Nitrosyl Iron Complexes with Leghemoglobin
3. Discussion
4. Materials and Methods
4.1. Cultivation of E. coli Cells and Treatment with Redox-Active Compounds
4.2. Spectra of the Whole Cells
4.3. Total Protein Extraction
4.4. Determination of Lb Concentration
4.5. Determination of Peroxidase Activity
4.6. Isolation and Purification of Lb from E. coli Cells
4.7. Measurement of Lb Peroxidase Activity in Relation to t-BOOH
4.8. Synthesis of Dinitrosyl Iron Complexes
4.9. Registration of DNICs by EPR Method
4.10. Registration of Dinitrosyl Iron Complexes in Whole Cells
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Thony-Meyer, L.; Preisig, O.; Zufferey, R.; Hennecke, H. The role of a microaerobically induced cb-type cytochrome oxidase in symbiotic nitrogen fixation. In Nitrogen Fixation: Fundamentals and Applications; Tikhonovich, I.A., Provorov, N.A., Romanov, V.I., Newton, W.E., Eds.; Springer: Berlin/Heidelberg, Germany, 1995; pp. 383–388. [Google Scholar]
- Hebelstrup, K.H.; Igamberdiev, A.U.; Hill, R.D. Metabolic effects of hemoglobin gene expression in plants. Gene 2007, 398, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Varma, A. Structure, function, and estimation of leghemoglobin. In Rhizobium Biology and Biotechnology; Hansen, A.P., Choudhary, D.K., Agrawal, P.K., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 309–330. [Google Scholar]
- Uheda, E.; Syono, K. Effects of leghaemoglobin components on nitrogen fixation and oxygen consumption. Plant Cell Physiol. 1982, 23, 85–90. [Google Scholar] [CrossRef]
- Appleby, C.A.; Bradbury, J.H.; Morris, R.J.; Wittenberg, B.A.; Wittenberg, J.B.; Wright, P.E. Leghemoglobin. Kinetic, nuclear magnetic resonance, and optical studies of pH dependence of oxygen and carbon monoxide binding. J. Biol. Chem. 1983, 258, 2254–2259. [Google Scholar] [CrossRef]
- Kuzma, M.M.; Topunov, A.F.; Layzell, D.B. Effects of temperature on infected cell O2 concentration and adenylate levels in attached soybean nodules. Plant Physiol. 1995, 107, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Appleby, C.A. Leghemoglobin and Rhizobium respiration. Ann. Rev. Plant Physiol. 1984, 35, 443–478. [Google Scholar] [CrossRef]
- Topunov, A.F. Leghemoglobin and its role in regulation of oxygen concentration in nitrogen-fixing legume nodules. Biochemistry (Moscow) 1995, 60, 45–49. [Google Scholar]
- Layzell, D.B.; Atkins, C.A. The physiology and biochemistry of legume N2 fixation. In Plant Metabolism, 2nd ed.; Dennis, D.T., Turpin, D.H., Lefebvre, D.D., Layzell, D.B., Eds.; Longman: Harlow, UK, 1997; pp. 495–505. [Google Scholar]
- Larrainzar, E.; Villar, I.; Rubio, M.C.; Pérez-Rontomé, C.; Huertas, R.; Sato, S.; Mun, J.-H.; Becana, M. Hemoglobins in the legume–Rhizobium symbiosis. New Phytol. 2020, 228, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.P.S.; Bal, A.K. Intracellular site of synthesis and localization of leghemoglobin in root nodules. Proc. Natl. Acad. Sci. USA 1976, 73, 3843–3847. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J.; Mathieu, C.; Puppo, A. Leghemoglobin: Properties and reactions. Adv. Inorg. Chem. 1999, 46, 495–542. [Google Scholar] [CrossRef]
- Robertson, J.B.; Wells, B.; Bisseling, T.; Farndon, K.J.F.; Johnston, A.W.B. Immuno-gold localization of leghaemoglobin in cytoplasm in nitrogen-fixing root nodules of pea. Nature 1984, 311, 254–256. [Google Scholar] [CrossRef]
- Vainstein, B.K.; Kuranova, I.P.; Harutyunyan, E.G.; Egorov, T.A. Leghemoglobin II from yellow lupin - the peculiarities of its structure as compared with sperm whale myoglobin. Bioorg. Khimiya 1980, 6, 684–699. [Google Scholar]
- Herold, S.; Puppo, A. Kinetics and mechanistic studies of the reactions of metleghemoglobin, ferrylleghemoglobin, and nitrosylleghemoglobin with reactive nitrogen species. J. Biol. Inorg. Chem. 2005, 10, 946–957. [Google Scholar] [CrossRef][Green Version]
- Sievers, G.; Rönnberg, M. Study of pseudoperoxidatic activity ofsoybean leghemoglobin and sperm whale myoglobin. Biochim. Biophys. Acta 1978, 533, 293–301. [Google Scholar] [CrossRef]
- Herold, S.; Puppo, A. Oxyleghemoglobin scavenges nitrogen monoxide and peroxynitrite a possible role in functioning nodules? J. Biol. Chem. 2005, 10, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A. Legume haemoglobins: Symbiotic nitrogen fixation needs bloody nodules. Curr. Biol. 2005, 15, 196–198. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.; Davies, M.J.; Mathieu, C.; Herouart, D.; Puppo, A. Leghemoglobin-derived radicals. J. Biol. Chem. 1996, 271, 32557–32562. [Google Scholar] [CrossRef][Green Version]
- Puppo, A.; Halliwell, B. Generation of hydroxyl radicals by soybean nodule leghaemoglobin. Planta 1988, 173, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Nicola, N.A.; Minasian, E.; Appleby, C.A.; Leach, S.J. Circular dichroism studies of myoglobin and leghemoglobin. Biochemistry 1975, 14, 5141–5149. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, M.S.; Barry, J.K.; Brucker, E.A.; Berry, M.B.; Phillips, G.N., Jr.; Olson, J.S.; Arredondo-Peter, R.; Dean, J.M.; Klucas, R.V.; Sarath, G. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J. Mol. Biol. 1997, 266, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.Z.; Shitut, M.; Agrawal, P.; Mendes, O.; Klapholz, S. Safety evaluation of soy leghemoglobin protein preparation derived from Pichia pastoris, intended for use as a flavor catalyst in plant-based meat. Int. J. Toxicol. 2018, 37, 241–262. [Google Scholar] [CrossRef]
- Reyes, T.F.; Chen, Y.; Fraser, R.Z.; Chan, T.; Li, X. Assessment of the potential allergenicity and toxicity of Pichia proteins in a novel leghemoglobin preparation. Regul. Toxicol. Pharmacol. 2021, 119, e104817. [Google Scholar] [CrossRef] [PubMed]
- Zhiznevskaya, G.Y.; Kudryavtseva, N.N.; Genina, O.S.; Izmailov, S.F. Leghemoglobin content and stability, and nitrogen-fixing activity in lupine, soybean, and common bean nodules. Sov. Plant Physiol. 1990, 37, 219–224. [Google Scholar]
- Sikorski, M.M.; Topunov, A.F.; Strozycki, P.; Vorgias, C.E.; Wilson, K.S.; Legocki, A.B. Cloning and expression of plant leghemoglobin cDNA of Lupinus luteus in Escherichia coli and purification of the recombinant protein. Plant Sci. 1995, 108, 109–117. [Google Scholar] [CrossRef]
- Jones, D.K.; Badii, R.; Rosell, F.I.; Lloyd, E. Bacterial expression and spectroscopic characterization of soybean leghaemoglobin a. Biochem. J. 1998, 330 (Pt 2), 983–988. [Google Scholar] [CrossRef]
- Arredondo-Peter, R.; Moran, J.F.; Sarath, G.; Luan, P.; Klucas, R.V. Molecular cloning of the cowpea leghemoglobin II gene and expression of its cDNA in Escherichia coli. Purification and characterization of the recombinant protein. Plant Physiol. 1997, 114, 493–500. [Google Scholar] [CrossRef]
- Arredondo-Peter, R.; Hargrove, M.S.; Sarath, G.; Moran, J.F.; Lohrman, J.; Olson, J.S.; Klucas, R.V. Rice hemoglobins (Gene cloning, analysis, and O2-binding kinetics of a recombinant protein synthesized in Escherichia coli). Plant. Physiol. 1997, 115, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Topunov, A.F.; Talyzin, V.V.; Chekasina, E.V.; Kretovich, V.L. Metlegoglobin reductase activity of Rhizobium bacteria grown in a free culture. Dokl. Biochem. 1987, 296, 351–353. [Google Scholar]
- Talyzin, V.V.; Bashirova, N.F.; Kosmachevskaya, O.V.; Punina, N.V.; Arabova, L.I.; Tikhomirova, N.V.; Topunov, A.F. Methemoglobin reductase of bacteria and bacteroids Bradyrhizobium lupini: Purification and properties. Appl. Biochem. Microbiol. 2018, 54, 26–33. [Google Scholar] [CrossRef]
- Uppal, S.; Khan, M.A.; Kundu, S. Identification and characterization of a recombinant cognate hemoglobin reductase from Synechocystis sp. PCC 6803. Int. J. Biol. Macromol. 2020, 162, 1054–1063. [Google Scholar] [CrossRef]
- Shandilya, M.; Kumar, G.; Gomkale, R.; Singh, S.; Khan, M.A.; Kateriya, S.; Kundu, S. Multiple putative methemoglobin reductases in C. reinhardtii may support enzymatic functions for its multiple hemoglobins. Int. J. Biol. Macromol. 2021, 171, 465–479. [Google Scholar] [CrossRef]
- Kretovich, W.L.; Melik-Sarkissyan, S.S.; Bashirova, N.F.; Topunov, A.F. Enzymatic reduction of leghemoglobin in lupin nodules. J. Appl. Biochem. 1982, 4, 209–217. [Google Scholar]
- Saari, L.L.; Klucas, R.V. Ferric leghemoglobin reductase from soybean root nodules. Arch. Biochem. Biophys. 1984, 231, 102–113. [Google Scholar] [CrossRef]
- Urarte, E.; Auzmendi, I.; Rol, S.; Ariz, I.; Aparicio-Tejo, P.M.; Arredondo-Peter, R.; Moran, J.F. A self-induction method to produce high quantities of recombinant functional flavo-leghemoglobin reductase. Methods Enzymol. 2008, 436, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Bykova, N.V.; Hill, R.D. Nitric oxide scavenging by barley hemoglobin is facilitated by a monodehydroascorbate reductase-mediated ascorbate reduction of methemoglobin. Planta 2006, 223, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Hill, R.D. Purification of class 1 plant hemoglobins and examination of their functional properties. Methods Enzymol. 2008, 436, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Becana, M.; Klucas, R.V. Oxidation and reduction of leghemoglobin in root nodules of leguminous plants. Plant Physiol. 1992, 98, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Lira-Ruan, K.; Sarath, G.; Klucas, R.V.; Arredondo-Peter, R. Characterization of leghemoglobin from a mimozoid legume, Leucaena esculenta, root nodules. Braz. J. Plant Physiol. 2000, 12, 37–44. [Google Scholar]
- Bird, C.L.; Kuhn, A.T. Electrochemistry of the Viologens. Chem. Soc. Rev. 1981, 10, 49–82. [Google Scholar] [CrossRef]
- Gorbunov, N.V.; Osipov, A.N.; Day, B.W.; Zayas-Rivera, B.; Kagan, V.E.; Elsayed, N.M. Reduction of ferrylmyoglobin and ferrylhaemoglobin by nitric oxide: A protection mechanism against ferryl hemoprotein-induced oxidation. Biochemistry 1995, 34, 6689–6699. [Google Scholar] [CrossRef] [PubMed]
- Padmaja, S.; Huie, R.E. The reaction of nitric oxide with organic peroxyl radicals. Biochem. Biophys. Res. Commun. 1993, 195, 539–544. [Google Scholar] [CrossRef]
- Gorbunov, N.V.; Yalowich, J.C.; Gaddam, A.; Thampatty, P.; Ritov, V.B.; Kisin, E.R.; Elsayed, N.M.; Kagan, V.E. Nitric oxide prevents oxidative damage produced by tert-butyl hydroperoxide in Erythroleukemia cells via nitrosylation of heme non-heme iron. J. Biochem. 1997, 272, 12328–12341. [Google Scholar] [CrossRef]
- Puppo, A.; Rigaud, J.; Job, D.; Ricard, J.; Zeba, B. Peroxidase content of soybean root nodules. Biochim. Biophys. Acta 1980, 614, 303–312. [Google Scholar] [CrossRef]
- Davies, M.J.; Puppo, A. Identification of the site of the globin-derived radical in leghaemoglobins. Biochim. Biophys. Acta 1993, 1202, 182–188. [Google Scholar] [CrossRef]
- Davies, M.J.; Puppo, A. Direct detection of a globin-derived radical in leghaemoglobin treated with peroxides. Biochem. J. 1992, 281 (Pt 1), 197–201. [Google Scholar] [CrossRef]
- Moreau, S.; Davies, M.J.; Puppo, A. Reaction of ferric leghemoglobin with H2O2: Formation of heme-protein cross-links and dimeric species. Biochim. Biophys. Acta 1995, 1251, 17–22. [Google Scholar] [CrossRef]
- Singh, R.; Wiseman, B.; Deemagarn, T.; Jha, V.; Switala, J.; Loewen, P.C. Comparative study of catalase-peroxidases (KatGs). Arch. Biochem. Biophys. 2008, 471, 207–214. [Google Scholar] [CrossRef]
- Puppo, A.; Monny, C.; Davies, M.J. Glutathione–dependent conversion of ferryl leghaemoglobin into the ferric form: A potential protective process in soybean (Glycine max) root nodules. Biochem. J. 1993, 289, 435–438. [Google Scholar] [CrossRef][Green Version]
- Saari, L.L.; Klucas, R.V. Nonenzymatic reduction of ferric leghemoglobin. Biochim. Biophys. Acta 1987, 912, 198–202. [Google Scholar] [CrossRef]
- Becana, M.; Salin, M.L.; Ji, L.; Klucas, R.V. Flavin-mediated reduction of ferric leghemoglobin from soybean nodules. Planta 1991, 183, 575–583. [Google Scholar] [CrossRef][Green Version]
- Mannino, M.H.; Patel, R.S.; Eccardt, A.M.; Perez Magnelli, R.A.; Robinson, C.L.C.; Janowiak, B.E.; Warren, D.E.; Fisher, J.S. Myoglobin as a versatile peroxidase: Implications for a more important role for vertebrate striated muscle in antioxidant defense. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 234, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Vanin, A.F.; van Faassen, E. DNICs: Physico-chemical properties and their observation in cells and tissues. In Radicals for Life: The Various Forms of Nitric Oxide; Van Faassen, E., Vanin, A.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 17–74. [Google Scholar]
- Shumaev, K.B.; Gubkin, A.A.; Serezhenkov, V.A.; Lobysheva, I.I.; Kosmachevskaya, O.V.; Ruuge, E.K.; Lankin, V.Z.; Topunov, A.F.; Vanin, A.F. Interaction of reactive oxygen and nitrogen species with albumin- and hemoglobin-bound dinitrosyl iron complexes. Nitric Oxide 2008, 18, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Shumaev, K.B.; Kosmachevskaya, O.V.; Timoshin, A.A.; Vanin, A.F.; Topunov, A.F. Dinitrosyl iron complexes bound with haemoglobin as markers of oxidative stress. Methods Enzymol. 2008, 436, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Shumaev, K.B.; Kosmachevskaya, O.V.; Chumikina, L.V.; Topunov, A.F. Dinitrosyl iron complexes and other physiological metabolites of nitric oxide: Multifarious role in plants. Nat. Prod. Commun. 2016, 11, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Vanin, A.F. Dinitrosyl iron complexes and S-nitrosothiols are two possible forms for stabilization and transport of nitric oxide in biological systems. Biochemistry (Moscow) 1998, 63, 782–793. [Google Scholar]
- Muller, B.; Kleschyov, A.L.; Alencar, J.L.; Vanin, A.F.; Stoclet, J.C. Nitric oxide tronsport and storage in the cardiovascular system. Ann. N. Y. Acad. Sci. 2002, 962, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Lok, H.C.; Sahni, S.; Jansson, P.J.; Kovacevic, Z.; Hawkins, C.L.; Richardson, D.R. A nitric oxide storage and transport system that protects activated macrophages from endogenous nitric oxide cytotoxicity. J. Biol. Chem. 2016, 291, 27042–27061. [Google Scholar] [CrossRef]
- Kosmachevskaya, O.B.; Nasybullina, E.I.; Shumaev, K.B.; Chumikina, L.V.; Arabova, L.I.; Yaglova, N.V.; Obernikhin, S.S.; Topunov, A.F. Dinitrosyl iron complexes with glutathione ligands intercept peroxynitrite and protect hemoglobin from oxidative modification. Appl. Biochem. Microbiol. 2021, 57, 411–420. [Google Scholar] [CrossRef]
- Kosmachevskaya, O.V.; Nasybullina, E.I.; Shumaev, K.B.; Novikova, N.N.; Topunov, A.F. Effect of iron–nitric oxide complexes on the reactivity of hemoglobin cysteines. Appl. Biochem. Microbiol. 2020, 56, 512–520. [Google Scholar] [CrossRef]
- Shumaev, K.B.; Petrova, N.E.; Zabbarova, I.V.; Vanin, A.F.; Topunov, A.F.; Lankin, V.Z.; Ruuge, E.K. Interaction of oxoferrylmyoglobin and dinitrosyl-iron complexes. Biochemistry (Moscow) 2004, 69, 569–574. [Google Scholar] [CrossRef]
- Jucket, M.; Zheng, Y.; Yuan, H.; Pastor, T.; Antholine, W.; Weber, M.; Vercellotti, G. Heme and the endothelium. Effects of nitric oxide on catalytic iron and heme degradation by heme oxygenase. J. Biol. Chem. 1998, 273, 23388–23397. [Google Scholar] [CrossRef]
- Gow, A.J.; Luchsinger, B.P.; Pawloski, J.R.; Singel, D.J.; Stamler, J.S. The oxyhemoglobin reaction of nitric oxide. Biochemistry 1999, 96, 9027–9032. [Google Scholar] [CrossRef]
- Sundberg, R.J.; Martin, R.B. Interactions of histidine and other imidazole derivatives with transition metal ions in chemical and biological systems. Chem. Rev. 1974, 74, 471–517. [Google Scholar] [CrossRef]
- Demple, B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon. Gene 1996, 179, 153–157. [Google Scholar] [CrossRef]
- Kaur, S.; Benov, L.T. Methylene blue induces the soxRS regulon of Escherichia coli. Chem. Biol. Interact. 2020, 329, e109222. [Google Scholar] [CrossRef]
- Mongkolsuk, S.; Helmann, J.D. Regulation of inducible peroxide stress responses. Mol. Microbiol. 2002, 45, 9–15. [Google Scholar] [CrossRef]
- Ding, H.; Demple, B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. USA 2000, 97, 5146–5150. [Google Scholar] [CrossRef]
- Vasil’eva, S.V.; Stupakova, M.V.; Lobysheva, I.I.; Mikoyan, V.D.; Vanin, A.F. Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry (Moscow) 2001, 66, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Zhang, N.; Yang, J.; Ding, H. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol. Microbiol. 2008, 70, 953–964. [Google Scholar] [CrossRef]
- Lo, F.-C.; Lee, J.-F.; Liaw, W.-F.; Hsu, I.-J.; Tsai, Y.-F.; Chan, S.I.; Yu, S.S.-F. The metal core structures in the recombinant Escherichia coli transcriptional factor SoxR. Chemistry 2012, 18, 2565–2577. [Google Scholar] [CrossRef]
- Cruz-Ramos, H.; Crack, J.; Wu, G.; Hughes, M.N.; Scott, C.; Thomson, A.J.; Green, J.; Poole, R.K. NO sensing by FNR: Regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002, 21, 3235–3244. [Google Scholar] [CrossRef]
- Boccini, F.; Herold, S. Mechanistic studies of the oxidation oxyhemoglobin by peroxynitrite. Biochemistry 2004, 43, 16393–16404. [Google Scholar] [CrossRef]
- Herold, S.; Kalinga, S.; Matsui, T.; Watanabe, Y. Mechanistic studies of the isomerization of peroxynitrite to nitrate catalyzed by distal histidine metmyoglobin mutants. J. Am. Chem. Soc. 2004, 126, 6945–6955. [Google Scholar] [CrossRef] [PubMed]
- Kosmachevskaya, O.V.; Shumaev, K.B.; Arredondo-Peter, R.; Topunov, A.F. Influence of tert-butyl hydroperoxide and nitrosoglutathione on Escherichia coli cells expressing leghemoglobin. J. Stress Physiol. Biochem. 2007, 3, 18–24. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein due binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Riggs, A. Preparation of blood hemoglobins of vertebrates. Methods Enzymol. 1981, 76, 5–29. [Google Scholar] [CrossRef]
- Kosmachevskaya, O.V.; Topunov, A.F. Method of determination of the content of hemoglobin-like proteins in heterogenic mixtures. Appl. Biochem. Microbiol. 2007, 43, 313–319. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosmachevskaya, O.V.; Nasybullina, E.I.; Shumaev, K.B.; Topunov, A.F. Expressed Soybean Leghemoglobin: Effect on Escherichia coli at Oxidative and Nitrosative Stress. Molecules 2021, 26, 7207. https://doi.org/10.3390/molecules26237207
Kosmachevskaya OV, Nasybullina EI, Shumaev KB, Topunov AF. Expressed Soybean Leghemoglobin: Effect on Escherichia coli at Oxidative and Nitrosative Stress. Molecules. 2021; 26(23):7207. https://doi.org/10.3390/molecules26237207
Chicago/Turabian StyleKosmachevskaya, Olga V., Elvira I. Nasybullina, Konstantin B. Shumaev, and Alexey F. Topunov. 2021. "Expressed Soybean Leghemoglobin: Effect on Escherichia coli at Oxidative and Nitrosative Stress" Molecules 26, no. 23: 7207. https://doi.org/10.3390/molecules26237207
APA StyleKosmachevskaya, O. V., Nasybullina, E. I., Shumaev, K. B., & Topunov, A. F. (2021). Expressed Soybean Leghemoglobin: Effect on Escherichia coli at Oxidative and Nitrosative Stress. Molecules, 26(23), 7207. https://doi.org/10.3390/molecules26237207