Sequence Dependent Repair of 1,N6-Ethenoadenine by DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB
Abstract
1. Introduction
2. Synthesis of Oligonucleotides Containing εA and Purification of Proteins
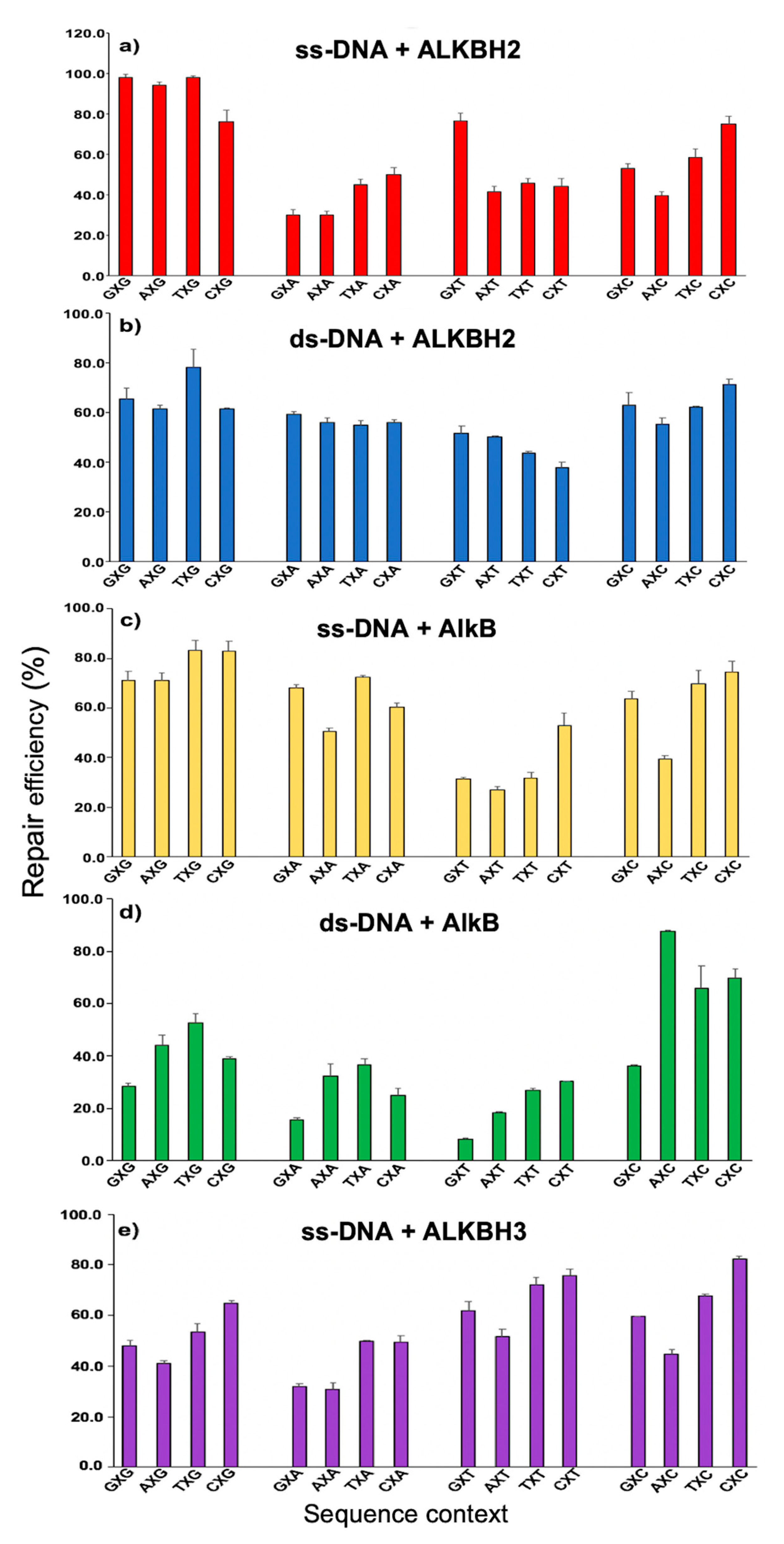
3. Repair of εA in Different Sequence Contexts by the AlkB Family Enzymes
4. Results from High Resolution LC-MS Analysis on εA Reactions
5. Sequence-/Enzyme-/Strand Context-Dependent Repair of εA
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| εA | 1,N6 ethenoadenine |
| ROS | reactive oxygen species |
| UV | ultraviolet |
| LPO | lipid peroxidation |
| VC | vinyl chloride |
| CAA | chloroacetaldehyde |
| 2OG-2 | 2-oxoglutarate |
| ss-DNA | single-strand-DNA |
| ds-DNA | double-strand-DNA |
| m/z | mass/charge |
| Mag | methyladenine DNA glycosylase |
| hOGG1 | human 8-oxoguanine glycosylase 1 |
References
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, B.I.; Essigmann, J.M. Impact of DNA lesion repair, replication and formation on the mutational spectra of environmental carcinogens: Aflatoxin B1 as a case study. DNA Repair 2018, 71, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Chawanthayatham, S.; Valentine, C.C.; Fedeles, B.I.; Fox, E.J.; Loeb, L.A.; Levine, S.S.; Slocum, S.L.; Wogan, G.N.; Croy, R.G.; Essigmann, J.M. Mutational spectra of aflatoxin B1 in vivo establish biomarkers of exposure for human hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, E3101–E3109. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.H. Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair. DNA Repair 2018, 71, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Maura, F.; Degasperi, A.; Nadeu, F.; Leongamornlert, D.; Davies, H.; Moore, L.; Royo, R.; Ziccheddu, B.; Puente, X.S.; Avet-Loiseau, H.; et al. A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun. 2019, 10, 2969. [Google Scholar] [CrossRef] [PubMed]
- Kucab, J.E.; Zou, X.; Morganella, S.; Joel, M.; Nanda, A.S.; Nagy, E.; Gomez, C.; Degasperi, A.; Harris, R.; Jackson, S.P.; et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019, 177, 821–836.e16. [Google Scholar] [CrossRef] [PubMed]
- Linhart, K.-B.; Glassen, K.; Peccerella, T.; Waldherr, R.; Linhart, H.; Bartsch, H.; Seitz, H.K. The generation of carcinogenic etheno-DNA adducts in the liver of patients with nonalcoholic fatty liver disease. Hepatob. Surg. Nutr. 2015, 4, 117–123. [Google Scholar]
- Bartsch, H.; Nair, J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: Role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch. Surg. 2006, 391, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Rioux, K.L.; Delaney, S. 1,N6-Ethenoadenine: From Molecular to Biological Consequences. Chem. Res. Toxicol. 2020, 33, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.C.; Smeester, L.; Wong, C.; Frick, L.E.; Taghizadeh, K.; Wishnok, J.S.; Drennan, C.L.; Samson, L.D.; Essigmann, J.M. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005, 12, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Yang, C.-G.; He, C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 2005, 127, 14594–14595. [Google Scholar] [CrossRef] [PubMed]
- Zdżalik, D.; Domańska, A.; Prorok, P.; Kosicki, K.; van den Born, E.; Falnes, P.Ø.; Rizzo, C.J.; Guengerich, F.P.; Tudek, B. Differential repair of etheno-DNA adducts by bacterial and human AlkB proteins. DNA Repair 2015, 30, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bian, K.; Tang, Q.; Fedeles, B.I.; Singh, V.; Humulock, Z.T.; Essigmann, J.M.; Li, D. Oncometabolites d- and l-2-Hydroxyglutarate Inhibit the AlkB Family DNA Repair Enzymes under Physiological Conditions. Chem. Res. Toxicol. 2017, 30, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tang, Q.; Bian, K.; Humulock, Z.T.; Yang, X.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Li, D. Adaptive Response Enzyme AlkB Preferentially Repairs 1-Methylguanine and 3-Methylthymine Adducts in Double-Stranded DNA. Chem. Res. Toxicol. 2016, 29, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Lingaraju, G.M.; Kartalou, M.; Meira, L.B.; Samson, L.D. Substrate specificity and sequence-dependent activity of the Saccharomyces cerevisiae 3-methyladenine DNA glycosylase (Mag). DNA Repair 2008, 7, 970–982. [Google Scholar] [CrossRef] [PubMed][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, R.; Bian, K.; Chen, F.; Tang, Q.; Zhou, X.; Li, D. Sequence Dependent Repair of 1,N6-Ethenoadenine by DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB. Molecules 2021, 26, 5285. https://doi.org/10.3390/molecules26175285
Qi R, Bian K, Chen F, Tang Q, Zhou X, Li D. Sequence Dependent Repair of 1,N6-Ethenoadenine by DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB. Molecules. 2021; 26(17):5285. https://doi.org/10.3390/molecules26175285
Chicago/Turabian StyleQi, Rui, Ke Bian, Fangyi Chen, Qi Tang, Xianhao Zhou, and Deyu Li. 2021. "Sequence Dependent Repair of 1,N6-Ethenoadenine by DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB" Molecules 26, no. 17: 5285. https://doi.org/10.3390/molecules26175285
APA StyleQi, R., Bian, K., Chen, F., Tang, Q., Zhou, X., & Li, D. (2021). Sequence Dependent Repair of 1,N6-Ethenoadenine by DNA Repair Enzymes ALKBH2, ALKBH3, and AlkB. Molecules, 26(17), 5285. https://doi.org/10.3390/molecules26175285






