Novel Glycerophospholipid, Lipo- and N-acyl Amino Acids from Bacteroidetes: Isolation, Structure Elucidation and Bioactivity
Abstract
1. Introduction
2. Results
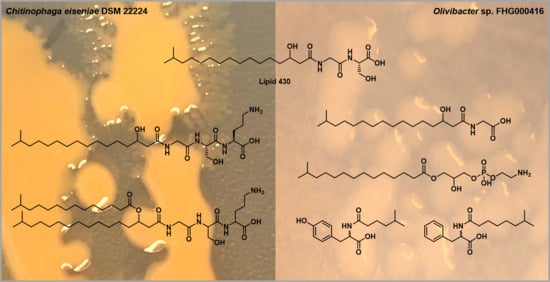
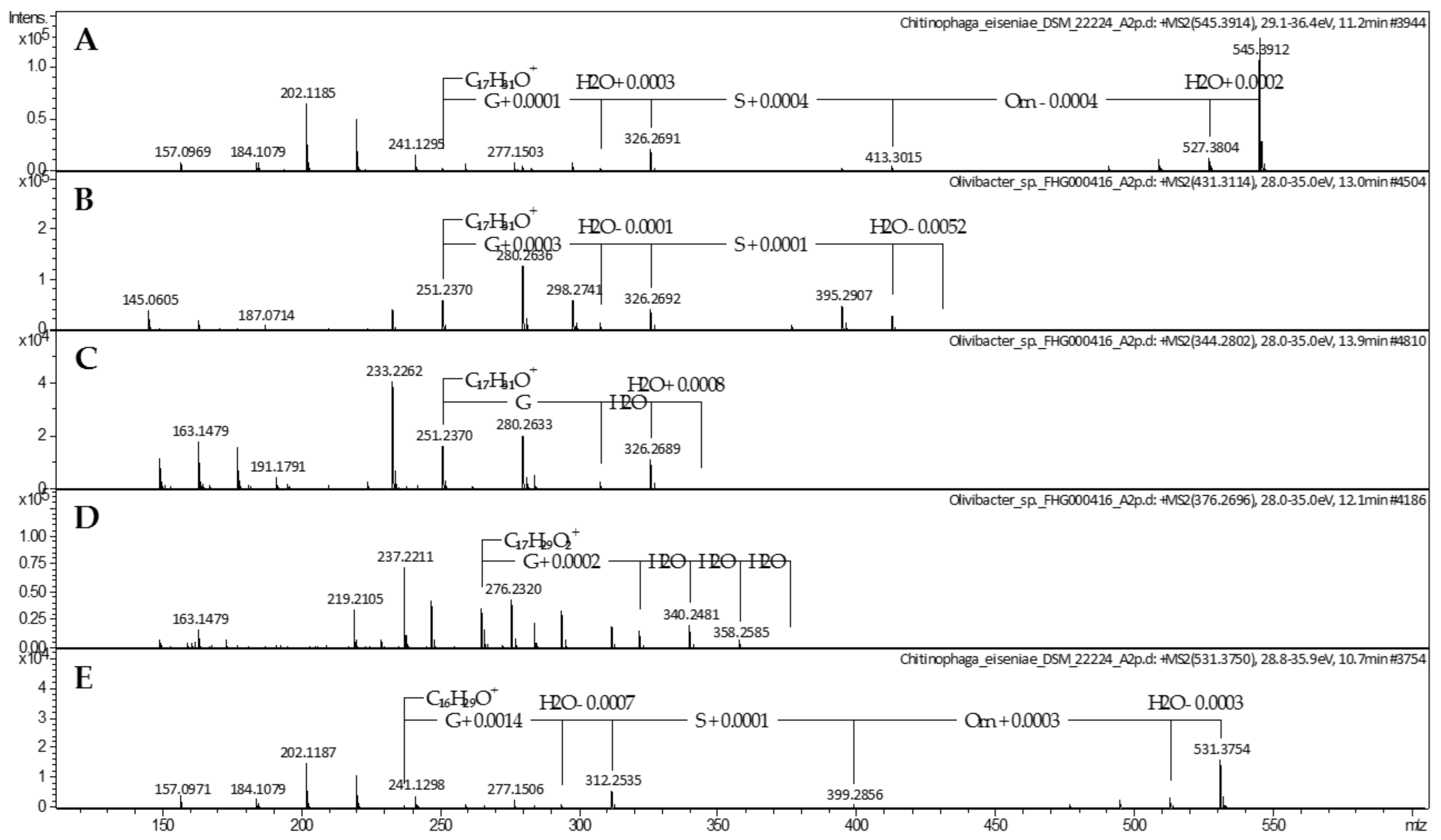
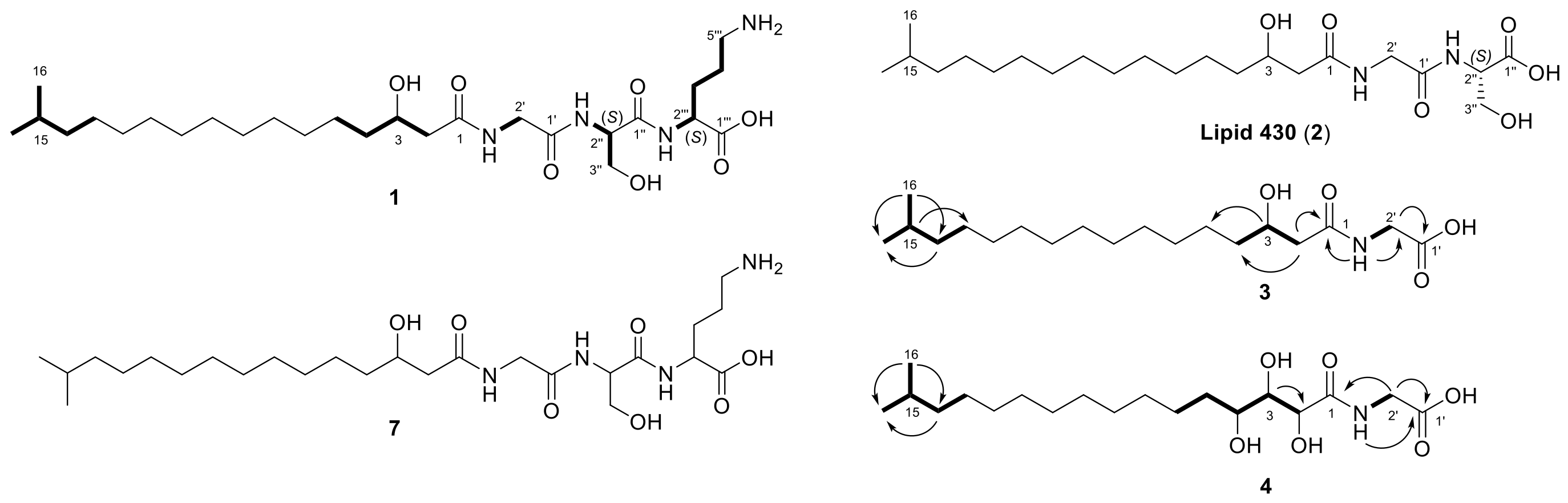
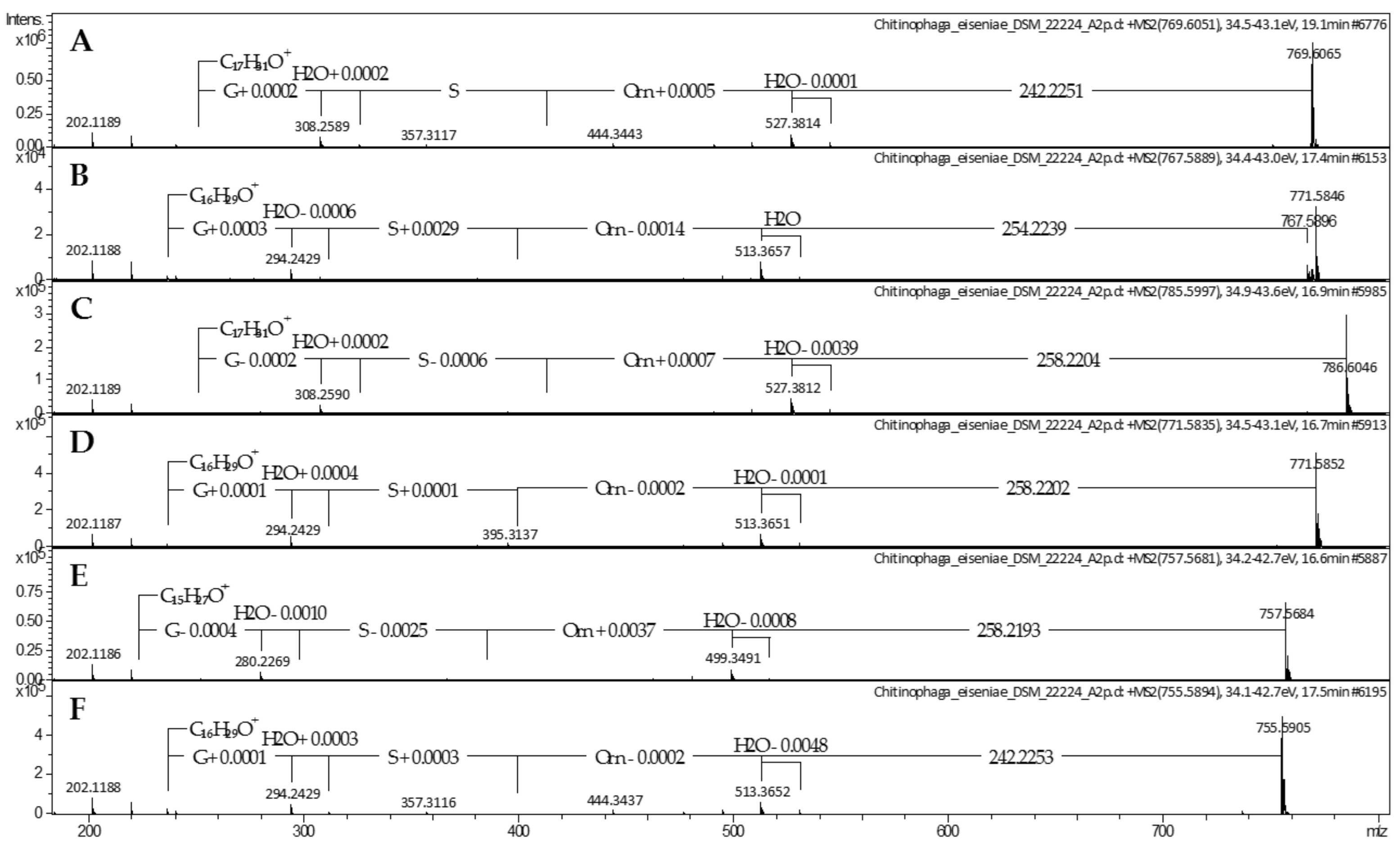
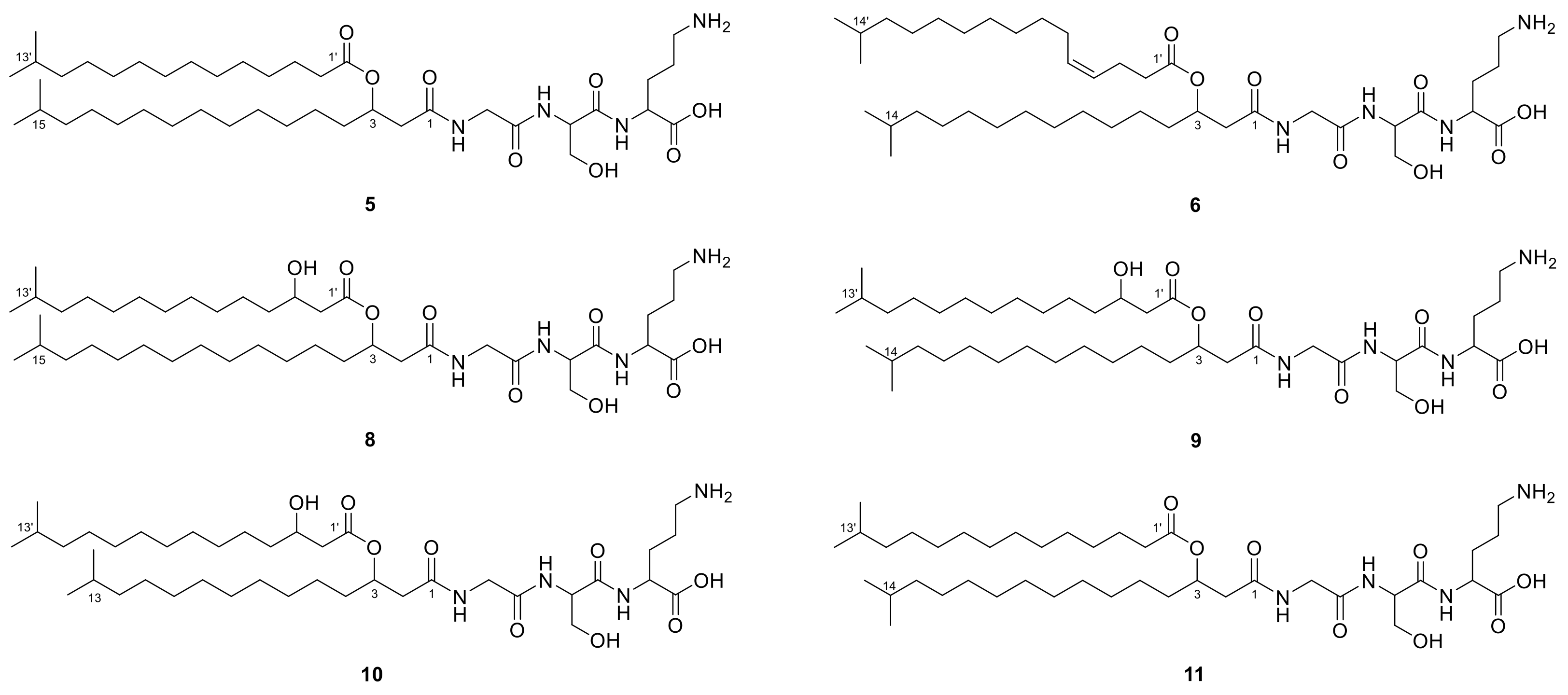
2.1. Lipoamino Acids
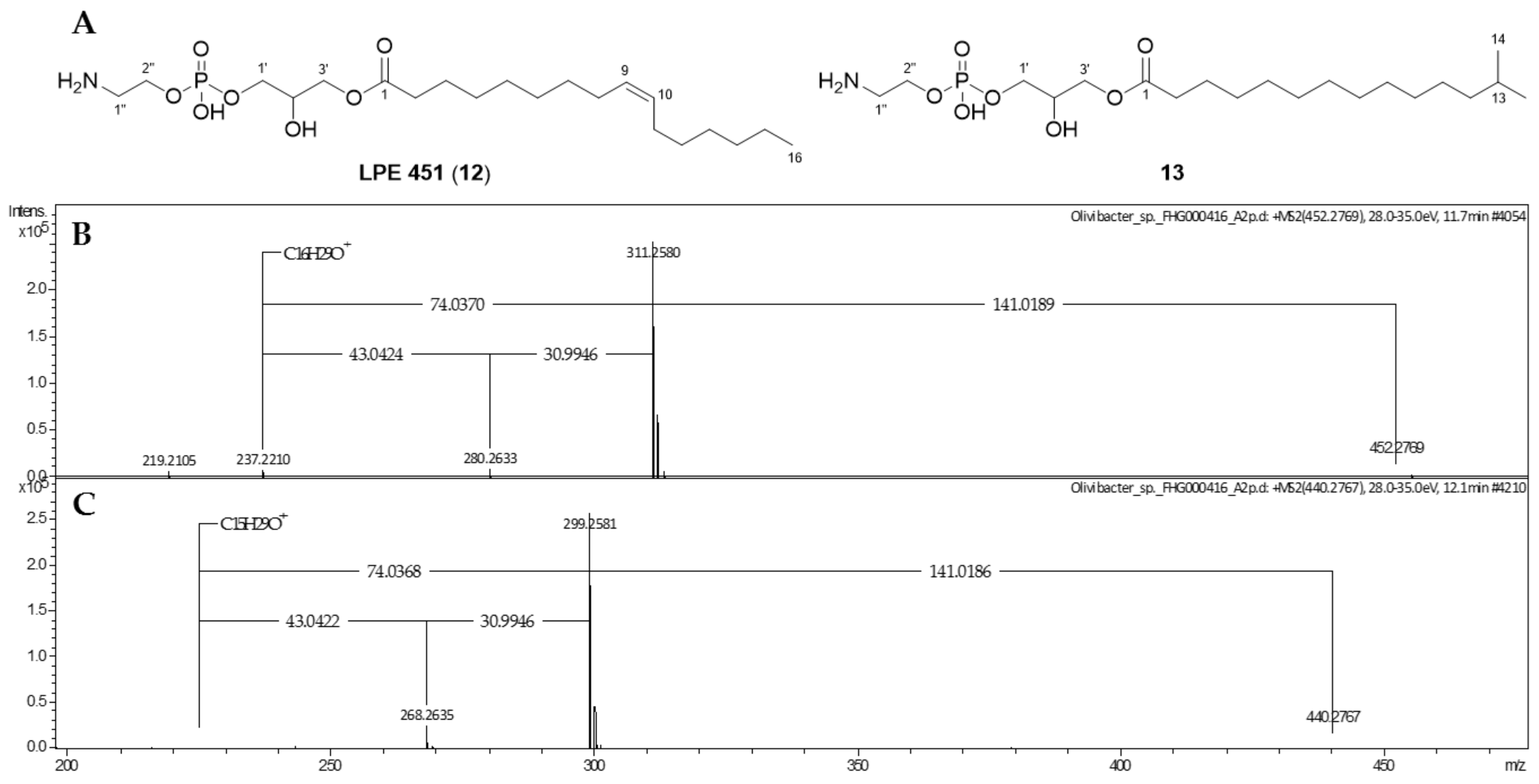
2.2. Phospholipids
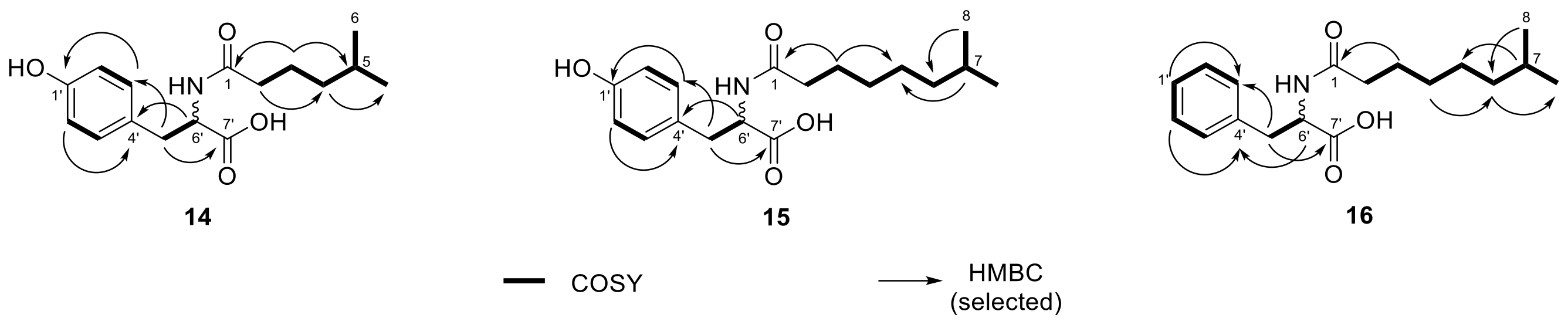
2.3. N-acyl Amino Acids from Olivibacter sp. FHG000416
2.4. Antimicrobial Activity of Lipids Isolated from Bacteroidetes
3. Discussion
4. Materials and Methods
4.1. Isolation of Olivibacter sp. FHG000416
4.2. Mass Spectrometric Analysis
4.3. Isolation of Lipids
NMR Studies
4.4. Optical Rotation
4.5. Advanced Marfey’s Analysis
4.6. Minimal Inhibitory Concentration (MIC)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Snijder, H.J.; Dijkstra, B.W. Bacterial phospholipase A: Structure and function of an integral membrane phospholipase. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2000, 1488, 91–101. [Google Scholar] [CrossRef]
- Asselineau, J. Bacterial Lipids Containing Amino Acids or Peptides Linked by Amide Bonds. In Fortschritte der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products; Asselineau, J., Kagan, J., Eds.; Springer: Vienna, Austria, 1991; pp. 1–85. ISBN 978-3-7091-9084-5. [Google Scholar]
- Batrakov, S.G.; Nikitin, D.I.; Mosezhnyi, A.E.; Ruzhitsky, A.O. A glycine-containing phosphorus-free lipoaminoacid from the gram-negative marine bacterium Cyclobacterium marinus WH. Chem. Phys. Lipids 1999, 99, 139–143. [Google Scholar] [CrossRef]
- Yoshida, K.; Iwami, M.; Umehara, Y.; Nishikawa, M.; Uchida, I.; Kohsaka, M.; Aoki, H.; Imanaka, H. Studies on WB-3559 A, B, C and D, new potent fibrinolytic agents. I. Discovery, identification, isolation and characterization. J. Antibiot. 1985, 38, 1469–1475. [Google Scholar] [CrossRef]
- Uchida, I.; Yoshida, K.; Kawai, Y.; Takase, S.; Itoh, Y.; Tanaka, H.; Kohsaka, M.; Imanaka, H. Studies on WB-3559 A, B, C and D, new potent fibrinolytic agents. II. Structure elucidation and synthesis. J. Antibiot. 1985, 38, 1476–1486. [Google Scholar] [CrossRef]
- Tahara, Y.; Kameda, M.; Yamada, Y.; Kondo, K. A new lipid the ornithine and taurine-containing “Cerilipin”. Agric. Biol. Chem. 1976, 40, 243–244. [Google Scholar] [CrossRef][Green Version]
- Kawai, Y.; Yano, I.; Kaneda, K. Various kinds of lipoamino acids including a novel serine-containing lipid in an opportunistic pathogen Flavobacterium. Their structures and biological activities on erythrocytes. Eur. J. Biochem. 1988, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Akagawa, K. Macrophage activation by an ornithine-containing lipid or a serine-containing lipid. Infect. Immun. 1989, 57, 2086–2091. [Google Scholar] [CrossRef] [PubMed]
- Kawazoe, R.; Okuyama, H.; Reichardt, W.; Sasaki, S. Phospholipids and a novel glycine-containing lipoamino acid in Cytophaga johnsonae Stanier strain C21. J. Bacteriol. 1991, 173, 5470–5475. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Sato, A.; Hisamoto, M.; Oda, T.; Matsuda, K.; Ishii, A.; Kodama, K. N-type calcium channel blockers from a marine bacterium, Cytophaga sp. SANK 71996. J. Antibiot. 1997, 50, 457–468. [Google Scholar] [CrossRef]
- Chianese, G.; Esposito, F.P.; Parrot, D.; Ingham, C.; de Pascale, D.; Tasdemir, D. Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp. Mar. Drugs 2018, 16, 187. [Google Scholar] [CrossRef]
- Gomi, K.; Kawasaki, K.; Kawai, Y.; Shiozaki, M.; Nishijima, M. Toll-like receptor 4-MD-2 complex mediates the signal transduction induced by flavolipin, an amino acid-containing lipid unique to Flavobacterium meningosepticum. J. Immunol. 2002, 168, 2939–2943. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; O’Dell, D.K.; Yu, Y.W.; Monn, M.F.; Hughes, H.V.; Burstein, S.; Walker, J.M. Identification of endogenous acyl amino acids based on a targeted lipidomics approach. J. Lipid Res. 2010, 51, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Clardy, J. Long-Chain N-Acyl Amino Acid Antibiotics Isolated from Heterologously Expressed Environmental DNA. J. Am. Chem. Soc. 2000, 122, 12903–12904. [Google Scholar] [CrossRef]
- Brady, S.F.; Chao, C.J.; Clardy, J. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 2002, 124, 9968–9969. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Clardy, J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J. Nat. Prod. 2004, 67, 1283–1286. [Google Scholar] [CrossRef]
- Brady, S.F.; Clardy, J. N-acyl derivatives of arginine and tryptophan isolated from environmental DNA expressed in Escherichia coli. Org. Lett. 2005, 7, 3613–3616. [Google Scholar] [CrossRef]
- Clardy, J.; Brady, S.F. Cyclic AMP directly activates NasP, an N-acyl amino acid antibiotic biosynthetic enzyme cloned from an uncultured beta-proteobacterium. J. Bacteriol. 2007, 189, 6487–6489. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Phoon, C.W.; Somanadhan, B.; Heng, S.C.H.; Ngo, A.; Ng, S.B.; Butler, M.S.; Buss, A.D.; Sim, M.M. Isolation and total synthesis of gymnastatin N, a POLO-like kinase 1 active constituent from the fungus Arachniotus punctatus. Tetrahedron 2004, 60, 11619–11628. [Google Scholar] [CrossRef]
- Touré, S.; Desrat, S.; Pellissier, L.; Allard, P.M.; Wolfender, J.L.; Dusfour, I.; Stien, D.; Eparvier, V. Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. Int. J. Mol. Sci. 2019, 20, 1083. [Google Scholar] [CrossRef]
- Vitale, G.A.; Sciarretta, M.; Cassiano, C.; Buonocore, C.; Festa, C.; Mazzella, V.; Núñez Pons, L.; D’Auria, M.V.; de Pascale, D. Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge. Int. J. Mol. Sci. 2020, 21, 6307. [Google Scholar] [CrossRef] [PubMed]
- Teng, O.; Ang, C.K.E.; Guan, X.L. Macrophage-Bacteria Interactions-A Lipid-Centric Relationship. Front. Immunol. 2017, 8, 1836. [Google Scholar] [CrossRef] [PubMed]
- Farrokhi, V.; Nemati, R.; Nichols, F.C.; Yao, X.; Anstadt, E.; Fujiwara, M.; Grady, J.; Wakefield, D.; Castro, W.; Donaldson, J.; et al. Bacterial lipodipeptide, Lipid 654, is a microbiome-associated biomarker for multiple sclerosis. Clin. Transl. Immunol. 2013, 2, e8. [Google Scholar] [CrossRef]
- Yoon, B.K.; Jackman, J.A.; Valle-González, E.R.; Cho, N.J. Antibacterial Free Fatty Acids and Monoglycerides: Biological Activities, Experimental Testing, and Therapeutic Applications. Int. J. Mol. Sci. 2018, 19, 1114. [Google Scholar] [CrossRef]
- Alves, E.; Dias, M.; Lopes, D.; Almeida, A.; Domingues, M.D.R.; Rey, F. Antimicrobial Lipids from Plants and Marine Organisms: An Overview of the Current State-of-the-Art and Future Prospects. Antibiotics 2020, 9, 441. [Google Scholar] [CrossRef]
- Shiozaki, M.; Deguchi, N.; Mochizuki, T.; Wakabayashi, T.; Ishikawa, T.; Haruyama, H.; Kawai, Y.; Nishijima, M. Revised structure and synthesis of flavolipin. Tetrahedron 1998, 54, 11861–11876. [Google Scholar] [CrossRef]
- Nemoto, T.; Ojika, M.; Takahata, Y.; Andoh, T.; Sakagami, Y. Structures of topostins, DNA topoisomerase I inhibitors of bacterial origin. Tetrahedron 1998, 54, 2683–2690. [Google Scholar] [CrossRef]
- Olsen, I.; Nichols, F.C. Are Sphingolipids and Serine Dipeptide Lipids Underestimated Virulence Factors of Porphyromonas gingivalis? Infect. Immun. 2018, 86, e00035-18. [Google Scholar] [CrossRef] [PubMed]
- Mirucki, C.S.; Abedi, M.; Jiang, J.; Zhu, Q.; Wang, Y.-H.; Safavi, K.E.; Clark, R.B.; Nichols, F.C. Biologic Activity of Porphyromonas endodontalis complex lipids. J. Endod. 2014, 40, 1342–1348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.-H.; Nemati, R.; Anstadt, E.; Liu, Y.; Son, Y.; Zhu, Q.; Yao, X.; Clark, R.B.; Rowe, D.W.; Nichols, F.C. Serine dipeptide lipids of Porphyromonas gingivalis inhibit osteoblast differentiation: Relationship to Toll-like receptor 2. Bone 2015, 81, 654–661. [Google Scholar] [CrossRef]
- Nichols, F.C.; Housley, W.J.; O’Conor, C.A.; Manning, T.; Wu, S.; Clark, R.B. Unique lipids from a common human bacterium represent a new class of Toll-like receptor 2 ligands capable of enhancing autoimmunity. Am. J. Pathol. 2009, 175, 2430–2438. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.B.; Cervantes, J.L.; Maciejewski, M.W.; Farrokhi, V.; Nemati, R.; Yao, X.; Anstadt, E.; Fujiwara, M.; Wright, K.T.; Riddle, C.; et al. Serine lipids of Porphyromonas gingivalis are human and mouse Toll-like receptor 2 ligands. Infect. Immun. 2013, 81, 3479–3489. [Google Scholar] [CrossRef]
- Schneider, Y.K.-H.; Hansen, K.Ø.; Isaksson, J.; Ullsten, S.; H Hansen, E.; Hammer Andersen, J. Anti-Bacterial Effect and Cytotoxicity Assessment of Lipid 430 Isolated from Algibacter sp. Molecules 2019, 24, 3991. [Google Scholar] [CrossRef]
- Nemati, R.; Dietz, C.; Anstadt, E.J.; Cervantes, J.; Liu, Y.; Dewhirst, F.E.; Clark, R.B.; Finegold, S.; Gallagher, J.J.; Smith, M.B.; et al. Deposition and hydrolysis of serine dipeptide lipids of Bacteroidetes bacteria in human arteries: Relationship to atherosclerosis. J. Lipid Res. 2017, 58, 1999–2007. [Google Scholar] [CrossRef]
- Blanksby, S.J.; Mitchell, T.W. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. [Google Scholar] [CrossRef]
- Hancock, S.E.; Poad, B.L.J.; Batarseh, A.; Abbott, S.K.; Mitchell, T.W. Advances and unresolved challenges in the structural characterization of isomeric lipids. Anal. Biochem. 2017, 524, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Swiezewska, E.; Wójcik, J. NMR of carbohydrates, lipids and membranes. In Nuclear Magnetic Resonance: A Review of the Lterature Published between January 2009 and May 2010; Kamieńska-Trela, K., Aliev, A.E., Eds.; RSC Publications: Cambridge, UK, 2011; pp. 344–390. ISBN 978-1-84973-147-8. [Google Scholar]
- Swiezewska, E.; Wójcik, J. NMR of lipids and membranes. In Nuclear Magnetic Resonance; Kamienska-Trela, K., Wojcik, J., Eds.; Royal Society of Chemistry: Cambridge, UK, 2012; pp. 320–347. ISBN 978-1-84973-373-1. [Google Scholar]
- Pikula, S.; Bandorowicz-Pikula, J.; Groves, P. NMR of lipids. In Nuclear Magnetic Resonance; Wójcik, J., Kamieńska-Trela, K., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2013; pp. 362–382. ISBN 978-1-84973-577-3. [Google Scholar]
- Pikula, S.; Bandorowicz-Pikula, J.; Groves, P. NMR of lipids. In Nuclear Magnetic Resonance; Kamieńska-Trela, K., Aliev, A.E., Bandorowicz-Pikula, J., Buda, S., D’Errico, G., de Dios, A.C., Eds.; Royal Society of Chemistry: Cambridge, UK, 2015; pp. 385–406. ISBN 978-1-78262-052-5. [Google Scholar]
- Pikula, S.; Bandorowicz-Pikula, J.; Groves, P. Recent Advances in NMR Studies of Lipids. In Annual Reports on NMR Spectroscopy; Webb, G.A., Ed.; Elsevier Science: Burlington, NJ, USA, 2015; pp. 195–246. ISBN 9780128030905. [Google Scholar]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High Resolution NMR Spectroscopy as a Structural and Analytical Tool for Unsaturated Lipids in Solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, S.; Kurz, M.; Patras, M.A.; Hartwig, C.; Marner, M.; Leis, B.; Billion, A.; Kleiner, Y.; Bauer, A.; Toti, L.; et al. Genomic and chemical decryption of the Bacteroidetes phylum for its potential to biosynthesize natural products. BioRxiv 2021. [Google Scholar] [CrossRef]
- Yasir, M.; Chung, E.J.; Song, G.C.; Bibi, F.; Jeon, C.O.; Chung, Y.R. Chitinophaga eiseniae sp. nov., isolated from vermicompost. Int. J. Syst. Evol. Microbiol. 2011, 61, 2373–2378. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nobre, M.F.; Nunes, O.C.; Manaia, C.M. Pseudosphingobacterium domesticum gen. nov., sp. nov., isolated from home-made compost. Int. J. Syst. Evol. Microbiol. 2007, 57, 1535–1538. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Liu, Q.; Lee, S.Y.; Choi, K.D.; Im, W.-T. Olivibacter ginsenosidimutans sp nov., with ginsenoside converting activity isolated from compost, and reclassification of Pseudosphingobacterium domesticum as Olivibacter domesticus comb. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Brückner, H. Marfey’s reagent for chiral amino acid analysis: A review. Amino Acids 2004, 27, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Woznica, A.; Cantley, A.M.; Beemelmanns, C.; Freinkman, E.; Clardy, J.; King, N. Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc. Natl. Acad. Sci. USA 2016, 113, 7894–7899. [Google Scholar] [CrossRef]
- Kayganich, K.A.; Murphy, R.C. Fast atom bombardment tandem mass spectrometric identification of diacyl, alkylacyl, and alk-1-enylacyl molecular species of glycerophosphoethanolamine in human polymorphonuclear leukocytes. Anal. Chem. 1992, 64, 2965–2971. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.C.; Harrison, K.A. Fast atom bombardment mass spectrometry of phospholipids. Mass Spectrom. Rev. 1994, 13, 57–75. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Makovitzki, A.; Avrahami, D.; Shai, Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Gotoh, N.; Tanaka, S.; Nishino, T. Supersusceptibility to hydrophobic antimicrobial agents and cell surface hydrophobicity in Branhamella catarrhalis. FEMS Microbiol. Lett. 1989, 59, 211–213. [Google Scholar] [CrossRef]
- Fomsgaard, J.S.; Fomsgaard, A.; Høiby, N.; Bruun, B.; Galanos, C. Comparative immunochemistry of lipopolysaccharides from Branhamella catarrhalis strains. Infect. Immun. 1991, 59, 3346–3349. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, H.; Gotoh, N.; Nishino, T. Diffusion of macrolide antibiotics through the outer membrane of Moraxella catarrhalis. J. Infect. Chemother. 1999, 5, 196–200. [Google Scholar] [CrossRef]
- Oberpaul, M.; Zumkeller, C.M.; Culver, T.; Spohn, M.; Mihajlovic, S.; Leis, B.; Glaeser, S.P.; Plarre, R.; McMahon, D.P.; Hammann, P.; et al. High-Throughput Cultivation for the Selective Isolation of Acidobacteria From Termite Nests. Front. Microbiol. 2020, 11, 597628. [Google Scholar] [CrossRef]
- Kämpfer, P.; Dott, W.; Martin, K.; Glaeser, S.P. Rhodococcus defluvii sp. nov., isolated from wastewater of a bioreactor and formal proposal to reclassify Corynebacterium hoagii and Rhodococcus equi as Rhodococcus hoagii comb. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 755–761. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix. [CrossRef]
- Arendrup, M.C.; Guinea, J.; Cuenca-Estrella, M.; Meletiadia, J.; Mouton, J.W.; Lagrou, K.; Howard, S.J.; The Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). Eucast definitive document EDef 7.1: Method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef]
| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| Position | δH, (J in Hz) | δC, Type a | δH, (J in Hz) | δC, Type | δH, (J in Hz) | δC, Type |
| 1 | 171.1, C | 175.0, C | 171.1, C | |||
| 2 | 2.20, d (6.1) | 43.3, CH2 | 2.41, dd (14.0, 4.3), 2.34, dd (14.0, 8.6) | 44.8, CH2 | 2.19, d (6.4) | 43.6, CH2 |
| 3 | 3.82–3.75, m | 67.2, CH | 4.02–3.94, m | 70.0, CH | 3.79–3.74, m | 67.4, CH |
| 4 | 1.39–1.31, m | 36.7, CH2 | 1.53–1.46, m | 38.4, CH2 | 1.40–1.34, m, 1.33–1.27, m | 36.8, CH2 |
| 5 | 1.39–1.31, m b, 1.31–1.26, m b | 24.8, CH2 | 1.50–1.43, m, 1.38–1.32, m | 26.7, CH2 | 1.40–1.35, m, 1.31–1.25, m | 25.1, CH2 |
| 6 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 7 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 8 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 9 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 10 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 11 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 12 | 1.26–1.22, m b | 28.0–28.9 c, CH2 | 1.36–1.25, m e | 30.7–31.0 f, CH2 | 1.26–1.21, m g | 29.0–29.3 h, CH2 |
| 13 | 1.26–1.22, m b | 26.4, CH2 | 1.36–1.25, m e | 28.5, CH2 | 1.26–1.21, m g | 26.8, CH2 |
| 14 | 1.16–1.11, m | 38.1, CH2 | 1.17, q (6.7) | 40.2, CH2 | 1.15–1.11, m | 38.5, CH2 |
| 15 | 1.50, non (6.6) | 27.0, CH | 1.50, sept (6.6) | 29.2, CH | 1.49, non (6.6) | 27.4, CH |
| 16 | 0.85, d (6.6) | 22.2, CH3 | 0.88, d (6.7) | 23.0, CH3 | 0.84, d (6.6) | 22.5, CH3 |
| 1′ | 168.6, C | 171.6, C | n.o. d | |||
| 2′ | 3.73, d (4.6) | 41.5, CH2 | 3.98, d (16.8), 3.90, d (16.7) | 43.5, CH2 | 3.70, dd (17.4, 5.7), 3.65, dd (17.4, 5.7) | 41.0, CH2 |
| 2-NH | 8.10–8.04, m | 8.30, t (5.6) | 8.01, t (5.7) | |||
| 1′′′ | n.o. d | 173.4, C | ||||
| 2′′ | 4.25, dd (12.6, 6.2) | 54.7, CH | 4.50, t (4.3) | 56.2, CH | ||
| 2′′-NH | 7.91–7.83, m | 8.04, d (7.7) | ||||
| 3′′ | 3.56, m, 3.46, m | 62.0, CH2 | 3.91, dd (11.5, 4.5), 3.83, dd (11.3, 3.8) | 63.0, CH2 | ||
| 1′′′ | n.o. d | |||||
| 2′′′ | 3.82–3.75, m | 53.6, CH | ||||
| 2′′′-NH | 7.66–7.59, m | |||||
| 3′′′ | 1.80–1.71, m, 1.69–1.61, m | n.o. d | ||||
| 4′′′ | 1.66–1.56, m | 24.0, CH2 | ||||
| 5′′′ | 2.80–2.73, m | 38.4, CH2 | ||||
| 14 | 15 | 16 | ||||
|---|---|---|---|---|---|---|
| Position | δH, (J in Hz) | δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | δC, Type |
| 1′ | 157.3, C | 157.2, C | 7.23–7.17, m | 127.8, CH | ||
| 2′ | 6.69, dd (6.6, 1.9) | 116.1, CH | 6.68, dd (6.5, 2.1) | 116.1, CH | 7.30–7.22, m | 129.4, CH |
| 3′ | 7.04, dd (6.6, 1.9) | 131.2, CH | 7.03, dd (6.6, 1.5) | 131.3, CH | 7.26–7.21, m | 130.3, CH |
| 4′ | 129.2, C | 129.4, C | 138.7, C | |||
| 5′ | 3.11, dd (14.0, 4.8), 2.83, dd (14.0, 9.3) | 37.7, CH2 | 3.11, dd (14.0, 4.9), 2.84, dd (13.9, 8.9) | 37.9, CH2 | 3.22, dd (13.9, 4.8), 2.93, dd (13.9, 9.5) | 38.5, CH2 |
| 6′ | 4.59, dd (9.3, 4.6) | 55.3, CH | 4.57, dd (8.9, 4.9) | 55.7, CH | 4.66, dd (9.4, 4.8) | 55.1, CH |
| 7′ | 175.3, C | 175.9, C | 175.2, C | |||
| 1 | 176.1, C | 175.9, C | 176.1, C | |||
| 2 | 2.13, t (7.5) | 37.1, CH2 | 2.15, t (7.4) | 37.0, CH2 | 2.14, t (7.4) | 36.9, CH2 |
| 3 | 1.55–1.47, m | 24.8, CH2 | 1.56–1.48, m | 27.0, CH2 | 1.55–1.46, m | 26.9, CH2 |
| 4 | 1.14–1.05, m | 39.4, CH2 | 1.32–1.24, m | 28.2, CH2 | 1.32–1.23, m | 28.2, CH2 |
| 5 | 1.52–1.46, m | 29.0, CH | 1.24–1.18, m | 30.4, CH2 | 1.23–1.14, m | 30.4, CH2 |
| 6 | 0.86, d (6.6) | 22.8/22.9, CH3 | 1.20–1.12, m | 40.0, CH2 | 1.22–1.10, m | 40.0, CH2 |
| 7 | 1.57–1.49, m | 29.1, CH | 1.55–1.49, m | 29.1, CH | ||
| 8 | 0.88, d (6.6) | 23.0, CH3 | 0.87, d (6.6) | 23.1/23.0, CH3 | ||
| Compounds | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 12 | 13 | 14 | 15 | 16 | |
| E. coli ATCC 35218 (MH-II) | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| E. coli ATCC 35218 (MHC) | 64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| E. coli ATCC 25922 ΔTolC | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| P. aeruginosa ATCC 27853 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| K. pneumoniae ATCC 13883 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| M. catarrhalis ATCC 25238 | 4–8 | 16–32 | 8 | >64 | 4–8 | 16 | 64 | >64 | >64 |
| A. baumannii ATCC 19606 | n.d. | n.d. | >64 | >64 | n.d. | n.d. | >64 | >64 | >64 |
| B. subtilis DSM 10 | 8 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| S. aureus ATCC 25923 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| M. luteus DSM 20030 | 64 | 32–64 | >64 | >64 | 16 | 16 | 64 | >64 | >64 |
| L. monocytogenes DSM 20600 | n.d. | n.d. | >64 | >64 | >64 | n.d. | >64 | >64 | >64 |
| M. smegmatis ATCC 607 | >64 | n.d. | >64 | n.d. | >64 | n.d. | >64 | >64 | >64 |
| C. albicans FH2173 | 64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bill, M.-K.; Brinkmann, S.; Oberpaul, M.; Patras, M.A.; Leis, B.; Marner, M.; Maitre, M.-P.; Hammann, P.E.; Vilcinskas, A.; Schuler, S.M.M.; et al. Novel Glycerophospholipid, Lipo- and N-acyl Amino Acids from Bacteroidetes: Isolation, Structure Elucidation and Bioactivity. Molecules 2021, 26, 5195. https://doi.org/10.3390/molecules26175195
Bill M-K, Brinkmann S, Oberpaul M, Patras MA, Leis B, Marner M, Maitre M-P, Hammann PE, Vilcinskas A, Schuler SMM, et al. Novel Glycerophospholipid, Lipo- and N-acyl Amino Acids from Bacteroidetes: Isolation, Structure Elucidation and Bioactivity. Molecules. 2021; 26(17):5195. https://doi.org/10.3390/molecules26175195
Chicago/Turabian StyleBill, Mona-Katharina, Stephan Brinkmann, Markus Oberpaul, Maria A. Patras, Benedikt Leis, Michael Marner, Marc-Philippe Maitre, Peter E. Hammann, Andreas Vilcinskas, Sören M. M. Schuler, and et al. 2021. "Novel Glycerophospholipid, Lipo- and N-acyl Amino Acids from Bacteroidetes: Isolation, Structure Elucidation and Bioactivity" Molecules 26, no. 17: 5195. https://doi.org/10.3390/molecules26175195
APA StyleBill, M.-K., Brinkmann, S., Oberpaul, M., Patras, M. A., Leis, B., Marner, M., Maitre, M.-P., Hammann, P. E., Vilcinskas, A., Schuler, S. M. M., & Schäberle, T. F. (2021). Novel Glycerophospholipid, Lipo- and N-acyl Amino Acids from Bacteroidetes: Isolation, Structure Elucidation and Bioactivity. Molecules, 26(17), 5195. https://doi.org/10.3390/molecules26175195