The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA
Abstract
:1. Introduction
2. Results
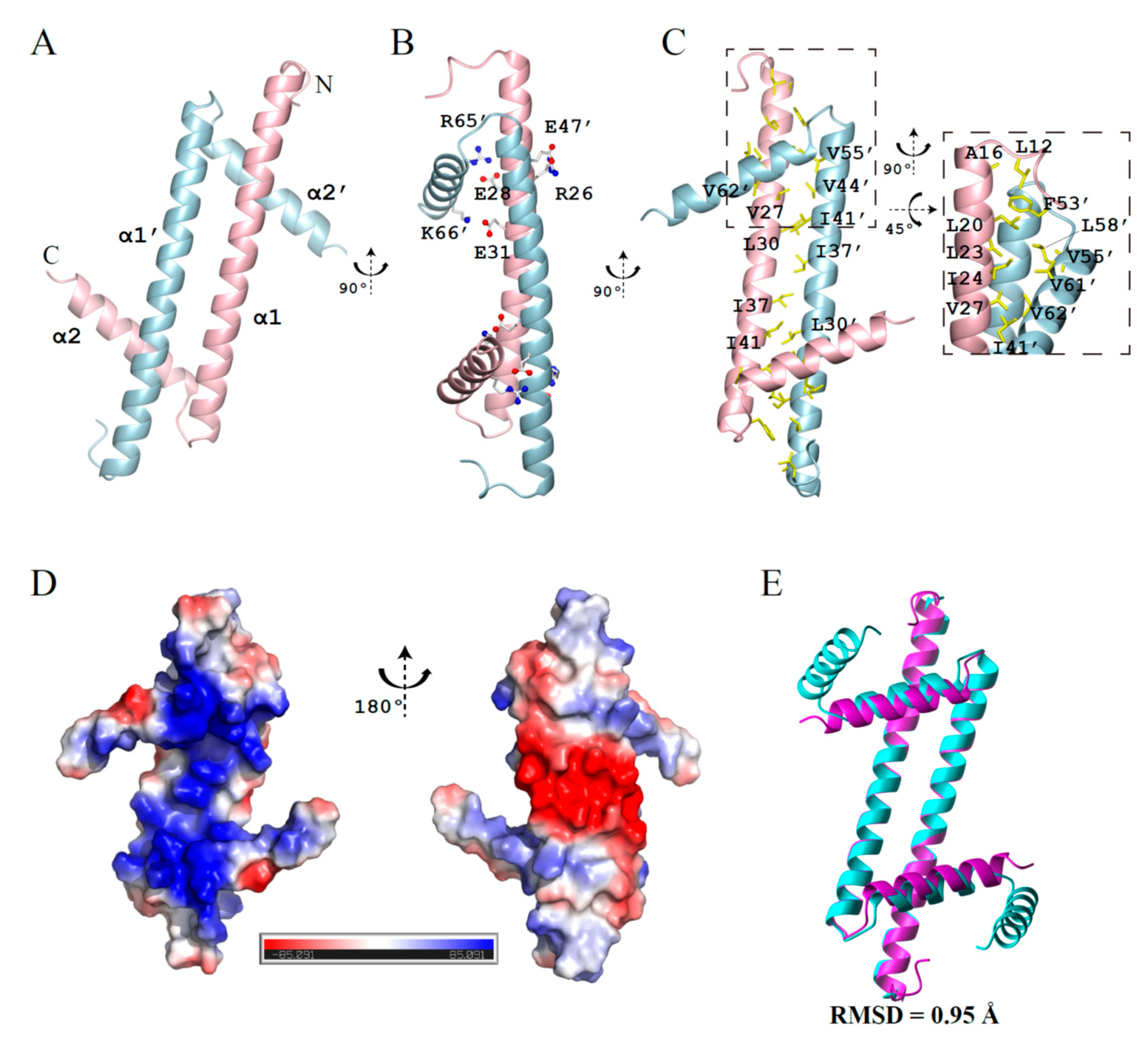
2.1. Dimeric GapRΔC17 Has the Same Fold as the Dimer Units of Full-Length Tetramer GapR
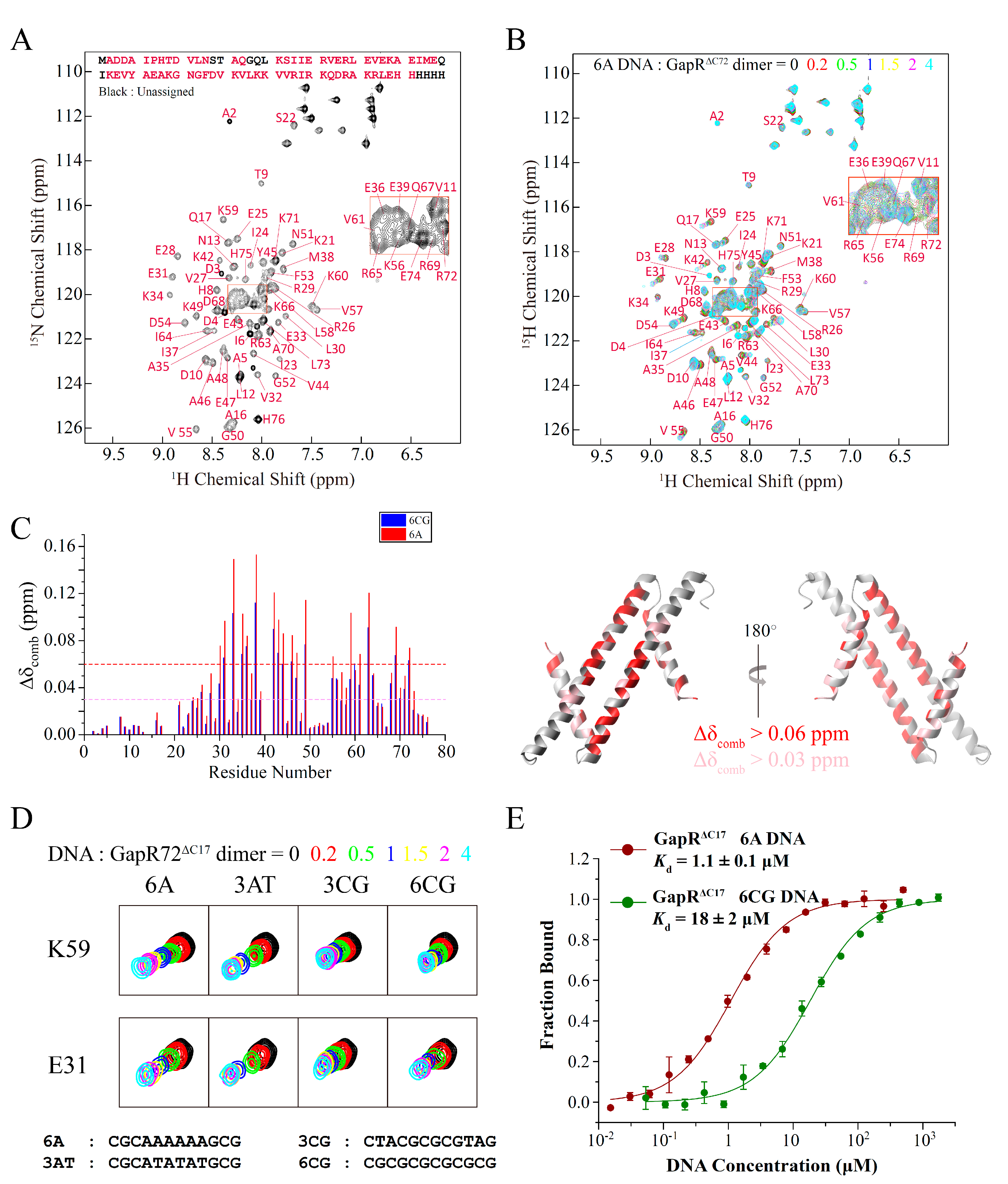
2.2. GapRΔC17 Can Bind to DNA with a Preference towards AT-rich Sequences
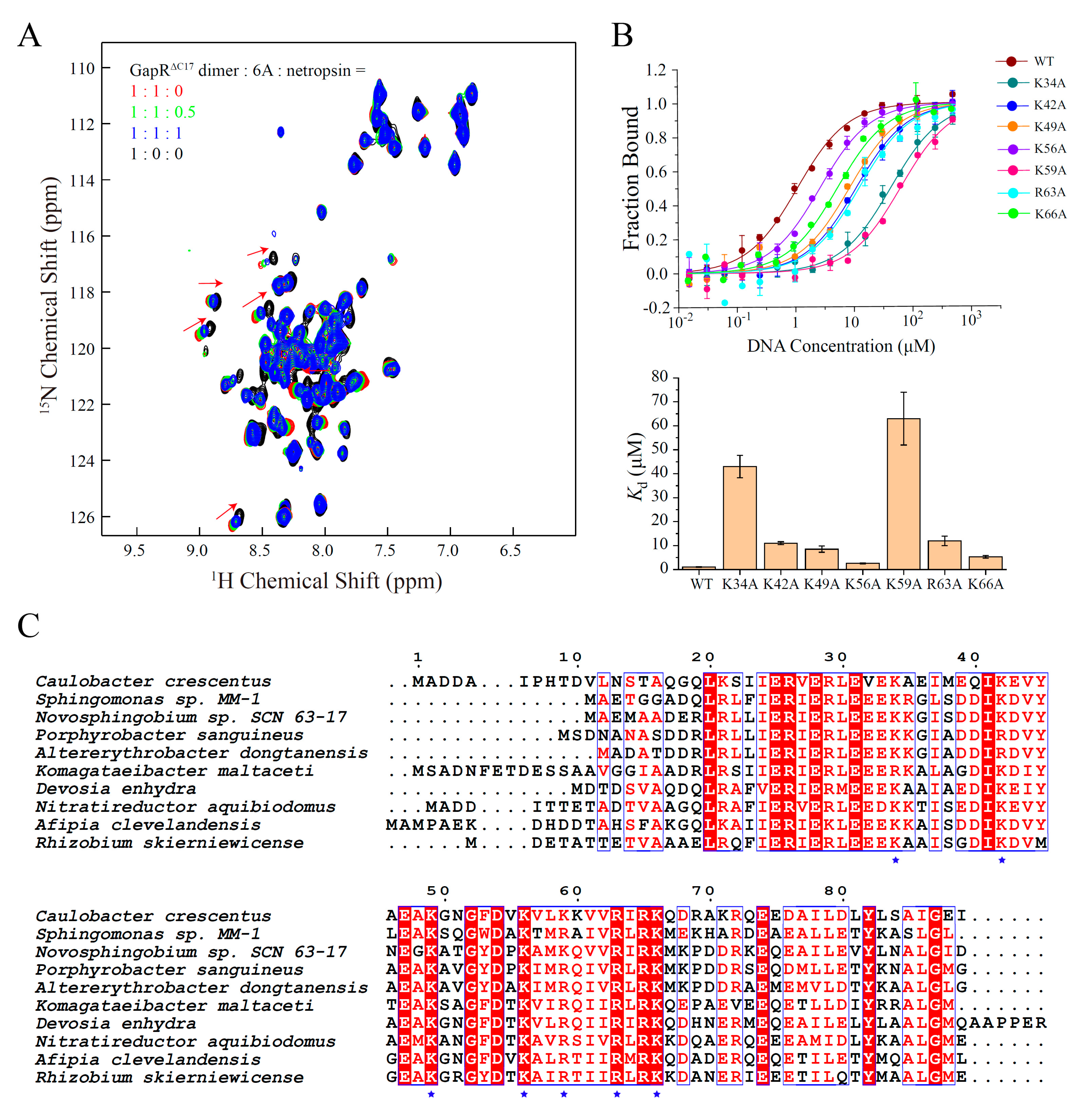
2.3. GapRΔC17 Binds DNA at the Minor Groove through Electrostatic Interactions between Its Lysine/Arginine Network and DNA Phosphate Groups
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Protein Crystallization
4.3. Crystallographic Structure Determination
4.4. Resonance Assignment of Backbone NH Signals of GapRΔC17
4.5. NMR Titration Experiments
4.6. MST Assay
4.7. Generation of Haddock Models of GapRΔC17 and DNA
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ricci, D.P.; Melfi, M.D.; Lasker, K.; Dill, D.L.; McAdams, H.H.; Shapiro, L. Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition. Proc. Natl. Acad. Sci. USA 2016, 113, 5952–5961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.A.; Panis, G.; Viollier, P.H.; Marczynski, G.T. A novel nucleoid-associated protein coordinates chromosome replication and chromosome partition. Nucleic Acids Res. 2017, 45, 8916–8929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.S.; Haakonsen, D.L.; Zeng, W.; Schumacher, M.A.; Laub, M.T. A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication. Cell 2018, 175, 583–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arias-Cartin, R.; Dobihal, G.S.; Campos, M.; Surovtsev, I.V.; Parry, B.; Jacobs-Wagner, C. Replication fork passage drives asymmetric dynamics of a critical nucleoid-associated protein in Caulobacter. EMBO J. 2017, 36, 301–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.M.; Dröge, P. Chromatin Architectural Factors as Safeguards against Excessive Supercoiling during DNA Replication. Int. J. Mol. Sci. 2020, 21, 4504. [Google Scholar] [CrossRef] [PubMed]
- Tarry, M.J.; Harmel, C.; Taylor, J.A.; Marczynski, G.T.; Schmeing, T.M. Structures of GapR reveal a central channel which could accommodate B-DNA. Sci. Rep. 2019, 9, 16679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Duan, B.; Dong, X.; Fan, S.; Xia, B. GapR binds DNA through dynamic opening of its tetrameric interface. Nucleic Acids Res. 2020, 48, 9372–9386. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, R.F.; Saurabh, S.; Herrmann, J.; Wakatsuki, S.; Shapiro, L. The Nucleoid-Associated Protein GapR Uses Conserved Structural Elements To Oligomerize and Bind DNA. mBio. 2020, 11, e00448. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, H.C.M.; Finch, J.T.; Luisi, B.F.; Klug, A. The structure of an oligo(dA)·oligo(dT) tract and its biological implications. Nature 1987, 330, 221–226. [Google Scholar] [CrossRef]
- El Hassan, M.A.; Calladine, C.R. Propeller-Twisting of Base-pairs and the Conformational Mobility of Dinucleotide Steps in DNA. J. Mol. Biol. 1996, 259, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.; Kohlhoff, K.; Zacharias, M. B-DNA Under Stress: Over- and Untwisting of DNA during Molecular Dynamics Simulations. Biophys. J. 2006, 91, 2956–2965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardwick, J.S.; Ptchelkine, D.; El-Sagheer, A.H.; Tear, I.; Singleton, D.; Phillips, S.E.V.; Lane, A.N.; Brown, T. 5-Formylcytosine does not change the global structure of DNA. Nat. Struct. Mol. Biol. 2017, 24, 544–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; Visscher, K.M.; Kastritis, P.L.; Bonvin, A.M. Solvated protein—DNA docking using HADDOCK. J. Biomol. NMR 2013, 56, 51–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| GapRΔC17 (SeMet) | GapRΔC17 (Native) | |
|---|---|---|
| Data collection | ||
| Space Group | I4,122 | I4,122 |
| Cell dimensions | ||
| a, b, c (Å) | 44.8, 44.8, 227.2 | 47.4, 47.4, 225.5 |
| α, β, γ (º) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 56.8~3.0 | 47.72~2.08 |
| Rmerge (%) | 11.6 (27.1) | 10.2 (53.3) |
| I/σI | 29.28 (3.2) | 48.8 (1.93) |
| Completeness (%) | 99 (94.6) | 98.06 (95.0) |
| Redundancy | 22.4 (11.9) | 22.2 (12.7) |
| Refinement | ||
| Resolution (Å) | 47.72~2.08 | |
| No. reflections | 8095 | |
| Rwork/Rfree (%) | 25.1/26 | |
| No. atoms | ||
| Protein | 532 | |
| B-factors | ||
| Protein | 37.0 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (º) | 0.742 | |
| Ramachandran plot statistics | ||
| Most favored (%) | 100 | |
| Additional allowed (%) | 0.0 | |
| Generously allowed (%) | 0.0 | |
| Disallowed (%) | 0.0 | |
| GapRΔC17/6A | GapRΔC17/3GC | |
|---|---|---|
| Haddock score (kcal/mol) | −344 ± 27 | −335 ± 42 |
| Cluster size | 39 | 31 |
| RMSD (Å) | 4.2 ± 1.2 | 3.5 ± 2.1 |
| Evdw (kcal/mol) | −43 ± 7 | −47 ± 9 |
| Eelec (kcal/mol) | −735 ± 76 | −686 ± 98 |
| Eair (kcal/mol) | 77 ± 43 | 109 ± 47 |
| BSA (Å2) | 1695 ± 138 | 1641 ± 186 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Duan, B.; Qu, Z.; Fan, S.; Xia, B. The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA. Molecules 2021, 26, 5776. https://doi.org/10.3390/molecules26195776
Huang Q, Duan B, Qu Z, Fan S, Xia B. The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA. Molecules. 2021; 26(19):5776. https://doi.org/10.3390/molecules26195776
Chicago/Turabian StyleHuang, Qian, Bo Duan, Zhi Qu, Shilong Fan, and Bin Xia. 2021. "The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA" Molecules 26, no. 19: 5776. https://doi.org/10.3390/molecules26195776
APA StyleHuang, Q., Duan, B., Qu, Z., Fan, S., & Xia, B. (2021). The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA. Molecules, 26(19), 5776. https://doi.org/10.3390/molecules26195776






