Profiling of Carnitine Shuttle System Intermediates in Gliomas Using Solid-Phase Microextraction (SPME)
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Chemical Biopsy (Solid-Phase Microextraction) Protocol
4.3. Instrumental Analysis
4.4. Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lin, H.; Patel, S.; Affeck, V.S.; Wilson, I.; Turnbull, D.M.; Joshi, A.R.; Maxwell, R.; Stoll, E.A. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro-Oncology 2017, 19, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Kesarwani, P.; Prabhu, A.; Graham, S.F.; Buelow, K.L.; Nakano, I.; Chinnaiyan, P. Enhanced fatty acid oxidation provides glioblastoma cells metabolic plasticity to accommodate to its dynamic nutrient microenvironment. Cell Death Dis. 2020, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Duman, C.; Yaqubi, K.; Hoffmann, A.; Acikgöz, A.A.; Korshunov, A.; Bendszus, M.; Herold-Mende, C.; Liu, H.K.; Alfonso, J. Acyl-CoA-Binding Protein Drives Glioblastoma Tumorigenesis by Sustaining Fatty Acid Oxidation. Cell Metab. 2019, 30, 274–289.e5. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity review-article. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef]
- Juraszek, B.; Czarnecka-Herok, J.; Nałęcz, K.A. Glioma cells survival depends both on fatty acid oxidation and on functional carnitine transport by SLC22A5. J. Neurochem. 2021, 156, 642–657. [Google Scholar] [CrossRef]
- Strickland, M.; Stoll, E.A. Metabolic reprogramming in glioma. Front. Cell Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef]
- Reyes-Garcés, N.; Gionfriddo, E.; Gómez-Ríos, G.A.; Alam, M.N.; Boyacı, E.; Bojko, B.; Singh, V.; Grandy, J.; Pawliszyn, J. Advances in Solid Phase Microextraction and Perspective on Future Directions. Anal. Chem. 2018, 90, 302–360. [Google Scholar] [CrossRef]
- Ngernsutivorakul, T.; Steyer, D.J.; Valenta, A.C.; Kennedy, R.T. In Vivo Chemical Monitoring at High Spatiotemporal Resolution Using Microfabricated Sampling Probes and Droplet-Based Microfluidics Coupled to Mass Spectrometry. Anal. Chem. 2018, 90, 10943–10950. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.; Zhang, Y.; Zhang, K.; Zhan, C.; Shi, X.; Li, Y.; Zhao, J.; Bai, Y.; Wang, Y.; et al. Metabolic profiling analysis upon acylcarnitines in tissues of hepatocellular carcinoma revealed the inhibited carnitine shuttle system caused by the downregulated carnitine palmitoyltransferase 2. Mol. Carcinog. 2019, 58, 749–759. [Google Scholar] [CrossRef]
- Yu, D.; Xuan, Q.; Zhang, C.; Hu, C.; Li, Y.; Zhao, X.; Liu, S.; Ren, F.; Zhang, Y.; Zhou, L.; et al. Metabolic alterations related to glioma grading based on metabolomics and lipidomics analyses. Metabolites 2020, 10, 478. [Google Scholar] [CrossRef]
- Bogusiewicz, J.; Goryńska, P.Z.; Gaca, M.; Chmara, K.; Goryński, K.; Jaroch, K.; Paczkowski, D.; Furtak, J.; Harat, M.; Bojko, B. On-Site Sampling and Extraction of Brain Tumors for Metabolomics and Lipidomics Analysis. J. Vis. Exp. 2020, 2020, 159. [Google Scholar] [CrossRef]
- Bogusiewicz, J.; Goryńska, P.Z.; Jaroch, K.; Goryński, K.; Paczkowski, D.; Furtak, J.; Harat, M.; Bojko, B. P13.05 Chemical Biopsy as an Alternative Sampling Method in Neurooncology. Neuro-Oncology 2019, 21, iii63. [Google Scholar] [CrossRef]
- Goryńska, P.Z.; Chmara, K.; Goryński, K.; Paczkowski, D.; Harat, M.; Bojko, B. A new strategy for brain tumour metabolomic analysis. Med. Res. J. 2018, 3, 15–22. [Google Scholar] [CrossRef][Green Version]
- Menyhárt, O.; Győrffy, B. Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis. Comput. Struct. Biotechnol. J. 2021, 19, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Diaz, A.K.; Shaw, T.I.; Li, Y.; Niu, M.; Cho, J.H.; Paugh, B.S.; Zhang, Y.; Sifford, J.; Bai, B.; et al. Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes. Nat. Commun. 2019, 10, 3718. [Google Scholar] [CrossRef] [PubMed]
- ter Veld, F.; Primassin, S.; Hoffmann, L.; Mayatepek, E.; Spiekerkoetter, U. Corresponding increase in long-chain acyl-CoA and acylcarnitine after exercise in muscle from VLCAD mice. J. Lipid Res. 2009, 50, 1556–1562. [Google Scholar] [CrossRef]
- Van Vlies, N.; Tian, L.; Overmars, H.; Bootsma, A.H.; Kulik, W.; Wanders, R.J.A.; Wood, P.A.; Vaz, F.M. Characterization of carnitine and fatty acid metabolism in the long-chain acyl-CoA dehydrogenase-deficient mouse. Biochem. J. 2005, 387, 185–193. [Google Scholar] [CrossRef]
- Galievsky, V.; Pawliszyn, J. Fluorometer for Screening of Doxorubicin in Perfusate Solution and Tissue with Solid-Phase Microextraction Chemical Biopsy Sampling. Anal. Chem. 2020, 92, 13025–13033. [Google Scholar] [CrossRef]
- Gómez-Ríos, G.A.; Mirabelli, M.F. Solid Phase Microextraction-mass spectrometry: Metanoia. TrAC Trends Anal. Chem. 2019, 112, 201–211. [Google Scholar] [CrossRef]
- Acquaro Junior, V.R.; Goméz-Ríos, G.A.; Tascon, M.; Queiroz, M.E.C.; Pawliszyn, J. Analysis of endocannabinoids in plasma samples by biocompatible solid-phase microextraction devices coupled to mass spectrometry. Anal. Chim. Acta 2019, 1091, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Looby, N.T.; Tascon, M.; Acquaro, V.R.; Reyes-Garcés, N.; Vasiljevic, T.; Gomez-Rios, G.A.; Wasowicz, M.; Pawliszyn, J. Solid phase microextraction coupled to mass spectrometry: Via a microfluidic open interface for rapid therapeutic drug monitoring. Analyst 2019, 144, 3721–3728. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Tominaga, K.; Sakashita, E.; Urabe, M.; Onuki, Y.; Gomi, A.; Yamaguchi, T.; Mieno, M.; Mizukami, H.; Kume, A.; et al. Comprehensive Metabolomic Analysis of IDH1 R132H Clinical Glioma Samples Reveals Suppression of β-oxidation due to Carnitine Deficiency. Sci. Rep. 2019, 9, 9787. [Google Scholar] [CrossRef] [PubMed]
- Yaligar, J.; Teoh, W.W.; Othman, R.; Verma, S.K.; Phang, B.H.; Lee, S.S.; Wang, W.W.; Toh, H.C.; Gopalan, V.; Sabapathy, K.; et al. Longitudinal metabolic imaging of hepatocellular carcinoma in transgenic mouse models identifies acylcarnitine as a potential biomarker for early detection. Sci. Rep. 2016, 6, 20299. [Google Scholar] [CrossRef] [PubMed]
- Zoni, E.; Minoli, M.; Bovet, C.; Wehrhan, A.; Piscuoglio, S.; Ng, C.K.Y.; Gray, P.C.; Spahn, M.; Thalmann, G.N.; Kruithof-De Julio, M. Preoperative plasma fatty acid metabolites inform risk of prostate cancer progression and may be used for personalized patient stratification. BMC Cancer 2019, 19, 1216. [Google Scholar] [CrossRef]
- Li, S.; Gao, D.; Jiang, Y. Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma. Metabolites 2019, 9, 36. [Google Scholar] [CrossRef]
- Reyes-Garcés, N.; Boyacı, E.; Gómez-Ríos, G.A.; Olkowicz, M.; Monnin, C.; Bojko, B.; Vuckovic, D.; Pawliszyn, J. Assessment of solid phase microextraction as a sample preparation tool for untargeted analysis of brain tissue using liquid chromatography-mass spectrometry. J. Chromatogr. A 2021, 1638, 11–13. [Google Scholar] [CrossRef]
- Stryjak, I.; Warmuzińska, N.; Bogusiewicz, J.; Łuczykowski, K.; Bojko, B. Monitoring of the influence of long-term oxidative stress and ischemia on the condition of kidneys using solid-phase microextraction chemical biopsy coupled with liquid chromatography–high-resolution mass spectrometry. J. Sep. Sci. 2020, 43, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Lendor, S.; Hassani, S.A.; Boyaci, E.; Singh, V.; Womelsdorf, T.; Pawliszyn, J. Solid Phase Microextraction-Based Miniaturized Probe and Protocol for Extraction of Neurotransmitters from Brains in Vivo. Anal. Chem. 2019, 91, 4896–4905. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Fack, F.; Tardito, S.; Hochart, G.; Oudin, A.; Zheng, L.; Fritah, S.; Golebiewska, A.; Nazarov, P.V.; Bernard, A.; Hau, A.; et al. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol. Med. 2017, 9, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Martinez-Ledesma, E.; Zheng, S.; Kim, H.; Barthel, F.; Jiang, T.; Hess, K.R.; Verhaak, R.G.W. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro-Oncology 2017, 19, 786–795. [Google Scholar] [CrossRef]
- Bogusiewicz, J.; Burlikowska, K.; Łuczykowski, K.; Jaroch, K.; Birski, M.; Furtak, J.; Harat, M.; Pawliszyn, J.; Bojko, B. New chemical biopsy tool for spatially resolved profiling of human brain tissue in vivo. Sci. Rep. 2021, 11, 19522. [Google Scholar] [CrossRef] [PubMed]
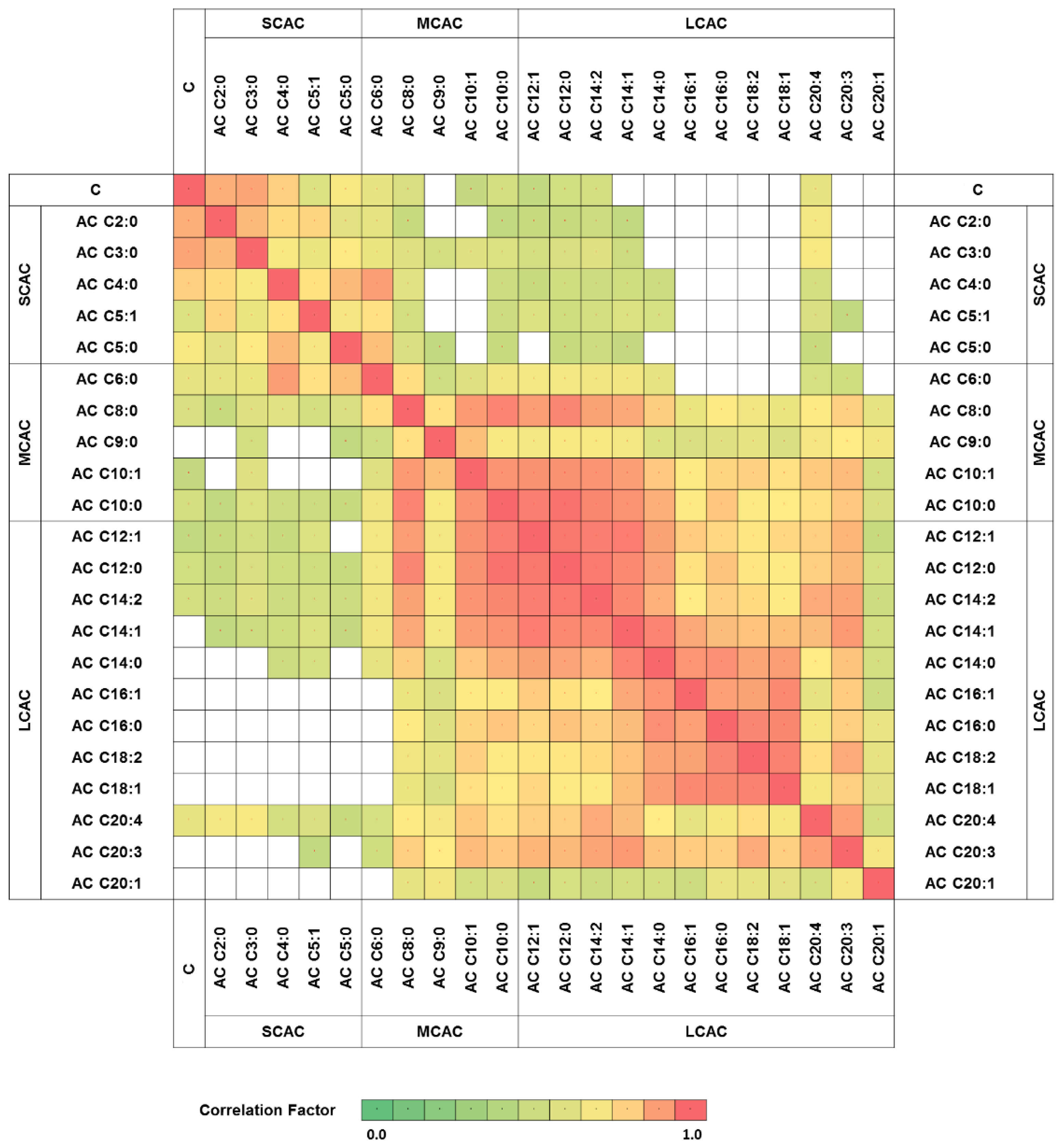
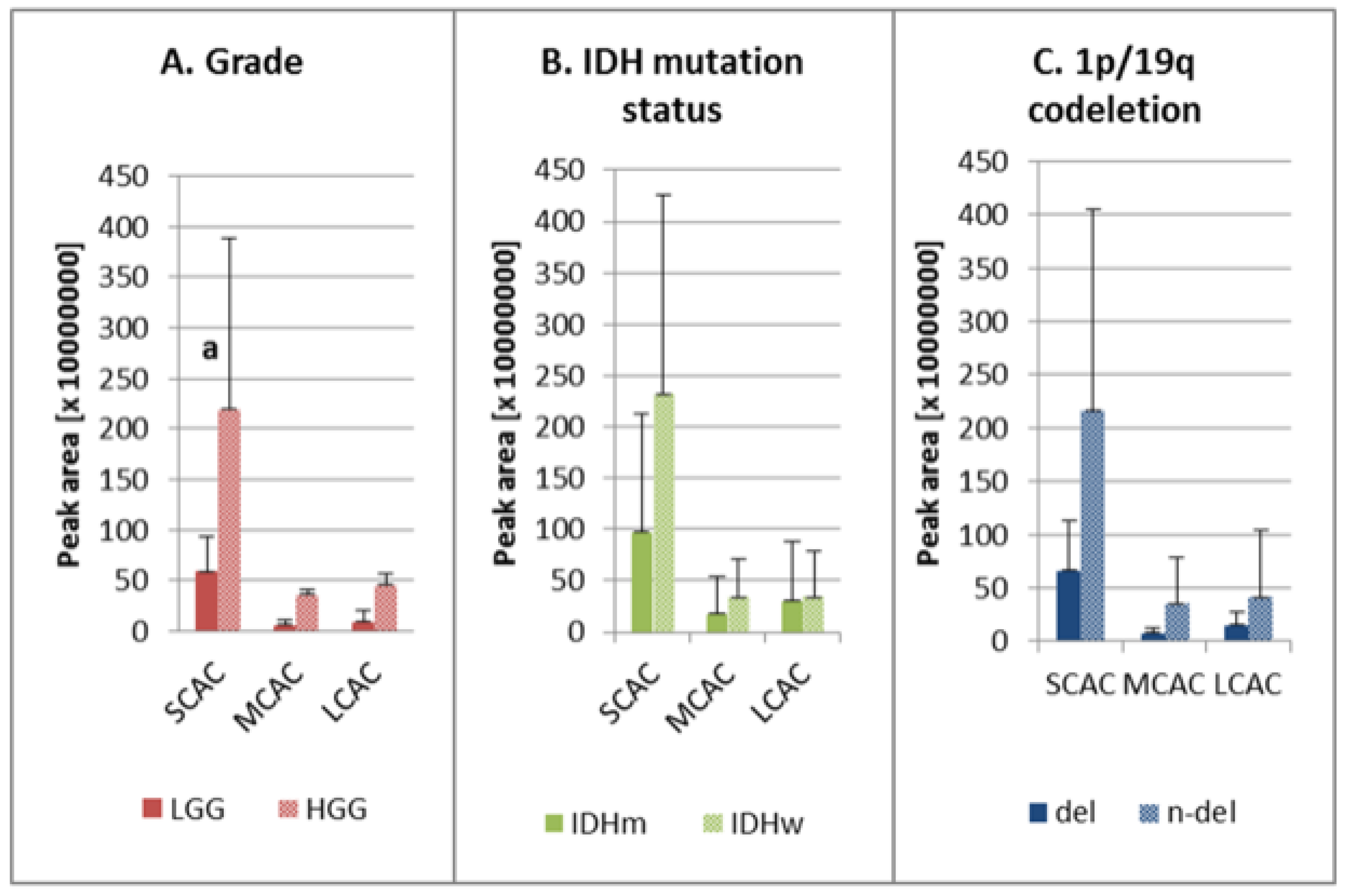
| Group | AC | Chemical Formula [M + H+] | M/Z [M + H+] | RT [min] | HGG: LGG | IDHw: IDHm | n-del: del |
|---|---|---|---|---|---|---|---|
| SCAC | AC C2:0 | C9H18O4N1 | 204.1231 | 12.46 | 0.87 | 0.97 | 0.93 |
| AC C3:0 | C10H20O4N1 | 218.1387 | 11.00 | 2.89 a | 1.68 | 1.68 | |
| AC C4:0 | C11H22O4N1 | 232.1543 | 9.72 | 0.85 | 0.94 | 0.84 | |
| AC C5:1 | C12H22O4N1 | 244.1543 | 9.28 | 0.95 | 0.78 | 0.83 | |
| AC C5:0 | C12H24O4N1 | 246.1700 | 8.96 | 1.38 | 1.00 | 1.22 | |
| MCAC | AC C6:0 | C13H26O4N1 | 260.1856 | 8.37 | 0.66 | 0.91 | 1.11 |
| AC C8:0 | C15H30O4N1 | 288.2169 | 7.79 | 1.20 | 1.04 | 0.95 | |
| AC C9:0 | C16H32O4N1 | 302.2325 | 7.62 | 29.98 a | 2.12 | 1.37 | |
| AC C10:1 | C17H32O4N1 | 314.2326 | 7.52 | 3.86 a | 1.18 | 0.74 | |
| AC C10:0 | C17H34O4N1 | 316.2484 | 7.48 | 1.53 | 1.09 | 0.93 | |
| LCAC | AC C12:1 | C19H36O4N1 | 342.2640 | 7.27 | 1.50 | 1.56 | 0.99 |
| AC C12:0 | C19H38O4N1 | 344.2796 | 7.23 | 1.34 | 1.66 b | 1.22 | |
| AC C14:2 | C21H38O4N1 | 368.2797 | 7.12 | 2.72 a | 1.94 | 1.33 | |
| AC C14:1 | C21H40O4N1 | 370.2953 | 7.07 | 1.06 | 1.19 | 1.02 | |
| AC C14:0 | C21H42O4N1 | 372.3108 | 7.10 | 0.89 | 1.08 | 0.96 | |
| AC C16:1 | C23H44O4N1 | 398.3266 | 6.96 | 0.82 | 0.60 b | 0.59 c | |
| AC C16:0 | C23H46O4N1 | 400.3423 | 6.96 | 0.92 | 0.96 | 1.06 | |
| AC C18:2 | C25H46O4N1 | 424.3422 | 6.89 | 1.05 | 0.79 | 0.86 | |
| AC C18:1 | C25H48O4N1 | 426.3579 | 6.84 | 0.87 | 0.79 | 1.00 | |
| AC C20:4 | C27H46O4N1 | 448.3424 | 6.80 | 2.43 | 1.77 | 1.73 | |
| AC C20:3 | C27H48O4N1 | 450.3578 | 6.78 | 13.89 a | 2.87 | 2.17 | |
| AC C20:1 | C27H50O4N1 | 454.3891 | 6.71 | 0.71 | 1.25 | 11.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogusiewicz, J.; Burlikowska, K.; Jaroch, K.; Gorynska, P.Z.; Gorynski, K.; Birski, M.; Furtak, J.; Paczkowski, D.; Harat, M.; Bojko, B. Profiling of Carnitine Shuttle System Intermediates in Gliomas Using Solid-Phase Microextraction (SPME). Molecules 2021, 26, 6112. https://doi.org/10.3390/molecules26206112
Bogusiewicz J, Burlikowska K, Jaroch K, Gorynska PZ, Gorynski K, Birski M, Furtak J, Paczkowski D, Harat M, Bojko B. Profiling of Carnitine Shuttle System Intermediates in Gliomas Using Solid-Phase Microextraction (SPME). Molecules. 2021; 26(20):6112. https://doi.org/10.3390/molecules26206112
Chicago/Turabian StyleBogusiewicz, Joanna, Katarzyna Burlikowska, Karol Jaroch, Paulina Zofia Gorynska, Krzysztof Gorynski, Marcin Birski, Jacek Furtak, Dariusz Paczkowski, Marek Harat, and Barbara Bojko. 2021. "Profiling of Carnitine Shuttle System Intermediates in Gliomas Using Solid-Phase Microextraction (SPME)" Molecules 26, no. 20: 6112. https://doi.org/10.3390/molecules26206112
APA StyleBogusiewicz, J., Burlikowska, K., Jaroch, K., Gorynska, P. Z., Gorynski, K., Birski, M., Furtak, J., Paczkowski, D., Harat, M., & Bojko, B. (2021). Profiling of Carnitine Shuttle System Intermediates in Gliomas Using Solid-Phase Microextraction (SPME). Molecules, 26(20), 6112. https://doi.org/10.3390/molecules26206112








