Abstract
Lipid A, the membrane-bound phosphoglycolipid component of bacteria, is held responsible for the clinical syndrome of gram-negative sepsis. In this study, the fragmentation behavior of a set of synthetic lipid A derivatives was studied by electrospray ionization multistage mass spectrometry (ESI-MSn), in conjunction with tandem mass spectrometry (MS/MS), using low-energy collision-induced dissociation (CID). Genealogical insight about the fragmentation pathways of the deprotonated 4’-monophosphoryl lipid A structural analogs led to proposals of a number of alternative dissociation routes that have not been reported previously. Each of the fragment ions was interpreted using various possible mechanisms, consistent with the principles of reactions described in organic chemistry. Specifically, the hypothesized mechanisms are: (i) cleavage of the C-3 primary fatty acid leaves behind an epoxide group attached to the reducing sugar; (ii) cleavage of the C-3’ primary fatty acid (as an acid) generates a cyclic phosphate connected to the nonreducing sugar; (iii) cleavage of the C-2’ secondary fatty acid occurs both in acid and ketene forms; iv) the C-2 and C-2’ primary fatty acids are eliminated as an amide and ketene, respectively; (v) the 0,2A2 cross-ring fragment contains a four-membered ring (oxetanose); (vi) the 0,4A2 ion is consecutively formed from the 0,2A2 ion by retro-aldol, retro-cycloaddition, and transesterification; and (vii) formations of H2PO4− and PO3− are associated with the formation of sugar epoxide. An understanding of the relation between 0,2A2 and 0,4A2-type sugar fragments and the different cleavage mechanisms of the two ester-linked primary fatty acids is invaluable for distinguishing lipid A isomers with different locations of a single ester-linked fatty acid (i.e., at C-3 or C-3’). Thus, in addition to a better comprehension of lipid A fragmentation processes in mass spectrometers, our observations can be applied for a more precise elucidation of naturally occurring lipid A structures.
1. Introduction
Lipid A is the active component of lipopolysaccharides (also called endotoxins) found in the outer membrane of most gram-negative bacteria [1]. On the one hand, lipid A has been known to affect the immune system, as it is an immunostimulant capable of inducing a protective, non-specific immune response in the host. On the other hand, depending on the dose and the structure of lipid A, the resulting immune response can lead to the uncontrolled production of inflammatory cytokines that may evoke fatal effects, such as high fever, septic shock, and multiorgan failure [2]. In this context, it is not surprising that several lipid A derivatives (synthetic or produced from bacteria) have been targets for developing effective adjuvants in vaccines and immunotherapy formulations [3,4].
Generally, bacterial lipid A comprises of a mixture of molecules, consisting of a β(1→6)-linked diglucosamine backbone, which is variably acylated with acyl or acyloxyacyl groups at the C-2, C-3, C-2’, and C-3’ positions, as well as phosphorylated at the C-1 and/or C-4’ positions [5]. The bisphosphoryl hexa-acyl species (carrying six fatty acyl chains in an asymmetric distribution (4 + 2) over the two glucosamine units), produced by E. coli, represents the highest endotoxic activity [6]. Alterations in the acylation and/or phosphorylation pattern, as well as post-translational modifications, significantly influence the immune system recognition (or evasion) [7] and potency of response [8] and may even modulate bacterial resistance to antibiotics [9].
In biological model systems in sepsis research, well-defined reference lipid A products are required to evaluate the cellular response after stimulation with lipid A. The most known synthetic lipid A standard is monophosphoryl lipid A (MPLA), which lacks the phosphate at the C-1 position and the acyl chain at the C-3 position, making it less toxic but with the retained activity of stimulating the adaptive immune response. This ligand is used as an adjuvant and it has been incorporated into several vaccines [10,11]. Besides, other synthetic MPLA structural analogs (such as those used in this study) containing a single molecular species are likely candidates in different adjuvant formulation development studies [4].
In order to accurately decipher the relationship between the functional groups and bioactivity of lipid A, a detailed characterization of its chemical structure is essential. Currently, tandem or multistage mass spectrometry (MS/MS, MSn) combined with soft ionization is the most effective method for enhancing structural information [12]. Complete structures of lipid A from a wide range of bacterial strains have been deducted through mass spectrometry alone (i.e., without prior separation or purification of the intrinsically heterogeneous lipid A samples) with the use of different mass analyzers [13,14,15,16,17,18,19,20,21]. Fragmentation rules, described for both negatively and positively charged lipid A molecules, greatly assist in the interpretation of the observed fragment ions [22,23,24,25]. Moreover, energy-resolved mass spectrometry (ERMS) may provide an opportunity to differentiate isobaric lipid A species [26].
Despite the fragmentation behavior of lipid A is reproducible and predictable, the general descriptions of the underlying mechanisms and dependence of fragmentation on specific linkages are rarely described or addressed. Mechanistic investigation, regarding the fragment ion formation by MSn, is essential to a better understanding of the ion chemistry and to facilitate the identification of specific product ions, such as cross-ring fragments or those formed by fatty acid eliminations. So far, there are only a few studies in the literature that contain information about the dissociation processes of 4’-monophosphoryl lipid A under low-energy CID conditions. Some of those studies deal with the elimination of the acyl groups closely situated to the phosphate anion [14,27] or chain cleavages taking place at a distance from the localized negative charge [16]. Furthermore, the mechanisms of cross-ring fragment formations for lipid A are scarcely detailed in the literature.
This study aimed to determine the multitude of ions formed during the fragmentation of four synthetic lipid A compounds with structural similarities using an ion trap and a triple quadrupole (QqQ) mass spectrometer with electrospray ionization (ESI), to discover more details about the specific dissociation events and fragmentation routes in the negative-ion mode. The explanation of characteristic ions in MSn mass spectra, obtained by low-energy collision-induced dissociation (CID), could help analysts better employ mass spectral observations for the structural elucidation of unknown lipid A.
2. Results and Discussion
2.1. MS2 Mass Spectra of the [M − H]− Lipid A Precursor Ions
For each synthetic standard, one major peak was detected in the mass spectrum, corresponding to the deprotonated 4’-monophosphoryl lipid A molecule [M − H]− (results not shown). Fragmentation patterns of each standard were studied at the MS2 stage, and results obtained with the two mass spectrometers at different RF amplitudes (ion trap) or collision energies (QqQ), respectively, were compared. As can be seen in the case of the deprotonated 3D-PHAD molecule (Figure S1 and S2 in Supplementary Information), there was a difference in the number, appearance, and intensity of fragment ions in the MS2 mass spectra from ion trap or QqQ. With the ion trap, mainly the first generation of product ions were produced from the selected precursor ion (m/z 1518); whereas, with the QqQ (especially when high collision energies were used), several other, higher generation product ions (resulting from a series of competing and consecutive reactions) were also formed. This also meant that sequential MSn analysis of ions, performed with the ion trap, was particularly useful for defining genealogical relationships; however, with the QqQ instrument, the genealogy between ions could not be established. On the other hand, with the QqQ instrument, fragment ions of m/z below 100 could be scanned; whereas, with the ion trap analyzer (which had a low-mass cut-off value), fragment ions were detected only from c.a. 27% of the precursor ion mass. So, the PO3− (m/z 79) and H2PO4− (m/z 97) fragment ions could not be observed during the sequential MSn analyses, even though they were formed.
MS2 mass spectra from ion trap of the [M − H]− precursor ions of the four lipid A standards are shown in Figure 1. The identity of fragment ions was determined by applying fragmentation rules (described previously) for the CID dissociation of lipid A molecules in the deprotonated form [23]. For each standard, the most intense peak (i.e., appearing at m/z 1290 for 3D-PHAD, m/z 1500 for 3D-(6-acyl)-PHAD, m/z 1516 for PHAD, and m/z 1488 for PHAD-504) was generated by releasing the fatty acid (C14:0, 228 u) from the C-3’ secondary position, known as the most labile cleavage site of 4’-monophosphoryl lipid A at low collision energies [23]. Next to this peak, another intense peak appeared for the 3-acyl standards (at m/z 1500 and m/z 1472 in Figure 1c,d, respectively), due to the elimination of a C14:0(3-OH) (Δm = 244 u) fatty acid from the C-3 primary position, known as the second most labile cleavage site [23]. Apart from these two, other types of single fatty acid loss from the precursor were not observed for the synthetic lipid A congeners used in this study. Meanwhile, in previous studies on bacterial-derived 4’-monophosphoryl lipid A species (such as from E. coli), usually, a third fragment was also detected arising from the fatty acid loss at the C-2’ secondary position from the precursor. If this were the case for the PHAD-504 standard, which has the same structure as the 4’-monophosphoryl lipid A species produced by E. coli (and the only structure among the standards in which the type of oxyacyl groups linked at C-2’ and C-3’were different), a fragment ion peak would be expected at m/z 1516. However, the lack of any signal at m/z 1516 in Figure 1d was a straightforward indication of the lack of release of the lauric acid (C12:0, 200 u) from the C-2’ branched position from the pure, synthetic sample. A conceivable explanation of the peak at m/z 1516, detected by others for various E. coli-type lipid A species [14,16], is that a structural isomer, i.e., the 1-monophosphoryl species may also have been present in the native samples. From that isomer, the fatty acid loss from the C-2’ secondary position is the most preferred cleavage process, based on previous observations on the CID fragmentation of lipid A phosphorylation isomers [24]. Furthermore, it should be noted that the complete lack of release of the C-2’ branched-chain in 4’-monophosphoryl lipid A may also have importance in the unambiguous identification of first-generation product ions from lipid A species having the same type of C-2’ and C-3’ secondary fatty acids, thus eliminating the theoretical possibility of multiple fragmentation routes, leading to the suggestion of various diastereomeric structures.
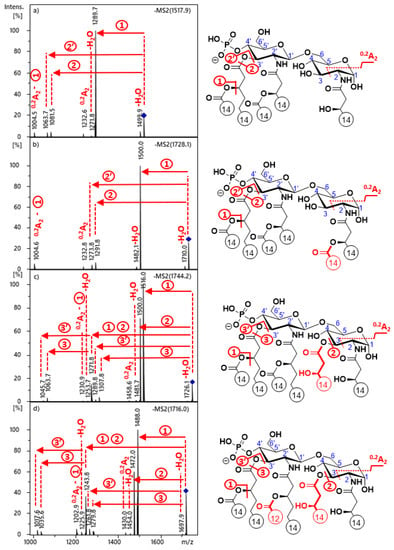
Figure 1.
ESI-ion trap MS2 mass spectra of the deprotonated 4’- monophosphoryl lipid A molecules of (a) 3D-PHAD, (b) 3D(6-acyl)-PHAD, (c) PHAD, and (d) PHAD-504, with the indication of cleavage sites in the structure. Fatty acyl chain lengths are given by numbers. The colored structural parts indicate the differences between the four lipid A congeners.
For the 3-acyl standards, another peak with moderate intensity appeared (at m/z 1272 and 1244 in Figure 1c,d, respectively), which derived from the combined losses of the C-3’ secondary and C-3 primary fatty acids (Δm = 472 u). From this latter fragment, further loss of a C14:1, as a ketene or an acid (Δm = 208 or 226 u, respectively) from the C-3’ primary position, resulted in pairs of low-intensity fragment ions (at m/z 1064 and m/z 1046 in Figure 1c and m/z 1036 and m/z 1018 in Figure 1d).
The loss of the C-3’ linked C14:0(3-O-C14:0) acyloxyacyl chain, both as a ketene or an acid (Δm = 436 or 454 u), gave rise to peak pairs, detected with very low intensities for all precursor ions (at m/z 1082/1064, m/z 1292/1274, m/z 1308/1290, and m/z 1280/1262 in Figure 1a–d, respectively). Water losses (Δm = 18 u) were detected with low intensities, both from the precursors and some acyl loss fragment ions. It should also be noted that neither the release of the C-2 secondary fatty acid nor releases of the amide-linked primary fatty acids were observed. Although, these were not surprising because losses of those substituents are usually less preferential at the MS2 stage [18,23].
In each MS2 mass spectrum, several 0,2A2 diagnostic cross-ring fragments appeared, although with relatively low intensities. As the nonreducing GlcN structures of the 3-deacyl standards were comparable, they presented the same 0,2A2 fragment ion peak pairs (at m/z 1233 and m/z 1005 in both Figure 1a,b), of which the latter was formed after the loss of a C14:0 from the C-3’ secondary position. On the other hand, different 0,2A2-type ions were detected for the 3-acyl standards (at m/z 1459 and m/z 1231 in Figure 1c and m/z 1431 and m/z 1203 in Figure 1d), preceding and proceeding the C-3’ secondary acyl loss, respectively.
Next, all the above-mentioned fragment ions were subjected to sequential mass analysis measurements in the ion trap instrument. The MSn mass spectra (n = 3–6) revealed further cleavage products, such as 0,4A2-type cross-ring fragments and those formed by the release of fatty acids from the C-2’ secondary and C-2’ and C-2 primary positions. In the following chapters, a selection of fragment ions will be used to elucidate details on the prominent fragmentation pathways and propose the mechanistic origin of the lipid A product ions detected in the negative-ion mode.
2.2. Proposed Mechanisms for the Release of the Secondary Fatty Acids
An alternative elimination mechanism of the C-3’ oxyacyl group (as free fatty acid), adjacent to the phosphate group, has already been explained by Madalinski et al. [16] using a charge-driven dissociation via the formation of an anion–molecule complex (Scheme 1). Namely, the phosphate group (basic site) removes a proton from the α-position (acidic site) of the primary fatty acyl chain linked at the C-3’ position, then an α–β elimination is followed by the formation of an intermediate anion–molecule complex.
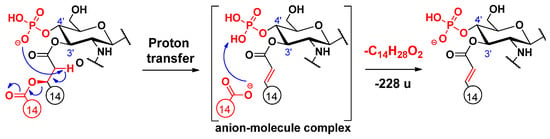
Scheme 1.
Proposed fragmentation mechanism for the loss of the C-3’ branched acyl chain [16].
Cleavages of the secondary fatty acids at C-2’ or C-2 were only observed in higher MSn stages (n = 4, 5, 6), after most of the labile ester-linked fatty acids had been released and/or cross-ring fragmentation(s) had occurred (for representative examples, see Figure S3 and S4 in Supplementary information). Similar to the previously described cleavage mechanism of the C-3’ oxyacyl group (vide supra), we propose similar processes for the eliminations of the C-2 and C-2’ secondary fatty acids (Scheme 2 and Scheme 3, respectively), proceeding with the release of the C-3’ primary fatty acid as a ketene. Only the proton transfer step is different, as the acidic site is at the α-position of the respective amide group, and the basic site is the phosphate ion at C-4’. Furthermore, according to the MSn measurements, the C-2’ secondary fatty acid was also eliminated in ketene form (Figure S5). The elimination can be explained with an intramolecular hydrogen transfer from the α-carbon of the ester group and the direct O–CO bond cleavage occurring without anion–molecule complex formation (Scheme 4) (for eliminations proceeding the release of the C-3’ primary fatty acid as an acid, see Schemes S1–S3). Apart from the product ions assigned to the secondary fatty acyl eliminations, the released secondary fatty acids could also be detected in deprotonated forms (e.g., m/z 227 for a C14:0 in Figure S2) but only with very low intensities and under relatively high-energy collisional conditions.
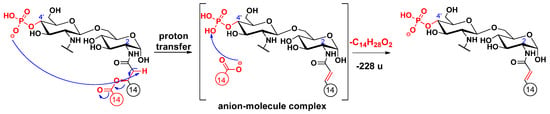
Scheme 2.
Proposed fragmentation mechanism for the loss of the C-2 branched acyl chain (as an acid).
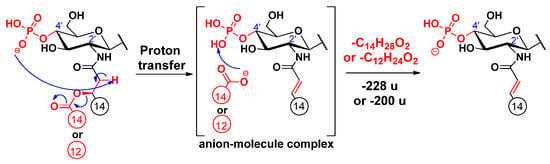
Scheme 3.
Proposed fragmentation mechanism for the loss of the C-2’ branched acyl chain (as an acid).
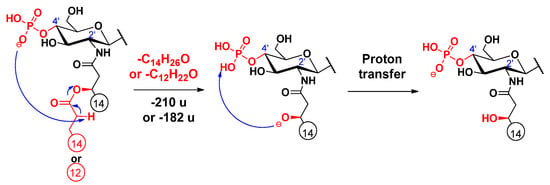
Scheme 4.
Proposed fragmentation mechanism for the loss of the C-2’ branched acyl chain (as a ketene).
Furthermore, it should be noted that the precursor ions (shown in Scheme 2, Scheme 3 and Scheme 4) may have even different structures. Either a double bond is present in the sugar ring (due to loss of the C-3’ linkage) and the negative charge is on the P–O oxygen atom or it involves a cyclic phosphate species carrying the negative charge, as suggested for the first time in this study (vide infra).
2.3. Proposed Mechanisms for the Release of the Primary Fatty Acids
Cleavage of the C-3 linked primary fatty acyl chain was observed for the 3-acyl lipid A molecules: PHAD and PHAD-504. Unlike the C-3’ linked primary fatty acid, which can leave both as an acid and a ketene (vide infra), the C-3 linked fatty acyl chain is lost only as an acid. The first step of this charge-driven process (induced by a long proton transfer, as proposed before [16]) is the formation of a reactive alkoxide group at the C-4 position. Next, either a ketone derivative can be formed (Scheme 5), as previously reported [16], or an epoxide derivative may develop (Scheme 6), as proposed here (it should be noted that epoxide sugars are versatile intermediates in organic synthesis [28]). While the former involves an anion–molecule complex formation by hydride transfer, the latter product ion assumes an anion–molecule complex, resulting from the anti-attack of the alkoxide ion at C-4 on the hydroxymyristic acid chain at C-3.
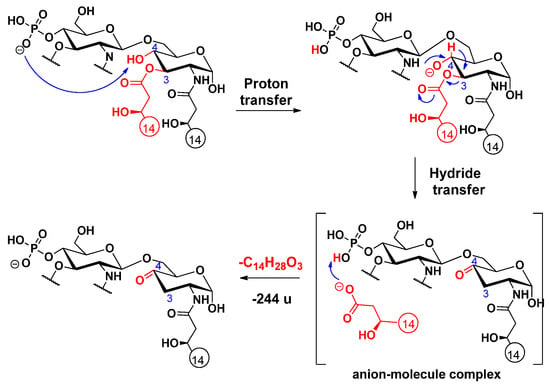
Scheme 5.
Proposed fragmentation mechanism for the loss of the C-3 primary fatty acid, leading to the formation of a ketone group in the sugar ring [16].
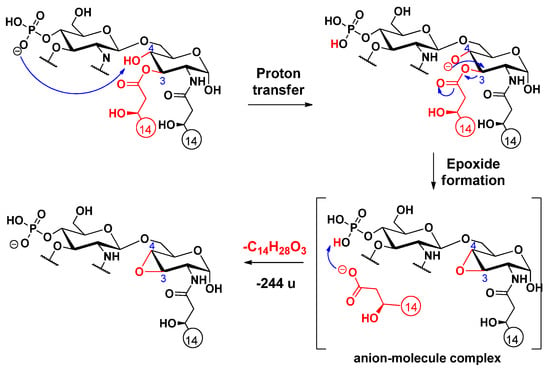
Scheme 6.
Proposed fragmentation mechanism for the loss of the C-3 primary fatty acid, leading to the formation of a sugar epoxide.
The primary fatty acid was eliminated from the C-3’ position as an unsaturated primary fatty acid (C14:1) in all standards (observe that the preceding elimination of the C-3’ branched-chain leaves behind a double bond). Regularly, the loss of this primary substituent occurs in two forms, as a ketene and an acid, by two competing processes, resulting in diagnostic peak pairs with 18 u apart in the mass spectrum.
For the ketene loss, a charge-driven fragmentation process has been suggested earlier [16]. In our case, this type of process is initiated with a proton removal (where the proton is uptaken by the anionic charge of the phosphate group) from the allylic position of the unsaturated chain; then, the conjugated carbanion promotes the direct O–CO bond cleavage, leading to the release of the unsaturated alkyl chain as ketene (Scheme 7). In addition, we hypothesize that a proton transfer takes place, and the negative charge is held by the phosphate group in the end.
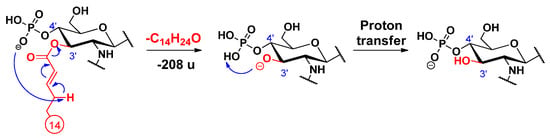
Scheme 7.
Proposed fragmentation mechanism for the loss of the C-3’ primary fatty acid residue as a ketene, along with a proton transfer.
For loss of the C-3’ primary chain as a free fatty acid, a charge-remote fragmentation process has been proposed [14], in which the removal of a hydrogen atom at C-4’ led to the formation of a double bond between the C-3’ and C-4’ carbon atoms (Scheme 8). Besides, we propose here an alternative, charge-driven dissociation pathway. Namely, a five-membered cyclic phosphate is generated in a process initiated by the attack of the anionic charge site of the phosphate group on the carbon C-3’, leading to the removal of the unsaturated alkyl chain as a free fatty acid (Scheme 9). In fact, this process is very similar to what was observed for cyclic phosphate anions of diacyl glycerophosphoethanolamines [29].
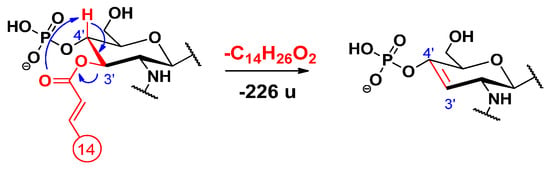
Scheme 8.
Proposed fragmentation mechanism for the loss of the C-3’ primary fatty acid residue as an acid (as described before [14]).
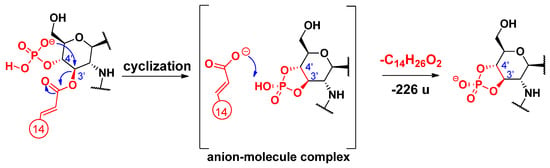
Scheme 9.
Proposed alternative fragmentation mechanism for the loss of the C-3’ primary fatty acid residue as an alkyl carboxylic acid.
Elimination of the amide-linked primary fatty acid at C-2’ was observed only in the case of 3D-PHAD, having the lowest molecular weight among the four standards. Here, a fragment ion at m/z 282 (Figure S5) was formed by eliminating a residual C14:1 from the C-2’ position as ketene (Δm = 208 u). The proposed mechanism starts with a proton transfer from the allylic position of the amide-linked unsaturated alkyl chain to the phosphate group, either to the cyclic one (as shown in Scheme 10) or to the noncyclic alternative (Scheme S4). Then, the conjugated carbanion promotes the direct N–CO bond cleavage.
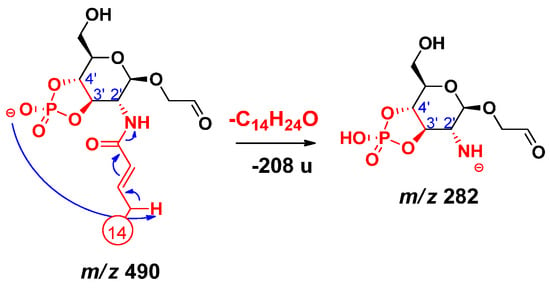
Scheme 10.
Proposed fragmentation mechanism leading to the release of the residual C-2’ primary fatty acid as ketene.
Cleavage of the amide-linked C-2 primary fatty acyl chain was observed for all standards. An example is shown for PHAD-504 (Figure S4), where the fragment at m/z 774 was formed by elimination of the hydroxymyristic acid amide (Δm = 243 u) from the ion at m/z 1017. The first step of this charge-driven process (induced by a long proton transfer) is the formation of a reactive alkoxide group at the C-1 position. Next, a ketone derivative is formed by a hydride transfer via the formation of an anion–molecule complex (Scheme 11).
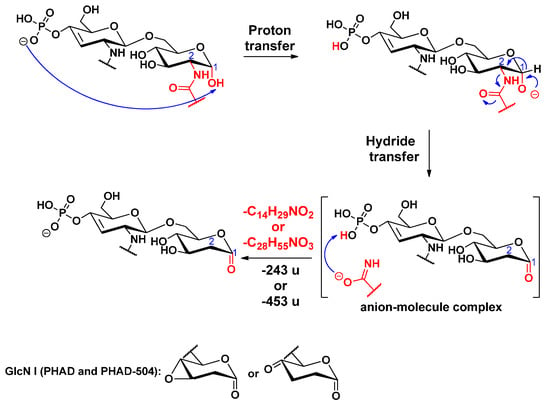
Scheme 11.
Proposed fragmentation mechanism for the loss of the C-2 primary fatty acid, leading to the formation of a ketone group in the sugar ring.
2.4. Cross-ring Fragmentations
In the MS3 spectra of selected precursors, m/z 1290 of 3D-PHAD (Figure 2a) and m/z 1500 of 3D(6-acyl)-PHAD (Figure S6a), both of which lack the C-3’ secondary acyl chain, two types of cross-ring fragment ions were detected: 0,2A2 (e.g., at m/z 1005) and 0,4A2 (at m/z 945). However, the 0,4A2 fragment ion was also present in the MS4 spectrum of the 0,2A2 ion selected as a precursor (see Figure 2b for 3D-PHAD and Figure S6b for 3D(6-acyl)-PHAD). This observation suggests that a structural relationship exists between the two cross-ring fragments. As the mechanisms of cross-ring fragment formations for lipid A are not very well detailed in the literature, we took these dissociations under investigation.
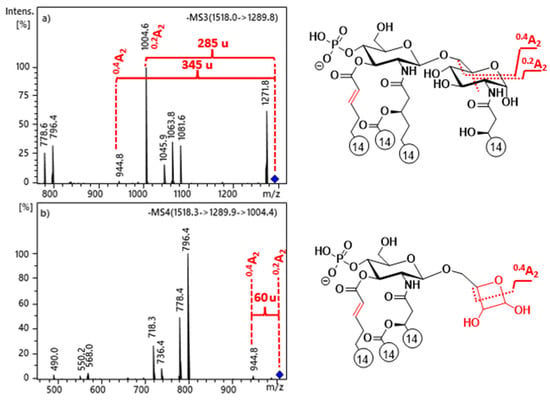
Figure 2.
ESI-ion trap mass spectra obtained at MS3 and MS4 stages of selected ions at (a) m/z 1290 and (b) m/z 1005 for 3D-PHAD, demonstrating the structural relationship between 0,2A2 and 0,4A2 cross-ring fragment ions in 3-deacyl lipid A.
Generally, 0,2A2 and 0,4A2 cross-ring fragments are simultaneously detected in the MS2 mass spectra of 4’-monophosphoryl lipid A species, in which the C-1 position is free [25]. Thus, it can be assumed that the first step in the mechanism of A-type cross-ring cleavages involves ring-opening [30] of the reducing sugar, in order to produce an equilibrium amount of the open-chain aldehyde, which is then capable of acting as a reducing agent. Consequently, this form can undergo a retro-aldol reaction [31], in order to yield a shorter chain of α,β-dihydroxy aldehyde (i.e., 0,2A2 fragment) via the neutral loss of the anchored enol (Scheme 12). For the further decomposition of the 0,2A2 product ion, we suggested two pathways: (i) either a second retro-aldol reaction takes place and leads directly to the 0,4A2 fragment ion by loss of an ethane-1,2-diol (Δm = 60 u); or (ii) the hydroxyl group attached to C-5 reacts with the formed aldehyde, which, by a second hemiacetal-aldehyde equilibrium, forms a four-membered ring, so-called oxetanose [32], which can undergo a [2 + 2] retrocycloaddition reaction [33] (Scheme 12). The predominance of the oxetanose derivative is also supported by the study of Lee et al. [34], in which they showed via synthesis of diacylglycerol analogs that a four-membered ring (oxetanose) was obtained in equilibrium, with its corresponding open-chain form (nonoxetanose) at a ratio of 5:1.
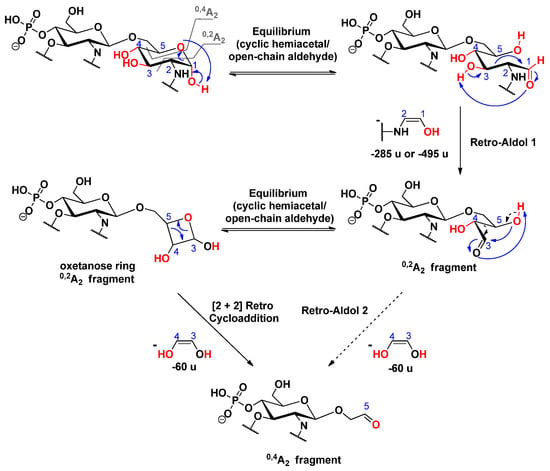
Scheme 12.
Proposed mechanisms for the consecutive formation of 0,2A2 and 0,4A2 fragment ions in 3-deacyl lipid A.
In the MS3 spectra of selected precursors, m/z 1488 of PHAD-504 (Figure 3a) and m/z 1516 of PHAD (Figure S7a), both of which had lost the C-3’ secondary acyl chain, signals originating from 0,2A2 and 0,4A2 cross-ring fragmentations (m/z 1203 and 917 for PHAD-504, and m/z 1231 and 944 for PHAD, respectively) appeared, as well. Furthermore, a relationship between the 0,2A2 and 0,4A2 ions was observed in the MS4 spectrum of the chosen 0,2A2 precursor ion (Figure 3b for PHAD-504, Figure S7b for PHAD). Due to the presence of an additional C14:0(3-OH) chain at C-3 in the reducing sugar of these molecules, the proposed mechanism of cross-ring cleavages is one step different from that described above for the 3-deacyl lipid A derivatives. Namely, following the first step (i.e., ring-opening of the reducing sugar), an intramolecular transesterification occurs between the hydroxyl group at C-4 and the ester chain at C-3, leading to a derivative that can undergo a retro-aldol reaction. Thus, it is suggested that the 0,4A2 fragment be formed, either from a second retro-aldol or a [2 + 2] retro-cycloaddition reaction (Scheme 13).
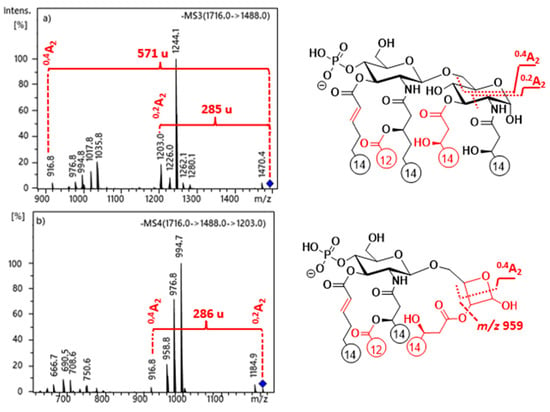
Figure 3.
ESI-ion trap mass spectra obtained at MS3 and MS4 stages of selected ions at (a) m/z 1488 and (b) m/z 1203 for PHAD-504, demonstrating the structural relationship between 0,2A2 and 0,4A2 cross-ring fragment ions in 3-acyl lipid A.
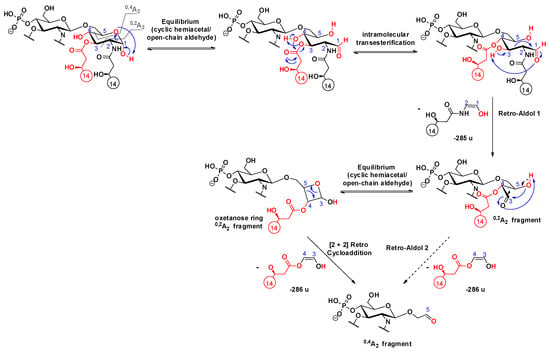
Scheme 13.
Proposed mechanisms for the consecutive formation of 0,2A2 and 0,4A2 fragment ions in 3-acyl lipid A.
In addition, a small signal appeared at m/z 959 in Figure 3b, resulting from the release of the fatty acyl chain from the oxetanose ring in the 0,2A2 fragment. Indeed, the cleavage of this fatty acyl chain (originally linked at C-3 in lipid A) may occur from the 0,2A2 fragment, but only with a small probability. Additionally, note that when the C-3 primary fatty acid had been eliminated (i.e., preceding the cross-ring fragmentation), the 0,2A2 fragment cannot be formed at all [14].
In summary, the observation that there is a structural relationship between the two A-type cross-ring fragments and those intensity ratios between them are different in the case of 3-acyl and 3-deacyl lipid A also implies that one may easily distinguish the simultaneous presence of C-3 or C-3’ acylation isomers in a single sample. Namely, the appearance of low-intensity 0,2A2 fragments in the mass spectrum is characteristic of a 3-acyl, 3’-deacyl lipid A; meanwhile, high-intensity 0,2A2 fragments appearing as peak pairs (with 18 u apart) are typical for a 3-deacyl, 3’-acyl lipid A. Without applying these “rules”, misleading information of some product ions in complex lipid A mixtures may be obtained, and some low abundance, but biologically important, acylation isomers can remain unexplored [23,35].
2.5. Dephosphorylation
Two signals, at m/z 79 and 97, corresponding to the monometaphosphate (PO3−) and dihydrogen phosphate (H2PO4−) ions, respectively, were observed in the MS2 mass spectra from QqQ of the four lipid A precursor ions (an example for 3D-PHAD is shown in Figure S2). As both fragment ions were related to the C-4’ phosphate moiety, we took mechanisms leading to their formation under investigation. In the literature, the mechanism of dephosphorylation reaction of lipid A under acidic conditions has only been proposed for the anomeric phosphate group (at position C-1) [16].
By comparing the MS2 mass spectra related to different collision energies in Figure S2, it could be seen that as the collision energy increased, the relative intensity of the peaks at m/z 796 and 778 decreased, while that of the peaks at m/z 79 and 97 increased. The peaks at m/z 796 and 778 each correspond to an 0,2A2 fragment, which had lost the C-3’ acyloxyacyl group either as a ketene or an acid, respectively. The formation of dihydrogen phosphate ion (H2PO4−, m/z 97) was considered from the fragment at m/z 796, by two possible pathways (Scheme 14). In the first way, an epoxidation takes place (involving the antiperiplanar relationships between the formed oxide ion at C-3’ and the phosphate group), which will then easily promote the removal of the H2PO4 that will hold the negative charge. In the second way, a hydride transfer takes place, leading to the formation of the H2PO4− ion. The formation of monometaphosphate ion (PO3−, m/z 79) was considered from the fragment at m/z 778, by two pathways, starting from the cyclic phosphate derivative (Scheme 15), from which the monometaphosphate ion could be considered as a good leaving group, either following a hydride transfer or an epoxide formation. However, the pathway starting from the non-cyclic phosphate derivative (Scheme 16) was not considered, as in that case, the PO3− would be formed via the removal of a proton from the detached neutral metaphosphoric acid (HPO3), which would lead to detection of the intermediate fragmentation product, i.e., the negatively charged lipid A, without the C-4’ phosphate group; albeit, such a phosphate-free product ion (m/z 698) was not seen in the MSn mass spectra.
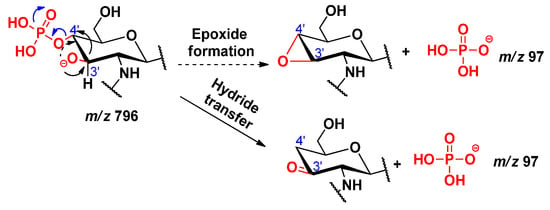
Scheme 14.
Proposed mechanisms for the formation of dihydrogen phosphate ion H2PO4−.
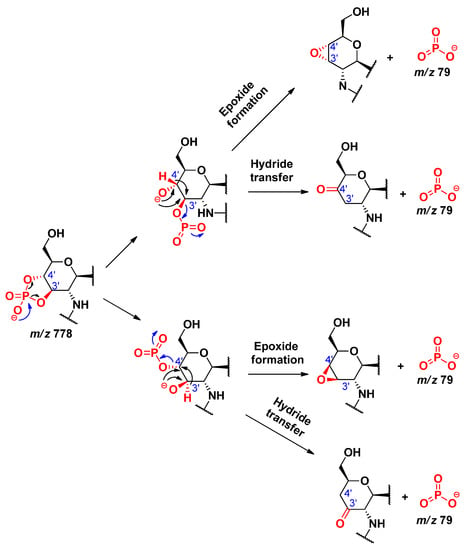
Scheme 15.
Proposed alternative routes for the formation of metaphosphate ion, PO3−, from the cyclic phosphate lipid A derivative.
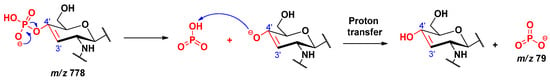
Scheme 16.
An unlikely mechanism for the formation of metaphosphate ion, PO3−, from the noncyclic phosphate lipid A derivative.
3. Experimental
3.1. Chemicals
Methanol and dichloromethane (LC-MS Chromasolv grade) were purchased from Sigma–Aldrich (Steinheim, Germany).
3.2. Lipid A Samples
Four types of synthetic monophosphoryl lipid A standards were purchased from Sigma–Aldrich, the exclusive Avanti Polar Lipids provider (Alabaster, AL, USA) in Hungary. The standards were monophosphoryl 3-deacyl penta-acyl lipid A (3D-PHAD, Pat No. 9,241,988), monophosphoryl 3-deacyl hexa-acyl lipid A (3D(6-acyl)-PHAD), monophosphoryl lipid A (PHAD), and monophosphoryl lipid A-504 (PHAD-504). The standards were purchased in their ammonium salt forms.
3.3. Lipid A Sample Preparation
A few amounts (0.1 mg in powder form) of each synthetic lipid A were introduced into Eppendorf tubes and suspended in a mixture of 900 μL methanol and 100 μL dichloromethane. The samples were vortexed and subsequently put in an ultrasonic bath (5 min) to be highly homogenous. Then, 100 μL of each sample was introduced into sealed glass vials, followed by the addition of 900 μL of methanol. After vortexing, the samples were ready for injection.
3.4. Mass Spectrometric Analysis
ESI-ion trap MSn analysis was performed in the negative-ion mode using an LC/MSD Trap XCT Plus mass spectrometer (Agilent Technologies, Waldbronn, Germany), controlled with the Agilent LC/MSD Trap Soft-ware 5.3. A syringe pump, set at a flow rate of 3 µL/min, was used for direct infusion of the samples into the ion source. The electrospray capillary high voltage was applied at 3500 V. Nitrogen was used both as nebulizer (15 psi, 5 L/min) and dry gas (at 325 °C). The smart parameter setting (SPS) of the software was utilized to simplify the ion-trap operation, i.e., the target mass was set at the mass of the deprotonated lipid A, and both compound stability and trap drive level were set at 100%. Mass spectra were scanned in the range of m/z 100–2200, with a scan speed of 8100 m/z per second. The mass isolation window for precursor ion selection was set to 2 m/z in the multiple-stage MS analyses. Each precursor ion was excited by resonant excitation voltage, fixed between 0.5 V and 1.5 V, according to the intensity of the parent ion signal. Negative-ion mass spectra, including collision-induced dissociation (CID) spectra, were averaged over 5–20 scans, depending on the relative abundances of the precursor ions.
ESI-QqQ measurements were performed in the negative-ion mode using a SCIEX Triple QuadTM 6500+ System (AB Sciex LP, Concord, ON, Canada), equipped with an integrated syringe pump set at a flow rate of 25 µL/min. Mass spectra were scanned in the range of m/z between 50 and 1600. The scan rate was 1000 for both Q1 MS and product ion MS2. The ion spray voltage and temperature were set at –4500 V and 200 °C, respectively. The curtain gas was set at a flow rate of 20 psi, gas 1 (nebulizer gas) was set at 3 psi, and gas 2 (heater gas) was set at 0 psi. The collision gas (CAD) was set at 8. Data were acquired using Analyst software 1.7.2.
3.5. Data Evaluation
The evaluation of the MSn mass spectra was made by considering the monoisotopic masses of the fragments ions and the neutral losses, based on the known composition and structure of the precursor ions. The following abbreviations were used to indicate the chemical compositions during the interpretation of mass spectra: GlcN (2-amino-2-deoxy-glucopyranose), C12:0 (dodecanoic acid or lauric acid), C14:0(3-OH) (3-hydroxytetradecanoic acid or hydroxymyristic acid), C14:0 (tetradecanoic acid or myristic acid), and C14:1 (tetradecenoic acid or unsaturated myristic acid). The classical nomenclature for glycoconjugates and cross-ring fragmentation nomenclature, described by Domon and Costello [36], was adopted to designate the fragment ions.
4. Conclusions
The objective of the present paper was to propose alternative dissociation mechanisms of deprotonated 4’-monophosphoryl lipid A species, as the generation of particular product ions in gas-phase has been scarcely explained in the literature. Mechanisms proposed here consider both single and multistep cleavages, with rearrangements occurring either by proton or hydride transfer and cyclization. Multistep fragmentation experiments (with ESI-ion trap) were especially useful for ascertaining the relationship between 0,2A2 and 0,4A2 cross-ring fragment ions, while ESI-QqQ (with varying energy levels) permitted the finding of a relationship between product ions related to dephosphorylation. By applying low-energy conditions, mainly charge-induced processes were considered for the release of the different ester- or amide-linked fatty acyl chains. Besides providing a better understanding of lipid A fragmentation processes, our observations may also have some potential applications in the interpretation of the mass spectra of naturally occurring lipid A compounds. For instance, the preferential loss of the secondary acyl group at C-3’ over that at C-2’ provides important information on the structure of 4’-monophosphoryl lipid A, carrying the same secondary substituents at these positions. Moreover, as first-generation product ions formed by the release of the C-2’ secondary fatty acyl chain are not favored in 4-monophosphoryl species (unlike 1-monophosphoryl lipid A); thus, the detection of both C-3’ and C-2’ deficient first-generation fragments in the mass spectrum indicates the presence of phosphorylation isomers in the sample. Furthermore, the detection of several pairs of peaks, with 18 u apart in the MS/MS or higher stage MSn mass spectra, in combination with relatively high-intensity 0,2A2 fragments, indicates the presence of a 3’-acyl, 3-deacyl lipid A compound in the sample.
Supplementary Materials
The following are available online, Figure S1: ESI-ion trap MS2 mass spectra of the deprotonated form of the 3D-PHAD lipid A molecule (precursor ion m/z 1518), applying different RF amplitudes; Figure S2: ESI-QqQ MS2 mass spectra of the deprotonated form of the 3D-PHAD lipid A molecule (precursor ion m/z 1518), applying different collision energies (CE); Figure S3: ESI-ion trap mass spectra, obtained at MS4, MS5, and MS6 stages for several fragment ions of 3D-PHAD, demonstrating the release of myristic acid (Δm = 228u) as an acid from the C-2’ secondary position; Figure S4: ESI-ion trap mass spectra, obtained at MS4 and MS5 stages for several fragment ions of PHAD-504, demonstrating the release of lauric acid (Δm = 200 u) as an acid from the C-2’ secondary position, and the release of hydroxymyristic acid amide from the C-2 primary position (Δm = 243 u); Figure S5: ESI-ion trap mass spectra, obtained at (a) MS4 stage of the selected ion at m/z 718 for 3D-PHAD, demonstrating the release of myristic acid (Δm = 210 u) as a ketene from the C-2’ secondary position, and the release of unsaturated myristic acid (Δm = 208 u) as a ketene from C-2’ primary position; and (b) at MS5 stage of the selected ion at m/z 690 for PHAD-504, demonstrating the release of lauric acid (Δm = 182 u) as a ketene from the C-2’ secondary position; Figure S6: ESI-ion trap mass spectra, obtained at MS3 and MS4 stages of selected precursor ions at (a) m/z 1500 and (b) m/z 1005 for 3D(6-acyl)-PHAD, demonstrating the structural relationship between 0,2A2 and 0,4A2 cross-ring fragment ions in 3-deacyl lipid A; Figure S7: ESI-ion trap mass spectra, obtained at MS3 and MS4 stages of selected precursor ions at (a) m/z 1516 and (b) m/z 1231 for PHAD, demonstrating the structural relationship between 0,2A2 and 0,4A2 cross-ring fragment ions in 3-acyl lipid A; Scheme S1: Alternative fragmentation pathways for the elimination of the C-2 branched acyl chain as an acid in lipid A; Scheme S2: Alternative fragmentation pathways for the elimination of the C-2’ branched acyl chain as an acid in lipid A; Scheme S3: Alternative fragmentation pathway for the C-2’ branched acyl chain as a ketene in lipid A; Scheme S4: Proposed mechanism leading to the release of the C-2’ primary fatty acid as ketene in lipid A.
Author Contributions
Experimental part and writing-original draft preparation, I.A.; supervisions, writing, and completing-final version, Á.D. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Research, Development and Innovation Office grant numbers FK-129038 and K-125275. The APC was funded by National Research, Development and Innovation Office grant number FK-129038.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
I.A. acknowledges the Stipendium Hungaricum Scholarship. The research was performed in collaboration with the Mass Spectrometry Core Facility at the Szentágothai Research Centre of the University of Pécs.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Caroff, M.; Novikov, A. Lipopolysaccharides: Structure, Function and Bacterial Identification. OCL: Oilseeds Fats, Crops Lipids 2020, 27, 31. [Google Scholar] [CrossRef]
- Cavaillon, J.M. Exotoxins and Endotoxins: Inducers of Inflammatory Cytokines. Toxicon 2018, 149, 45–53. [Google Scholar] [CrossRef]
- Baldridge, J.R.; Crane, R.T. Monophosphoryl Lipid A (MPL) Formulations for the Next Generation of Vaccines. Methods 1999, 19, 103–107. [Google Scholar] [CrossRef]
- Arenas, J. The Role of Bacterial Lipopolysaccharides as Immune Modulator in Vaccine and Drug Developement. Endocr. Metab. Immune Disord. Drug Targets 2012, 12, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Zahringer, U.; Lindner, B.; Rietschel, E.T. Molecular-Structure of Lipid-A, the Endotoxic Center of Bacterial Lipopolysaccharides. Adv. Carbohydr. Chem. Biochem. 1994, 50, 211–276. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di Lorenzo, F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of Lipid A: At the Heart of Innate Immunity. Chem. Eur. J. 2015, 21, 500–519. [Google Scholar] [CrossRef]
- Matsuura, M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front. Immunol. 2013, 4, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erridge, C.; Bennett-Guerrero, E.; Poxton, I.R. Structure and Function of Lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [Google Scholar] [CrossRef]
- Anandan, A.; Vrielink, A. Structure and Function of Lipid A-Modifying Enzymes. Ann. N. Y. Acad. Sci. 2020, 1459, 19–37. [Google Scholar] [CrossRef]
- Mata-Haro, V.; Cekic, C.; Martin, M.; Chilton, P.M.; Casella, C.R.; Mitchell, T.C. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science 2007, 316, 1628–1632. [Google Scholar] [CrossRef]
- Casella, C.R.; Mitchell, T.C. Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cellular and Molecular Life Sciences 2008, 65, 3231–3240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilár, A.; Dörnyei, Á.; Kocsis, B. Structural Characterization of Bacterial Lipopolysaccharides With Mass Spectrometry and on- and off-line Separation Techniques. Mass Spectrom. Rev. 2013, 32, 90–117. [Google Scholar] [CrossRef] [PubMed]
- Boue, S.M.; Cole, R.B. Confirmation of the Structure of Lipid A from Enterobacter Agglomerans by Electrospray Ionization Tandem Mass Spectrometry. J. Mass Spectrom. 2000, 35, 361–368. [Google Scholar] [CrossRef]
- Kussak, A.; Weintraub, A. Quadrupole Ion-Trap Mass Spectrometry to Locate Fatty Acids on Lipid A from Gram-Negative Bacteria. Anal. Biochem. 2002, 307, 131–137. [Google Scholar] [CrossRef]
- Bedoux, G.; Vallee-Rehel, K.; Kooistra, O.; Zahringer, U.; Haras, D. Lipid A Components from Pseudomonas Aeruginosa PAO1 (Serotype O5) and Mutant Strains Investigated by Electrospray Ionization Ion-Trap Mass Spectrometry. J. Mass Spectrom. 2004, 39, 505–513. [Google Scholar] [CrossRef]
- Madalinski, G.; Fournier, F.; Wind, F.L.; Afonso, C.; Tabet, J.C. Gram-Negative Bacterial Lipid A Analysis by Negative Electrospray Ion Trap Mass Spectrometry: Stepwise Dissociations of Deprotonated Species under Low Energy CID Conditions. Int. J. Mass Spectrom. 2006, 249, 77–92. [Google Scholar] [CrossRef]
- Schilling, B.; McLendon, M.K.; Phillips, N.J.; Apicella, M.A.; Gibson, B.W. Characterization of Lipid a Acylation Patterns in Francisella Tularensis, Francisella Novicida, and Francisella Philomiragia using Multiple-Stage Mass Spectrometry and Matrix-Assisted Laser Desorption/Ionization on an Intermediate Vacuum Source Linear Ion Trap. Anal. Chem. 2007, 79, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Silipo, A.; De Castro, C.; Lanzetta, R.; Molinaro, A.; Parrilli, M.; Vago, G.; Sturiale, L.; Messina, A.; Garozzo, D. Structural Characterizations of Lipids A by MS/MS of Doubly Charged Ions on a Hybrid Linear Ion Trap/Orbitrap Mass Spectrometer. J. Mass Spectrom. 2008, 43, 478–484. [Google Scholar] [CrossRef]
- Jones, J.W.; Cohen, I.E.; Turecek, F.; Goodlett, D.R.; Ernst, R.K. Comprehensive Structure Characterization of Lipid A Extracted from Yersinia Pestis for Determination of its Phosphorylation Configuration. J. Am. Soc. Mass Spectrom. 2010, 21, 785–799. [Google Scholar] [CrossRef] [Green Version]
- Morrison, L.J.; Parker, W.R.; Holden, D.D.; Henderson, J.C.; Boll, J.M.; Trent, M.S.; Brodbelt, J.S. UVliPiD: A UVPD-Based Hierarchical Approach for De Novo Characterization of Lipid A Structures. Anal. Chem. 2016, 88, 1812–1820. [Google Scholar] [CrossRef] [Green Version]
- Tawab, A.; Akbar, N.; Hasssan, M.; Habib, F.; Ali, A.; Rahman, M.; Jabbar, A.; Rauf, W.; Iqbal, M. Mass Spectrometric Analysis of Lipid A Obtained from the Lipopolysaccharide of Pasteurella Multocida. RSC Adv. 2020, 10, 30917–30933. [Google Scholar] [CrossRef]
- Sándor, V.; Kilár, A.; Kilár, F.; Kocsis, B.; Dörnyei, Á. Characterization of Complex, Heterogeneous Lipid A Samples using HPLC-MS/MS Technique II. Structural Elucidation of non-Phosphorylated Lipid A by Negative-Ion Mode Tandem Mass Spectrometry. J. Mass Spectrom. 2016, 51, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Sándor, V.; Dörnyei, Á.; Makszin, L.; Kilár, F.; Péterfi, Z.; Kocsis, B.; Kilár, A. Characterization of Complex, Heterogeneous Lipid A Samples using HPLC-MS/MS Technique I. Overall Analysis with Respect to Acylation, Phosphorylation and Isobaric Distribution. J. Mass Spectrom. 2016, 51, 1043–1063. [Google Scholar] [CrossRef] [PubMed]
- Sándor, V.; Kilár, A.; Kilár, F.; Kocsis, B.; Dörnyei, Á. Characterization of Complex, Heterogeneous Lipid A Samples using HPLC-MS/MS Technique III. Positive-Ion Mode Tandem Mass Spectrometry to Reveal Phosphorylation and Acylation Patterns of Lipid A. J. Mass Spectrom. 2018, 53, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Kilár, A.; Dörnyei, Á.; Sándor, V.; Kilár, F.; Kocsis, B. Phosphoglycolipid Profiling of Bacterial Endotoxins. Hung. J. Ind. Chem. 2018, 46, 7–11. [Google Scholar] [CrossRef]
- Crittenden, C.M.; Akin, L.D.; Morrison, L.J.; Trent, M.S.; Brodbelt, J.S. Characterization of Lipid A Variants by Energy-Resolved Mass Spectrometry: Impact of Acyl Chains. J. Am. Soc. Mass Spectrom. 2017, 28, 1118–1126. [Google Scholar] [CrossRef]
- El-Aneed, A.; Banoub, J. Elucidation of the Molecular Structure of Lipid A Isolated from Both a Rough Mutant and a Wild Strain of Aeromonas Salmonicida Lipopolysaccharides using Electrospray Ionization Quadrupole Time-Of-Flight Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 1683–1695. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Cheng, S.H.; Wu, Y.F.; Guo, D.J.; Fan, Q.H.; Wang, X.F.; Zhang, L.H.; Ye, X.S. Base-Promoted Rearrangement of Sugar Epoxides to Unsaturated Sugars. Org. Lett. 2005, 7, 5577–5579. [Google Scholar] [CrossRef]
- Hsu, F.F.; Turk, J. Charge-Remote and Charge-Driven Fragmentation Processes in Diacyl Glycerophosphoethanolamine upon Low-Energy Collisional Activation: A Mechanistic Proposal. J. Am. Soc. Mass Spectrom. 2000, 11, 892–899. [Google Scholar] [CrossRef]
- Spengler, B.; Dolce, J.W.; Cotter, R.J. Infrared-Laser Desorption Mass-spectrometry of Oligosaccharides-Fragmentation Mechanisms and Isomer Analysis. Anal. Chem. 1990, 62, 1731–1737. [Google Scholar] [CrossRef]
- Chiu, C.C.; Huynh, H.T.; Tsai, S.T.; Lin, H.Y.; Hsu, P.J.; Phan, H.T.; Karumanthra, A.; Thompson, H.; Lee, Y.C.; Kuo, J.L.; et al. Toward Closing the Gap between Hexoses and N-Acetlyhexosamines: Experimental and Computational Studies on the Collision-Induced Dissociation of Hexosamines. J. Phys. Chem. A 2019, 123, 6683–6700. [Google Scholar] [CrossRef] [PubMed]
- Hazelard, D.; Compain, P. Square sugars: Challenges and synthetic strategies. Org. Biomol. Chem. 2017, 15, 3806–3827. [Google Scholar] [CrossRef]
- Popsavin, V.; Radic, L.; Popsavin, M.; Cirin-Novta, V. Unexpected Cycloreversion of a Tosylated Tugar Oxetane Under E2 Conditions. The Facile Formation of 2-(2-Furanyl)-1,3-Dioxolane from a Novel 2,5:4,6-Dianhydro-L-Idose Derivative. J. Serb. Chem. Soc. 2004, 69, 117–122. [Google Scholar] [CrossRef]
- Lee, J.W.; Lewin, N.E.; Blumberg, P.M.; Marquez, V.E. Conformationally Constrained Analogs of Diacylglycerol.9. The Effect of Side-chain Orientation on the Protein-kinase-c (Pk-c) Binding-affinity of Delta-lactones. Bioorg. Med. Chem. Lett. 1994, 4, 2405–2410. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.J.; Altman, E. Structural Characterization of the Lipid A Region of Aeromonas Salmonicida Subsp Salmonicida Lipopolysaccharide. Carbohydr. Res. 2006, 341, 2816–2825. [Google Scholar] [CrossRef]
- Domon, B.; Costello, C.E. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).