Phytotoxicity of Schiekia timida Seed Extracts, a Mixture of Phenylphenalenones
Abstract
:1. Introduction
2. Results
2.1. Identification and Annotation of Phenylphenalenones
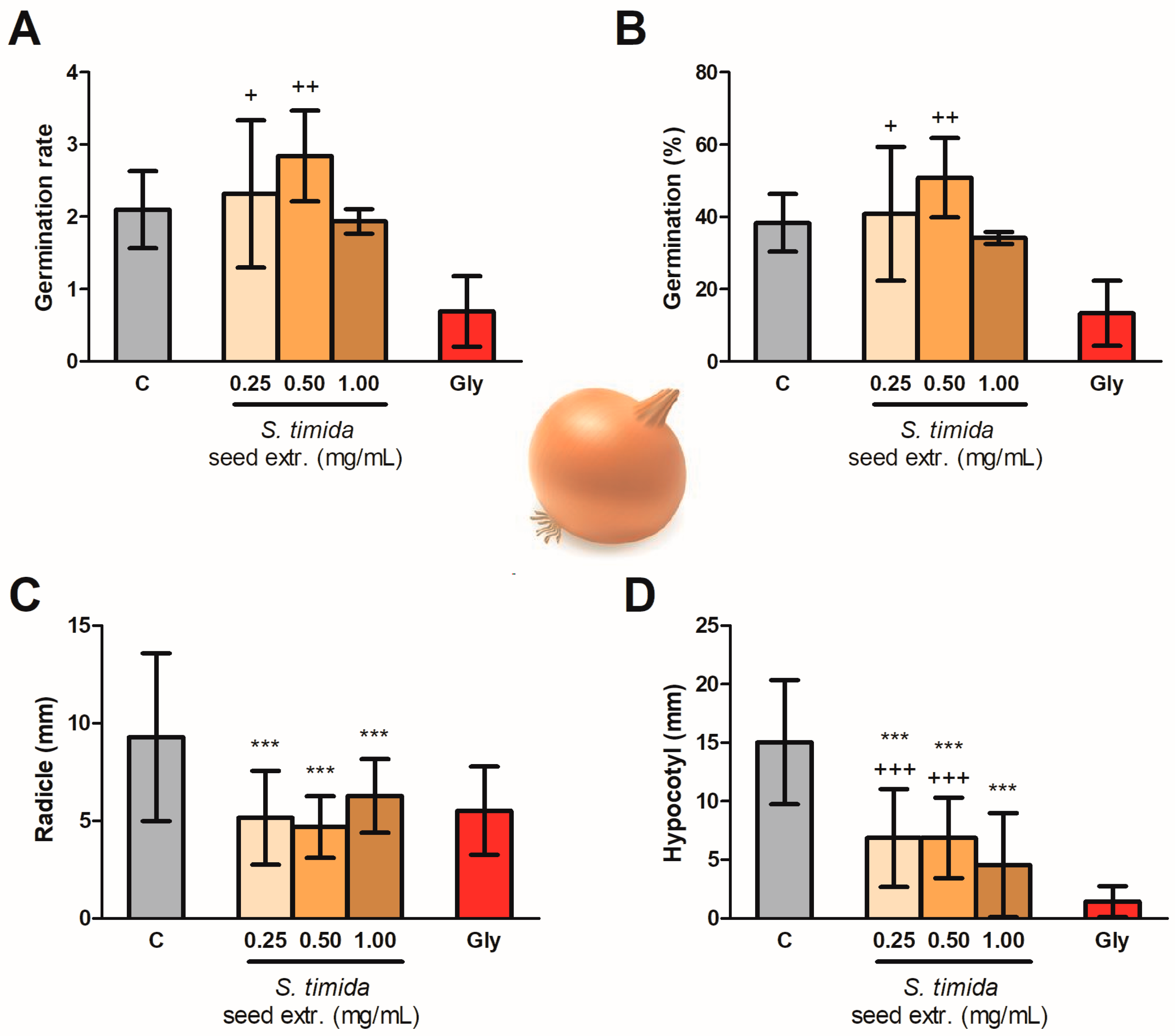
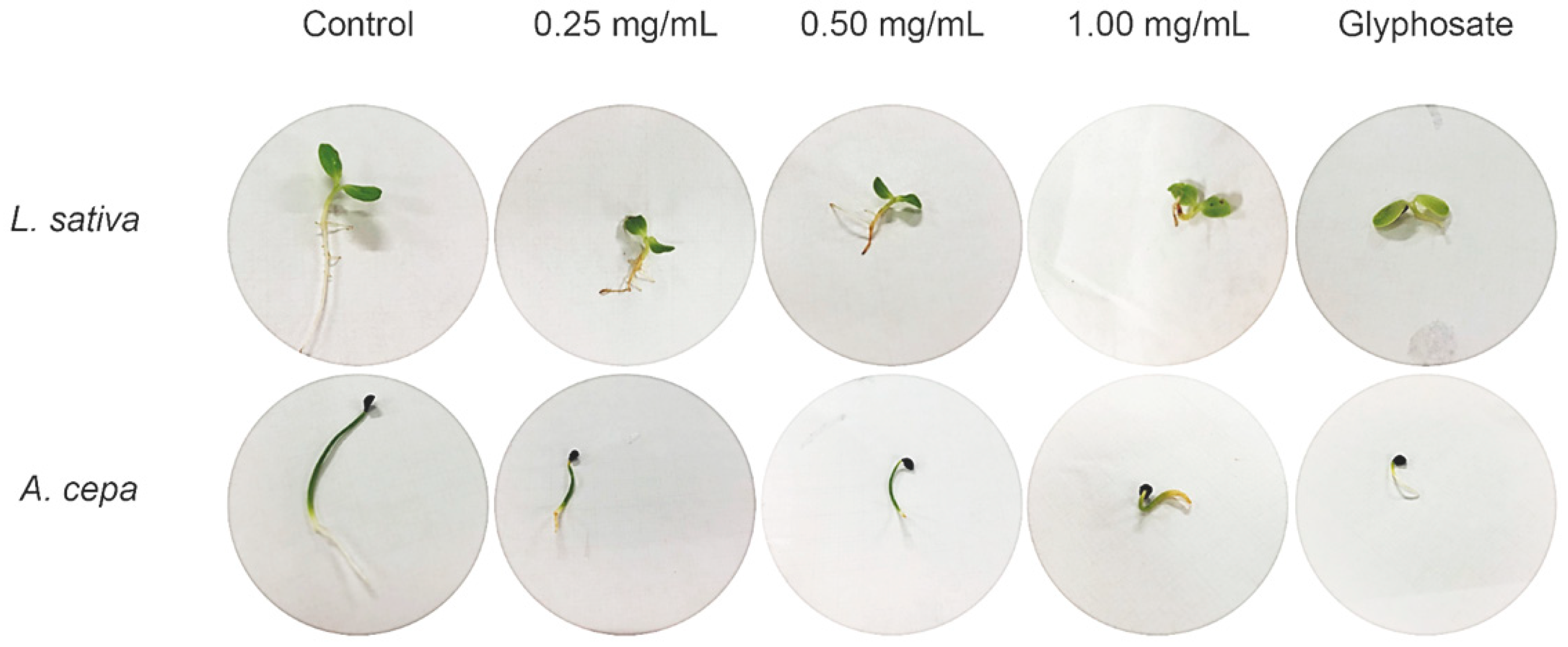
2.2. Seed Germination and Phytotoxicity
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Plant Material
4.3. Extraction and Identification of Natural Compounds
4.4. Phytotoxicity Bioassay
4.4.1. Germination Test
4.4.2. Seedling Growth Test
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Todero, I.; Confortin, T.C.; Luft, L.; Brun, T.; Ugalde, G.A.; de Almeida, T.C.; Arnemann, J.A.; Zabot, G.L.; Mazutti, M.A. Formulation of a bioherbicide with metabolites from Phoma sp. Sci. Hortic. 2018, 241, 285–292. [Google Scholar] [CrossRef]
- Beckie, H.J.; Ashworth, M.B.; Flower, K.C. Herbicide Resistance Management: Recent Developments and Trends. Plants 2019, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.P.; Baroni, A.C.M.; Carollo, C.A.; Demarque, D.P.; Pardo, L.F.L.; de Rezende, L.M.P.; dos Santos, F.J.L.; Lima, W.G.; de Siqueira, J.M. Synthesis, phytotoxic evaluation and in silico studies for the development of novel natural products-inspired herbicides. Biocatal. Agric. Biotechnol. 2020, 24, 101559. [Google Scholar] [CrossRef]
- Dayan, F.E.; Duke, S.O. Natural Compounds as Next-Generation Herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Kumar, V.; Dhanjal, D.S.; Singh, J. Herbicides and Plant Growth Regulators: Current Developments and Future Challenges. In Natural Bioactive Products in Sustainable Agriculture; Springer: Singapore, 2020; pp. 67–81. [Google Scholar]
- Andreotti, G.; Koutros, S.; Hofmann, J.N.; Sandler, D.P.; Lubin, J.H.; Lynch, C.F.; Lerro, C.C.; De Roos, A.J.; Parks, C.G.; Alavanja, M.C.; et al. Glyphosate Use and Cancer Incidence in the Agricultural Health Study. J. Natl. Cancer Inst. 2018, 110, 509–516. [Google Scholar] [CrossRef] [Green Version]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, F.; Dalla Valle, N.; Fogliatto, S.; Milan, M.; De Palo, F.; Tabacchi, M.; Ferrero, A. Rapid increase of herbicide resistance in Echinochloa spp. consequent to repeated applications of the same herbicides over time. Arch. Agron. Soil Sci. 2020, 1–13. [Google Scholar] [CrossRef]
- Shaner, D.L.; Beckie, H.J. The future for weed control and technology. Pest Manag. Sci. 2014, 70, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Bagavathiannan, M.V.; Davis, A.S. An ecological perspective on managing weeds during the great selection for herbicide resistance. Pest Manag. Sci. 2018, 74, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Perotti, V.E.; Larran, A.S.; Palmieri, V.E.; Martinatto, A.K.; Permingeat, H.R. Herbicide resistant weeds: A call to integrate conventional agricultural practices, molecular biology knowledge and new technologies. Plant Sci. 2020, 290, 110255. [Google Scholar] [CrossRef]
- Duke, S.O.; Dayan, F.E.; Romagni, J.G.; Rimando, A.M. Natural products as sources of herbicides: Current status and future trends. Weed Res. 2000, 40, 99–111. [Google Scholar] [CrossRef]
- Schulz, S.; Kubanek, J.; Piel, J. Editorial: Chemical Ecology. Nat. Prod. Rep. 2015, 32, 886–887. [Google Scholar] [CrossRef] [PubMed]
- Reigosa, M.; Gomes, A.S.; Ferreira, A.G.; Borghetti, F. Allelopathic research in Brazil. Acta Bot. Brasilica 2013, 27, 629–646. [Google Scholar] [CrossRef] [Green Version]
- Reigosa, M.J.; Sánchez-Moreiras, A.; González, L. Ecophysiological Approach in Allelopathy. Crit. Rev. Plant Sci. 1999, 18, 577–608. [Google Scholar] [CrossRef]
- Hidalgo, W.; Cano, M.; Arbelaez, M.; Zarrazola, E.; Gil, J.; Schneider, B.; Otálvaro, F. 4-Phenylphenalenones as a template for new photodynamic compounds against Mycosphaerella fijiensis. Pest Manag. Sci. 2016, 72, 796–800. [Google Scholar] [CrossRef]
- Brand, S.; Hölscher, D.; Schierhorn, A.; Svatos, A.; Schröder, J.; Schneider, B. A type III polyketide synthase from Wachendorfia thyrsiflora and its role in diarylheptanoid and phenylphenalenone biosynthesis. Planta 2006, 224, 413–428. [Google Scholar] [CrossRef]
- Chen, Y.; Paetz, C.; Menezes, R.C.; Schneider, B. Cultured roots of Xiphidium caeruleum: Phenylphenalenones and their biosynthetic and extractant-dependent conversion. Phytochemistry 2017, 133, 15–25. [Google Scholar] [CrossRef]
- Monakhova, Y.; Schneider, B. The intramolecular Diels-Alder reaction of diarylheptanoids--quantum chemical calculation of structural features favoring the formation of phenylphenalenones. Molecules 2014, 19, 5231–5242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duque, L.; Zapata, C.; Rojano, B.; Schneider, B.; Otálvaro, F. Radical Scavenging Capacity of 2,4-Dihydroxy-9-phenyl-1H-phenalen-1-one: A Functional Group Exclusion Approach. Org. Lett. 2013, 15, 3542–3545. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Flores, N.; Abad-Grillo, T.; McNaughton-Smith, G. Evaluation of substituted phenalenone analogues as antiplasmodial agents. Exp. Parasitol. 2013, 135, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Binks, R.H.; Greenham, J.R.; Luis, J.G.; Gowen, S.R. A phytoalexin from roots of Musa acuminata var. pisang sipulu. Phytochemistry 1997, 45, 47–49. [Google Scholar] [CrossRef]
- Ocampos, F.M.M.; Paetz, C.; Antar, G.M.; Menezes, R.C.; Miguel, O.G.; Schneider, B. Phytochemical profile of Schiekia orinocensis (Haemodoraceae). Phytochem. Lett. 2017, 21, 139–145. [Google Scholar] [CrossRef]
- Pellegrini, M.O.O.; Hickman, E.J.; Guttiérrez, J.E.; Smith, R.J.; Hopper, S.D. Revisiting the taxonomy of the Neotropical Haemodoraceae (Commelinales). PhytoKeys 2020, 169, 1–59. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal. Chem. 2014, 86, 5433–5440. [Google Scholar] [CrossRef] [PubMed]
- Riihinen, K.R.; Mihaleva, V.V.; Gödecke, T.; Soininen, P.; Laatikainen, R.; Vervoort, J.M.; Lankin, D.C.; Pauli, G.F. 1H-NMR Fingerprinting of Vaccinium vitis-idaea Flavonol Glycosides. Phytochem. Anal. 2013, 24, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Pauli, G.F.; Jaki, B.U.; Lankin, D.C. A Routine Experimental Protocol for qHNMR Illustrated with Taxol. J. Nat. Prod. 2007, 70, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Pauli, G.F.; Jaki, B.U.; Lankin, D.C. Quantitative 1H NMR: Development and Potential of a Method for Natural Products Analysis §. J. Nat. Prod. 2005, 68, 133–149. [Google Scholar] [CrossRef]
- Pauli, G.F.; Gödecke, T.; Jaki, B.U.; Lankin, D.C. Quantitative 1H NMR. Development and Potential of an Analytical Method: An Update. J. Nat. Prod. 2012, 75, 834–851. [Google Scholar] [CrossRef] [Green Version]
- Seger, C.; Sturm, S.; Stuppner, H. Mass spectrometry and NMR spectroscopy: Modern high-end detectors for high resolution separation techniques - state of the art in natural product HPLC-MS, HPLC-NMR, and CE-MS hyphenations. Nat. Prod. Rep. 2013, 30, 970–987. [Google Scholar] [CrossRef] [PubMed]
- Simmler, C.; Napolitano, J.G.; McAlpine, J.B.; Chen, S.-N.; Pauli, G.F. Universal quantitative NMR analysis of complex natural samples. Curr. Opin. Biotechnol. 2014, 25, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hölscher, D.; Schneider, B. HPLC-NMR analysis of phenylphenalenones and a stilbene from Anigozanthos flavidus. Phytochemistry 1999, 50, 155–161. [Google Scholar] [CrossRef]
- Cooke, R.; Thomas, R. Colouring Matters of Australian Plants. XVIII. Constituents of Anigozanthos rufus. Aust. J. Chem. 1975, 28, 1053–1057. [Google Scholar] [CrossRef]
- Duque, L.; Restrepo, C.; Sáez, J.; Gil, J.; Schneider, B.; Otálvaro, F. Synthesis of musafluorone: A naphthoxanthenone isolated from Musa acuminata. Tetrahedron Lett. 2010, 51, 4640–4643. [Google Scholar] [CrossRef]
- Hölscher, D.; Schneider, B. The Biosynthetic Origin of the Central One-Carbon Unit of Phenylphenalenones in Anigozanthos preissii. Nat. Prod. Lett. 1995, 7, 177–182. [Google Scholar] [CrossRef]
- Holscher, D.; Schneider, B. A diarylheptanoid intermediate in the biosynthesis of phenylphenalenones in Anigozanthos preissii. J. Chem. Soc. Chem. Commun. 1995, 525–526. [Google Scholar] [CrossRef]
- Echeverri, F.; Torres, F.; Quiñones, W.; Escobar, G.; Archbold, R. Phenylphenalenone phytoalexins, will they be a new type of fungicide? Phytochem. Rev. 2012, 11, 1–12. [Google Scholar] [CrossRef]
- Hidalgo, W.; Duque, L.; Saez, J.; Arango, R.; Gil, J.; Rojano, B.; Schneider, B.; Otálvaro, F. Structure–Activity Relationship in the Interaction of Substituted Perinaphthenones with Mycosphaerella fijiensis. J. Agric. Food Chem. 2009, 57, 7417–7421. [Google Scholar] [CrossRef]
- Brkljaca, R.; Urban, S.; Dahse, H.-M.; Voigt, K. Antimicrobial Evaluation of the Constituents Isolated From Macropidia fuliginosa (Hook.) Druce. Nat. Prod. Commun. 2019, 14, 1–6. [Google Scholar] [CrossRef]
- Späth, A.; Leibl, C.; Cieplik, F.; Lehner, K.; Regensburger, J.; Hiller, K.-A.; Bäumler, W.; Schmalz, G.; Maisch, T. Improving Photodynamic Inactivation of Bacteria in Dentistry: Highly Effective and Fast Killing of Oral Key Pathogens with Novel Tooth-Colored Type-II Photosensitizers. J. Med. Chem. 2014, 57, 5157–5168. [Google Scholar] [CrossRef]
- Holscher, D.; Dhakshinamoorthy, S.; Alexandrov, T.; Becker, M.; Bretschneider, T.; Buerkert, A.; Crecelius, A.C.; De Waele, D.; Elsen, A.; Heckel, D.G.; et al. Phenalenone-type phytoalexins mediate resistance of banana plants (Musa spp.) to the burrowing nematode Radopholus similis. Proc. Natl. Acad. Sci. USA 2014, 111, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Luque-Ortega, J.R.; Martínez, S.; Saugar, J.M.; Izquierdo, L.R.; Abad, T.; Luis, J.G.; Piñero, J.; Valladares, B.; Rivas, L. Fungus-Elicited Metabolites from Plants as an Enriched Source for New Leishmanicidal Agents: Antifungal Phenyl-Phenalenone Phytoalexins from the Banana Plant (Musa acuminata) Target Mitochondria of Leishmania donovani Promastigotes. Antimicrob. Agents Chemother. 2004, 48, 1534–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hölscher, D.; Schneider, B. Phenalenones from Strelitzia reginae. J. Nat. Prod. 2000, 63, 1027–1028. [Google Scholar] [CrossRef]
- López-Arencibia, A.; Reyes-Batlle, M.; Freijo, M.B.; Sifaoui, I.; Bethencourt-Estrella, C.J.; Rizo-Liendo, A.; Chiboub, O.; McNaughton-Smith, G.; Lorenzo-Morales, J.; Abad-Grillo, T.; et al. In vitro activity of 1H-phenalen-1-one derivatives against Leishmania spp. And evidence of programmed cell death. Parasit. Vectors 2019, 12, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Zhang, B.; Wang, Y.; Sun, J.; Ma, X.; Wang, G.; Fu, S.; Lin, C.; Li, Y. Three new acenaphthene derivatives from rhizomes of Musa basjoo and their cytotoxic activity. Nat. Prod. Res. 2019, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rosquete, L.I.; Cabrera-Serra, M.G.; Piñero, J.E.; Martín-Rodríguez, P.; Fernández-Pérez, L.; Luis, J.G.; McNaughton-Smith, G.; Abad-Grillo, T. Synthesis and in vitro antiprotozoal evaluation of substituted phenalenone analogues. Bioorg. Med. Chem. 2010, 18, 4530–4534. [Google Scholar] [CrossRef]
- Dayan, F.E.; Romagni, J.G.; Duke, S.O. Investigating the Mode of Action of Natural Phytotoxins. J. Chem. Ecol. 2000, 26, 2079–2094. [Google Scholar] [CrossRef]
- Busi, R.; Vila-Aiub, M.M.; Beckie, H.J.; Gaines, T.A.; Goggin, D.E.; Kaundun, S.S.; Lacoste, M.; Neve, P.; Nissen, S.J.; Norsworthy, J.K.; et al. Herbicide-resistant weeds: From research and knowledge to future needs. Evol. Appl. 2013, 6, 1218–1221. [Google Scholar] [CrossRef] [PubMed]
- Busi, R.; Powles, S.B.; Beckie, H.J.; Renton, M. Rotations and mixtures of soil-applied herbicides delay resistance. Pest Manag. Sci. 2020, 76, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Otálvaro, F.; Quiñones, W.; Echeverri, F.; Schneider, B. Synthesis of [phenyl-13C6]lachnanthocarpone and other 13C-labelled phenylphenalenones. J. Label. Compd. Radiopharm. 2004, 47, 147–159. [Google Scholar] [CrossRef]
- Nanclares, J.; Gil, J.; Rojano, B.; Saez, J.; Schneider, B.; Otálvaro, F. Synthesis of 4-methoxy-1H-phenalen-1-one: A subunit related to natural phenalenone-type compounds. Tetrahedron Lett. 2008, 49, 3844–3847. [Google Scholar] [CrossRef]
- Ospina, F.; Ramirez, A.; Cano, M.; Hidalgo, W.; Schneider, B.; Otálvaro, F. Synthesis of Positional Isomeric Phenylphenalenones. J. Org. Chem. 2017, 82, 3873–3879. [Google Scholar] [CrossRef]
- Ospina, F.; Hidalgo, W.; Cano, M.; Schneider, B.; Otálvaro, F. Synthesis of 8-phenylphenalenones: 2-hydroxy-8-(4-hydroxyphenyl)-1h-phenalen-1-one from Eichhornia crassipes. J. Org. Chem. 2016, 81, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-Z.; Ku, C.-F.; Si, T.-X.; Tsang, S.-W.; Lv, X.-M.; Li, X.-W.; Li, Z.-M.; Zhang, H.-J.; Chan, A.S.C. Concise Synthesis of Natural Phenylphenalenone Phytoalexins and a Regioisomer. J. Nat. Prod. 2018, 81, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Cano, M.; Rojas, C.; Hidalgo, W.; Sáez, J.; Gil, J.; Schneider, B.; Otálvaro, F. Improved synthesis of 4-phenylphenalenones: The case of isoanigorufone and structural analogs. Tetrahedron Lett. 2013, 54, 351–354. [Google Scholar] [CrossRef]
- Barrera, J.; Patiño, E.; Otálvaro, F. Improved synthesis of natural isomeric naphthoxanthenones. Tetrahedron Lett. 2020, 61, 151359. [Google Scholar] [CrossRef]
- Macías, F.A.; Castellano, D.; Molinillo, J.M.G. Search for a Standard Phytotoxic Bioassay for Allelochemicals. Selection of Standard Target Species. J. Agric. Food Chem. 2000, 48, 2512–2521. [Google Scholar] [CrossRef]
- Maguire, J.D. Speed of germination—Aid in selection and evaluation for seedling emergence and vigor. Crop Sci. 1962, 2. [Google Scholar] [CrossRef]
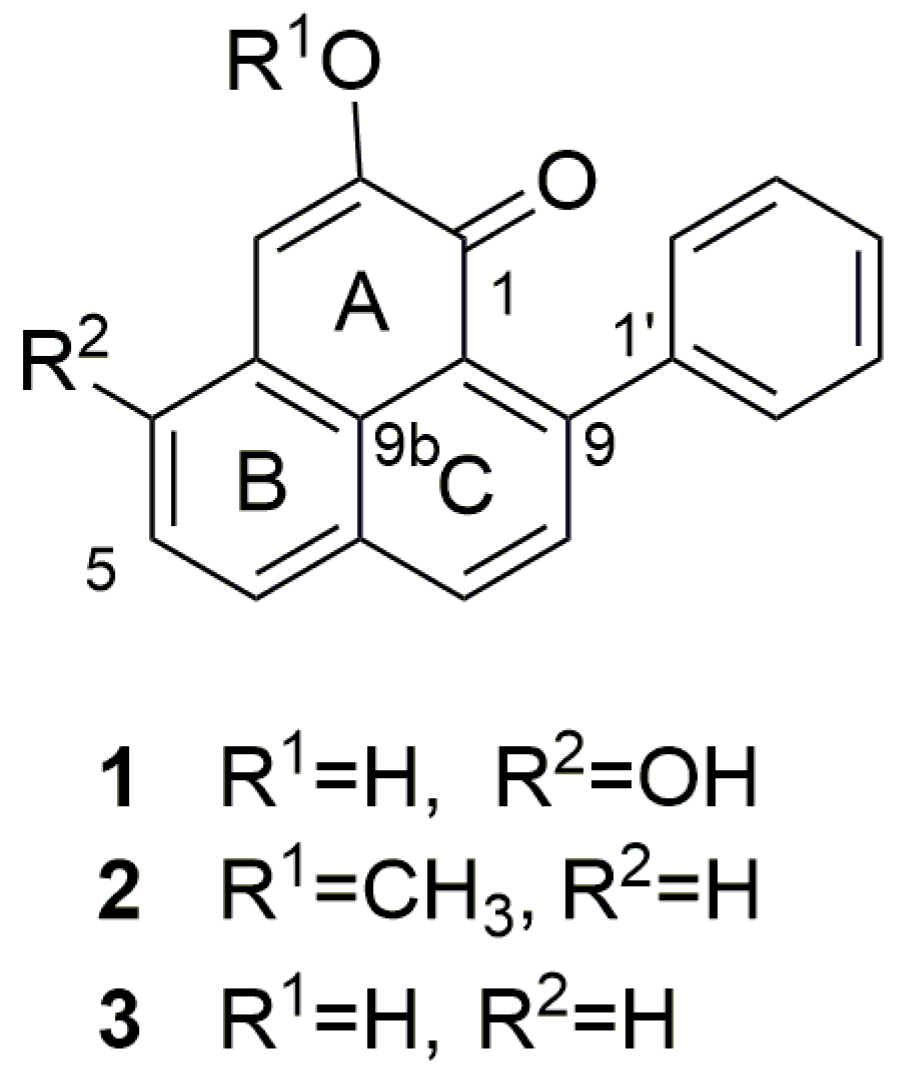
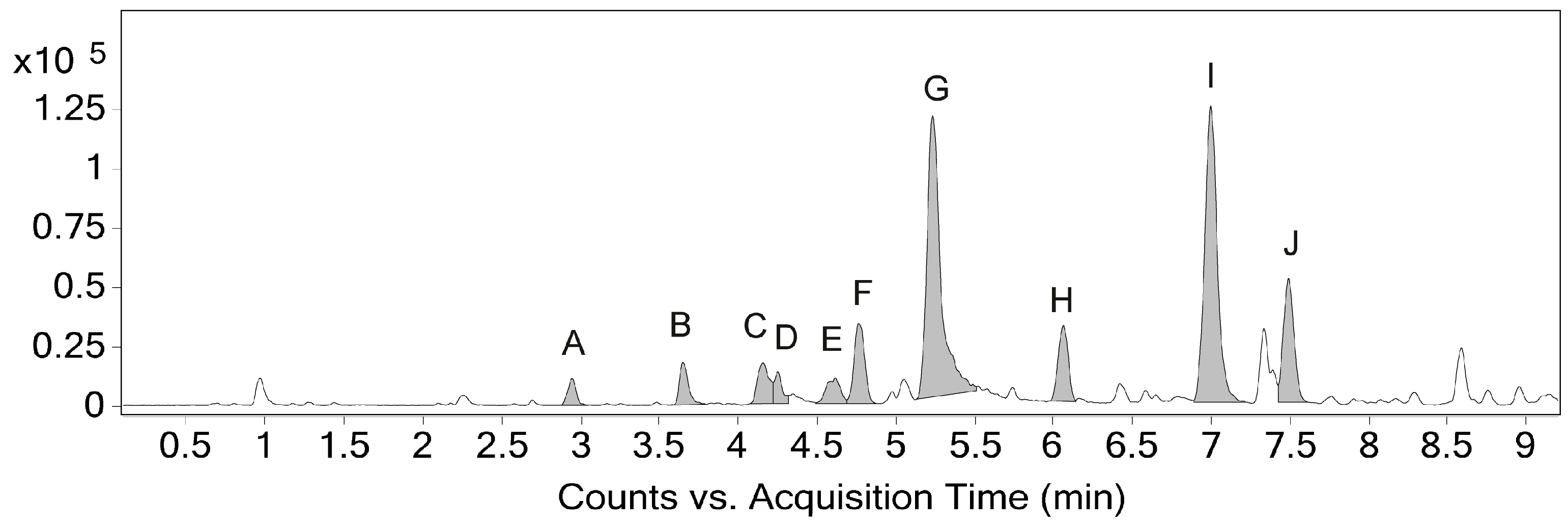

| Compound | Peak | RT (min) | Formula (M) | m/z Theoretical | m/z exp. | Error (ppm) |
|---|---|---|---|---|---|---|
| A | 2.95 | C19H14O4 | 307.0965 | 307.0968 | 0.98 | |
| B | 3.65 | C19H12O4 | 305.0808 | 305.0814 | 1.97 | |
| C | 4.16 | C19H12O4 | 305.0808 | 305.0814 | 1.97 | |
| D | 4.25 | C19H12O4 | 305.0808 | 305.0817 | 2.95 | |
| E | 4.62 | C20H12O4 | 319.0965 | 319.0970 | 1.57 | |
| F | 4.75 | C20H14O3 | 303.1016 | 303.1022 | 1.98 | |
| 1 | G | 5.23 | C19H12O3 | 289.0859 | 289.0869 | 3.46 |
| H | 6.06 | C19H12O4 | 305.0808 | 305.0816 | 2.62 | |
| 2 | I | 7.00 | C20H14O2 | 287.1067 | 287.1077 | 3.48 |
| 3 | J | 7.49 | C19H12O2 | 273.0910 | 273.0919 | 3.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocampos, F.M.M.; de Souza, A.J.B.; Antar, G.M.; Wouters, F.C.; Colnago, L.A. Phytotoxicity of Schiekia timida Seed Extracts, a Mixture of Phenylphenalenones. Molecules 2021, 26, 4197. https://doi.org/10.3390/molecules26144197
Ocampos FMM, de Souza AJB, Antar GM, Wouters FC, Colnago LA. Phytotoxicity of Schiekia timida Seed Extracts, a Mixture of Phenylphenalenones. Molecules. 2021; 26(14):4197. https://doi.org/10.3390/molecules26144197
Chicago/Turabian StyleOcampos, Fernanda Maria Marins, Ana Julia Borim de Souza, Guilherme Medeiros Antar, Felipe Christoff Wouters, and Luiz Alberto Colnago. 2021. "Phytotoxicity of Schiekia timida Seed Extracts, a Mixture of Phenylphenalenones" Molecules 26, no. 14: 4197. https://doi.org/10.3390/molecules26144197
APA StyleOcampos, F. M. M., de Souza, A. J. B., Antar, G. M., Wouters, F. C., & Colnago, L. A. (2021). Phytotoxicity of Schiekia timida Seed Extracts, a Mixture of Phenylphenalenones. Molecules, 26(14), 4197. https://doi.org/10.3390/molecules26144197







