The Tannins from Sanguisorba officinalis L. (Rosaceae): A Systematic Study on the Metabolites of Rats Based on HPLC–LTQ–Orbitrap MS2 Analysis
Abstract
:1. Introduction
2. Results
2.1. Establishment of the Analytical Strategy
2.2. Identification of Total Tannins Compounds Extracted from Sanguisorba Tannins
2.3. Identification of Metabolite of Sanguisorba Tannins In Vivo and In Vitro
2.4. Identification of Phase I Metabolites In Vivo
2.5. Identification of Phase II Metabolites In Vivo
2.6. The Identification and Characterization of Metabolites In Vitro
2.7. Identification of Phase II Metabolites
3. Discussion
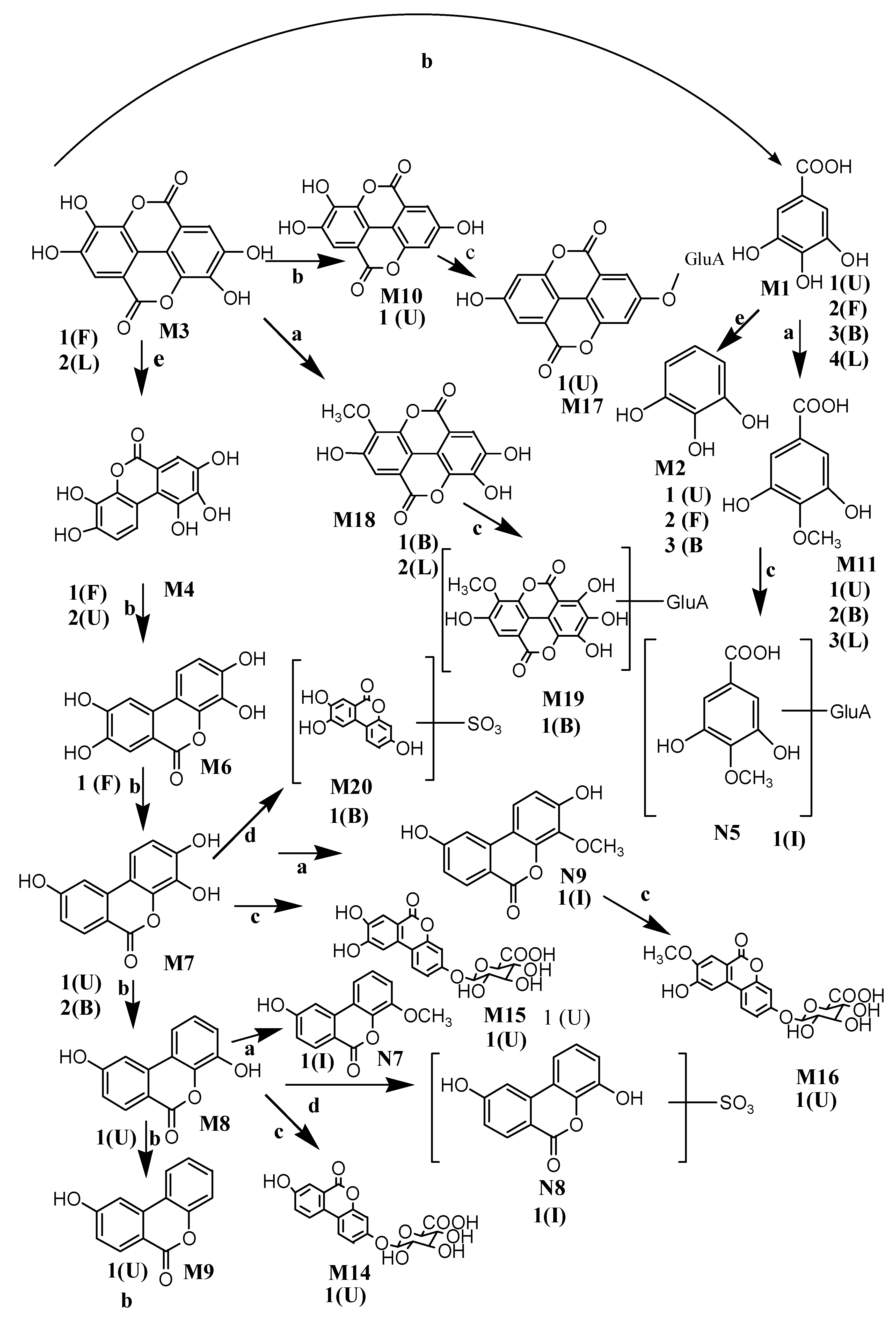
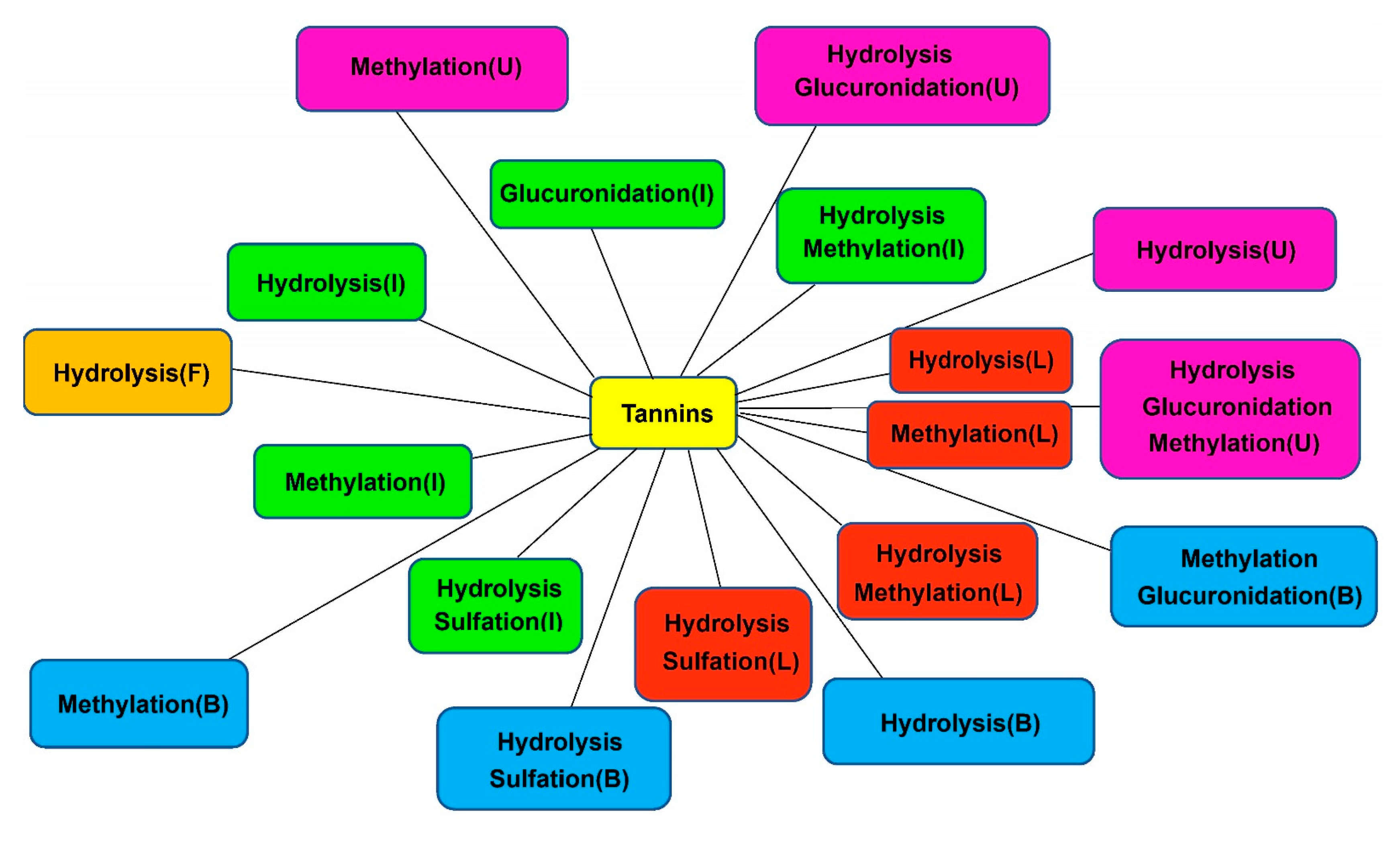
3.1. Metabolic Pathways of Sanguisorba Tannins
3.2. Comparison of Metabolites In Vitro and In Vivo
4. Materials and Methods
4.1. Plant Materials
4.2. Chemicals and Materials
4.3. Instrumentations and Investigation Conditions
4.4. Sample Preparation and Pretreatment
4.4.1. Extraction and Purification of the Total Tannins from Sanguisorba
4.4.2. Quantitation and Chemical Analysis of Total Tannins from Sanguisorba
4.5. Animal Experiments
4.6. Sample Preparation
4.6.1. Sample Collection and Pretreatment In Vivo
4.6.2. Sample Preparation and Treatment In Vitro
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zhao, Z.; He, X.; Zhang, Q.; Wei, X.; Huang, L.; Fang, J.C.; Wang, X.; Zhao, M.; Bai, Y.; Zheng, X. Traditional Uses, Chemical Constituents and Biological Activities of Plants from the Genus Sanguisorba L. Am. J. Chin. Med. 2017, 45, 199–224. [Google Scholar] [CrossRef]
- Karkanis, A.C.; Fernandes, Â.; Vaz, J.; Petropoulos, S.A.; Georgiou, E.; Ciric, A.; Sokovic, M.; Oludemi, T.; Barros, L.; Ferreira, I.C.F.R. Chemical composition and bioactive properties of Sanguisorba minor Scop. under Mediterranean growing conditions. Food Funct. 2019, 10, 1340–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karkanis, A.; Vellios, E.; Thomaidis, T.; Bilalis, D.; Efthimiadou, A.; Travlos, I. Phytochemistry and Biological Properties of Burnet Weed (Sanguisorba spp.): A Review. Not. Sci. Biol. 2014, 6, 395–398. [Google Scholar] [CrossRef] [Green Version]
- Shikov, A.N.; Narkevich, I.A.; Flisyuk, E.V.; Luzhanin, V.G.; Pozharitskaya, O.N. Medicinal plants from the 14th edition of the Russian Pharmacopoeia, recent updates. J. Ethnopharmacol. 2021, 268, 113685. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Rounds, B.V.; Bore, L.; Finlay, H.J.; Favaloro, F.G.; Suh, N.; Wang, Y.P.; Sporn, M.B.; Gribble, G.W. Synthetic oleanane and ursane triterpenoids with modified rings A and C: A series of highly active inhibitors of nitric oxide produc-tion in mouse macrophages. J. Med. Chem. 2000, 43, 4233–4246. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, C.-J.; Wang, L.-Q.; Wu, J.; Dai, C.; Yuan, Y.-M.; Li, G.Q.; Yao, M.-C. A tannin compound from Sanguisorba officinalis blocks Wnt/β-catenin signaling pathway and induces apoptosis of colorectal cancer cells. Chin. Med. 2019, 14, 22. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.; Taylor, J.R.N.; Belton, P.S.; Minnaar, A. Kafirin microparticle encapsulation of catechin and sorghum condensed tannins. J. Agric. Food Chem. 2009, 57, 7523–7528. [Google Scholar] [CrossRef]
- Yano, T.; Ohmori, K.; Takahashi, H.; Kusumi, T.; Suzuki, K. Unified approach to catechin hetero-oligomers: First total synthe-sis of trimer EZ–EG–CA isolated from Ziziphus jujube. Org. Biomol. Chem. 2012, 10, 7685–7688. [Google Scholar] [CrossRef]
- Cala, O.; Pinaud, N.; Simon, C.; Fouquet, E.; Laguerre, M.; Dufourc, E.J.; Pianet, I. NMR and molecular modeling of wine tan-nins binding to saliva proteins: Revisiting astringency from molecular and colloidal prospects. FASEB J. 2010, 24, 4281–4290. [Google Scholar] [CrossRef]
- Chen, H.F.; Tanaka, T.; Nonaka, G.I.; Fujioka, T.; Mihashi, K. Hydrolyzable tannins based on a triterpenoid glycoside core, from Castanopsis-hystrix. Phytochemistry 1993, 32, 1457–1460. [Google Scholar]
- Bae, Y.-S.; Burger, J.F.; Steynberg, J.P.; Ferreira, D.; Hemingway, R.W. Flavan and procyanidin glycosides from the bark of blackjack oak. Phytochemistry 1994, 35, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhou, M.; Tan, T.; Zhong, C.C.; Wang, Q.; Pan, L.L.; Luo, Y.Y.; Yang, S.L.; Feng, Y.L.; Ouyang, H. A Sample and Sen-sitive HPLC-MS/MS Method for Simultaneous Determination of Ziyuglycoside I and Its Metabolite Ziyuglycoside II in Rat Pharmacokinetics. Molecules 2018, 23, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yisimayili, Z.; Guo, X.; Liu, H.; Xu, Z.; Abdulla, R.; Aisa, H.A.; Huang, C. Metabolic profiling analysis of corilagin in vivo and in vitro using high-performance liquid chromatography quadrupole time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2019, 165, 251–260. [Google Scholar] [CrossRef]
- Ma, J.Y.; Zhou, X.L.; Fu, J.; He, C.Y.; Feng, R.; Huang, M.; Shou, J.W.; Zhao, Z.X.; Li, X.Y.; Zhang, L.Y. In vivo metabolite pro-filing of a purified ellagitannin isolated from Polygonum capitatum in rats. Molecules 2016, 21, 1110. [Google Scholar] [CrossRef] [Green Version]
- Saldan, N.C.; Almeida, R.T.R.; Avíncola, A.; Porto, C.; Galuch, M.B.; Magon, T.F.S.; Pilau, E.J.; Svidzinski, T.I.E.; Oliveira, C.C. Development of an analytical method for identification of Aspergillus flavus based on chemical markers using HPLC-MS. Food Chem. 2018, 241, 113. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.E.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.M.; Wu, Y.P.; Li, L. Intestinal micro-biota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.T.; Li, Y.Q.; Li, P.F.; Wang, M.; Wang, J.K.; Tang, Z.Y.; Wang, T.; Luo, L.; Wang, C.G.; Zhao, B.S. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed. Pharmacother. 2019, 117, 109–138. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Lou, Y.; Jiang, H.; Wang, X.; Chai, X.; Zeng, S. Metabolism of Ebracteolata Compound B Studied In Vitro with Human Liver Microsomes, HepG2 Cells, and Recombinant Human Enzymes. Drug Metab. Dispos. 2010, 38, 2157–2165. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.-Y.; Zhou, X.; Fu, J.; Hu, T.; Or, P.M.; Feng, R.; He, C.-Y.; Chen, W.-J.; Zhang, X.; Chen, Y.; et al. Metabolite profiling analysis of FR429, an ellagitannin purified from Polygonum capitatum, in rat and human liver microsomes, cytosol and rat primary hepatocytes In Vitro. Chem. Interact. 2014, 220, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ojanpera, S.; Ojanpera, L. Forensic drug screening by LC-MS using accurate mass measurement. LCGC Eur. 2005, 11, 607–614. [Google Scholar]
- Shahrzad, S.; Bitsch, I. Determination of gallic acid and its metabolites in human plasma and urine by high-performance liquid chromatography. J Chromatogr. B Biomed. Sci. AppI. 1998, 705, 87–95. [Google Scholar] [CrossRef]
- Landete, J. Ellagitannins, ellagic acid and their derived metabolites: A review about source, metabolism, functions and health. Food Res. Int. 2011, 44, 1150–1160. [Google Scholar] [CrossRef]
- Vallverdú-Queralt, A.; Meudec, E.; Eder, M.; Lamuela-Raventos, R.M.; Sommerer, N.; Cheynier, V. Targeted filtering reduces the complexity of UHPLC-Orbitrap-HRMS data to decipher polyphenol polymerization. Food Chem. 2017, 227, 255–263. [Google Scholar] [CrossRef]
- González-Barrio, R.; Truchado, P.; Ito, H.; Espín, J.C.; Tomás-Barberán, F.A. UV and MS Identification of Urolithins and Nasutins, the Bioavailable Metabolites of Ellagitannins and Ellagic Acid in Different Mammals. J. Agric. Food Chem. 2011, 59, 1152–1162. [Google Scholar] [CrossRef]
- Fu, J.; Ma, J.Y.; Zhang, X.F.; Wang, Y.; Feng, R.; Chen, Y.C.; Tan, X.S.; Zhang, Y.Y.; Sun, Y.P.; Zhou, Y. Identification of me-tabolites of FR429, a potential antitumor ellagitannin, transformed by rat intestinal bacteria in vitro, based on liquid chroma-tography–ion trap-time of flight mass spectrometry analysis. J. Pharm. Biomed. Anal. 2012, 71, 162–167. [Google Scholar] [CrossRef]
- Miao, X.; Wang, J.; Chen, L.; Peng, Z.; Chen, Y. Identification of in vivo and in vitro metabolites of 4,5-dimethoxycanthin-6-one by HPLC–Q-TOF-MS/MS. J. Chromatogr. B 2016, 1020, 78–84. [Google Scholar] [CrossRef]
- Scarth, J.P.; Spencer, H.A.; Timbers, S.E.; Hudson, S.C.; Hillyer, L.L. The use of in vitro technologies coupled with high reso-lution accurate mass LC-MS for studying drug metabolism in equine drug surveillance. Drug Test. Anal. 2010, 2, 1–10. [Google Scholar]
- Zhou, S.; Chan, S.Y.; Goh, B.C.; Chan, E.; Duan, W.; Huang, M.; McLeod, H.L. Mechanism-Based Inhibition of Cytochrome P450 3A4 by Therapeutic Drugs. Clin. Pharmacokinet. 2005, 44, 279–304. [Google Scholar] [CrossRef]
- Wilk-Zasadna, I.; Bernasconi, C.; Pelkonen, O.; Coecke, S. Biotransformation in vitro: An essential consideration in the quan-titative in vitro-to-in vivo extrapolation (QIVIVE) of toxicity data. Toxicology 2015, 332, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Luo, Z.; Tao, B.; Chen, C. Ultrasonic-assisted extraction and purifcation of phenolic compounds from sugarcane (Saccharum ofcinarum L.) rinds. LWT-Food Sci. Technol. 2015, 60, 970–976. [Google Scholar] [CrossRef]
- Li, Q.; Tu, J.; Zhou, B. The tannins from Punica granatum L, natural regulator of TGF-β1/Smad signaling activity improves nephrectomy and adriamycin induced focal segmental glomerulosclerosis in vivo. J. Funct. Foods 2019, 57, 361–372. [Google Scholar] [CrossRef]
- Kaur, C.; Kapoor, H.C. Anti-oxidant activity and total phenolic content of some Asian vegetables. Int. J. Food Sci. Technol. 2002, 37, 153–161. [Google Scholar] [CrossRef]
- Azuma, K.; Ippoushi, K.; Nakayama, M.; Ito, H.; Higashio, H.; Terao, J. Absorption of Chlorogenic Acid and Caffeic Acid in Rats after Oral Administration. J. Agric. Food Chem. 2000, 48, 5496–5500. [Google Scholar] [CrossRef]
- Prasain, J.K.; Jones, K.; Brissie, N.; Moore, R.; Wyss, A.J.M.; Barnes, S. Identification of Puerarin and Its Metabolites in Rats by Liquid Chromatography−Tandem Mass Spectrometry. J. Agric. Food Chem. 2004, 52, 3708–3712. [Google Scholar] [CrossRef]
- Zheng, D.; Sun, C.-C.; Su, H.; Zhang, Q.-F. Metabolism, Excretion, and Tissue Distribution of Astilbin–Zein Nanoparticles in Rats. J. Agric. Food Chem. 2019, 67, 8332–8338. [Google Scholar] [CrossRef]
- Zollner, P.; Jodlbauer, J.; Kleinova, M.; Kahlbacher, H.; Kuhn, T.; Hochsteiner, W.W. Lindner, Concentration levels of zeara-lenone and its metabolites in urine, muscle tissue, and liver samples of pigs fed with mycotoxin-contaminated oats. J. Agric. Food Chem. 2002, 50, 2494–2501. [Google Scholar] [CrossRef]
- Yoshihara, S.; Makishima, M.; Suzuki, N.; Ohta, S. Metabolic Activation of Bisphenol A by Rat Liver S9 Fraction. Toxicol. Sci. 2001, 62, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Ma, Y.H.; Zhou, Z.; Chen, Y.; Wang, Y.; Gao, X. Intestinal Absorption and Metabolism of Epimedium Flavonoids in Osteoporosis Rats. Drug Metab. Dispos. 2015, 43, 1590–1600. [Google Scholar] [CrossRef] [Green Version]
- Kang, I.; Kim, Y.; Tomás-Barberán, F.A.; Espín, J.C.; Chung, S. Urolithin A, C, and D, but not iso-urolithin A and urolithin B, attenuate triglyceride accumulation in human cultures of adipocytes and hepatocytes. Mol. Nutr. Food Res. 2016, 60, 1129–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Sarrías, A.; Miguel, V.; Merino, G.; Lucas, R.; Morales, J.C.; Tomás-Barberán, F.; Álvarez, A.I.; Espín, J.C. The Gut Microbiota Ellagic Acid-Derived Metabolite Urolithin A and Its Sulfate Conjugate Are Substrates for the Drug Efflux Transporter Breast Cancer Resistance Protein (ABCG2/BCRP). J. Agric. Food Chem. 2013, 61, 4352–4359. [Google Scholar] [CrossRef]
- Ishimoto, H.; Shibata, M.; Myojin, Y.; Ito, H.; Sugimoto, Y.; Tai, A.; Hatano, T. In vivo anti-inflammatory and antioxidant properties of ellagitannin metabolite urolithin A. Bioorganic Med. Chem. Lett. 2011, 21, 5901–5904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez-Bastida, J.A.; Gonzalez-Sarrias, A.M.; Larrosa, F.; Tomas-Barberan, J.C.; Espin, M.T.; Garcia-Conesa, M.T. Ellag-itannin metabolites, urolithin A glucuronide and its aglycone urolithin A, ameliorate TNF-α-induced inflammation and associated molecular markers in human aortic endothelial cells. Mol. Nutr. Food Res. 2012, 56, 784–796. [Google Scholar] [CrossRef]
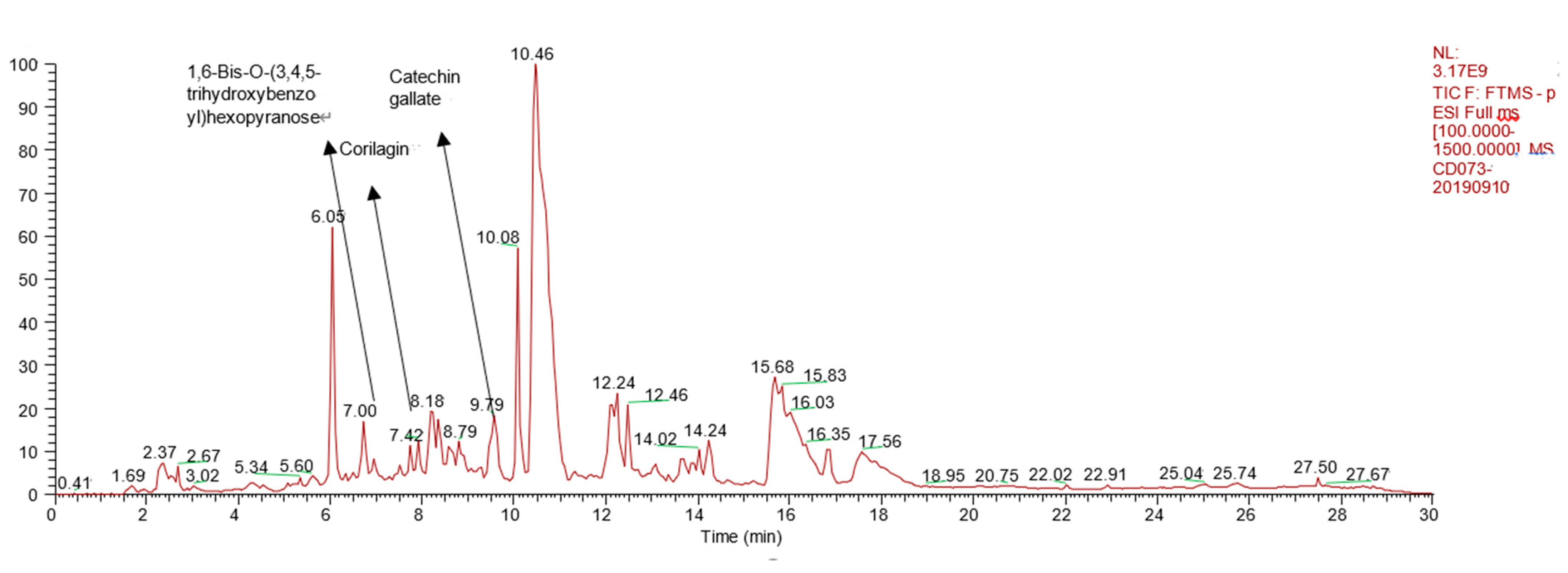

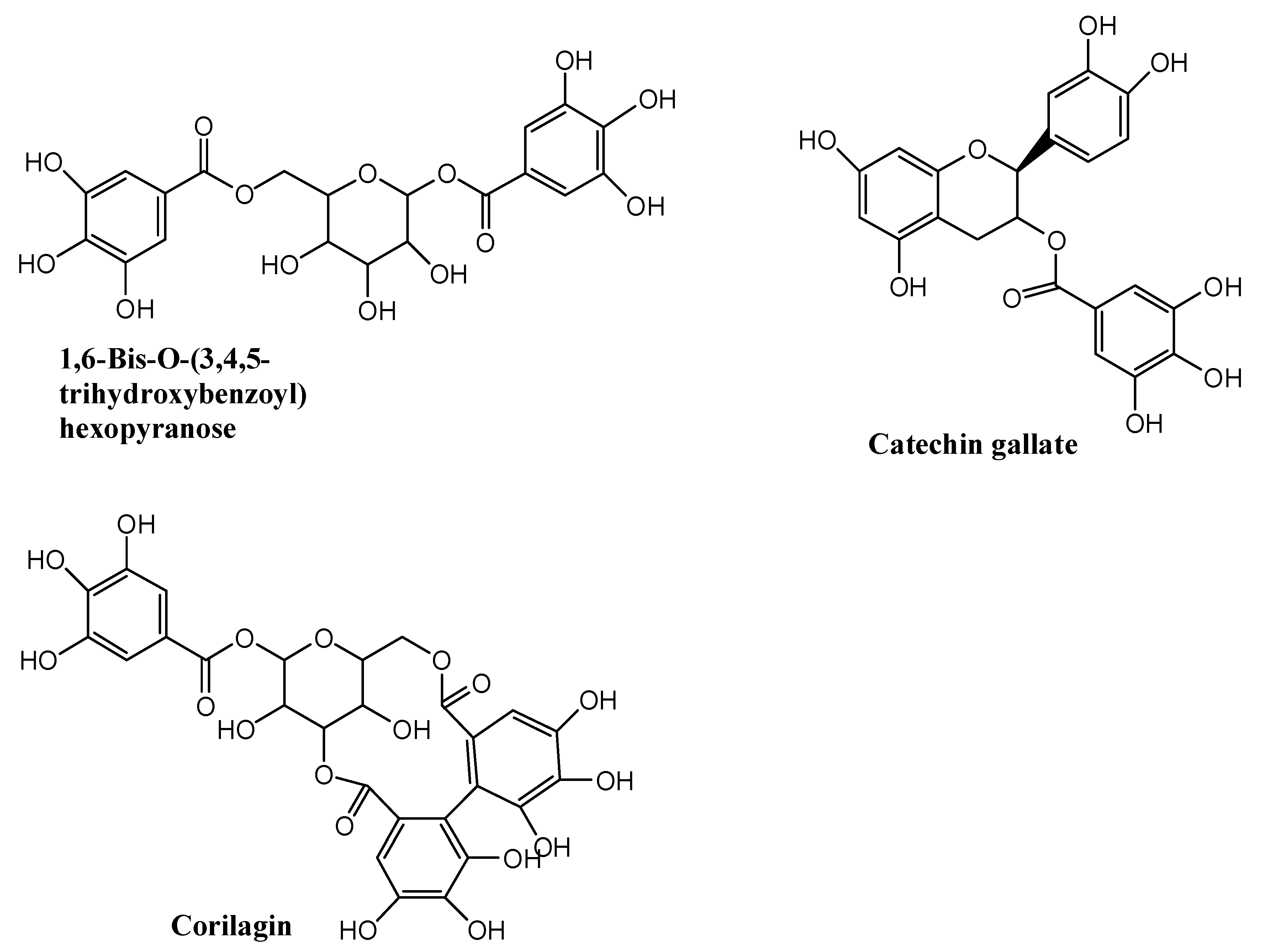
| NO. | Name | Formula | tR (min) | MS1 | MS2 | mzCloud Best Match |
|---|---|---|---|---|---|---|
| 1 | 1,6-Bis-O-(3,4,5-trihydroxybenzo yl)hexopyranose | C20H20O14 | 7.00 | 484.1111 | 153.0137 | 77.9 |
| 2 | Catechin gallate | C22H18O10 | 9.79 | 442.0894 | 151.0387 139.0380 123.0440 | 79.1 |
| 3 | Corilagin | C27H22O18 | 7.42 | 634.4729 | 463.0524 300.9995 | 89.3 |
| NO. | tR (min) | MS1 | Diff(ppm) | MS2 | Formula | Compound | Occurrence |
|---|---|---|---|---|---|---|---|
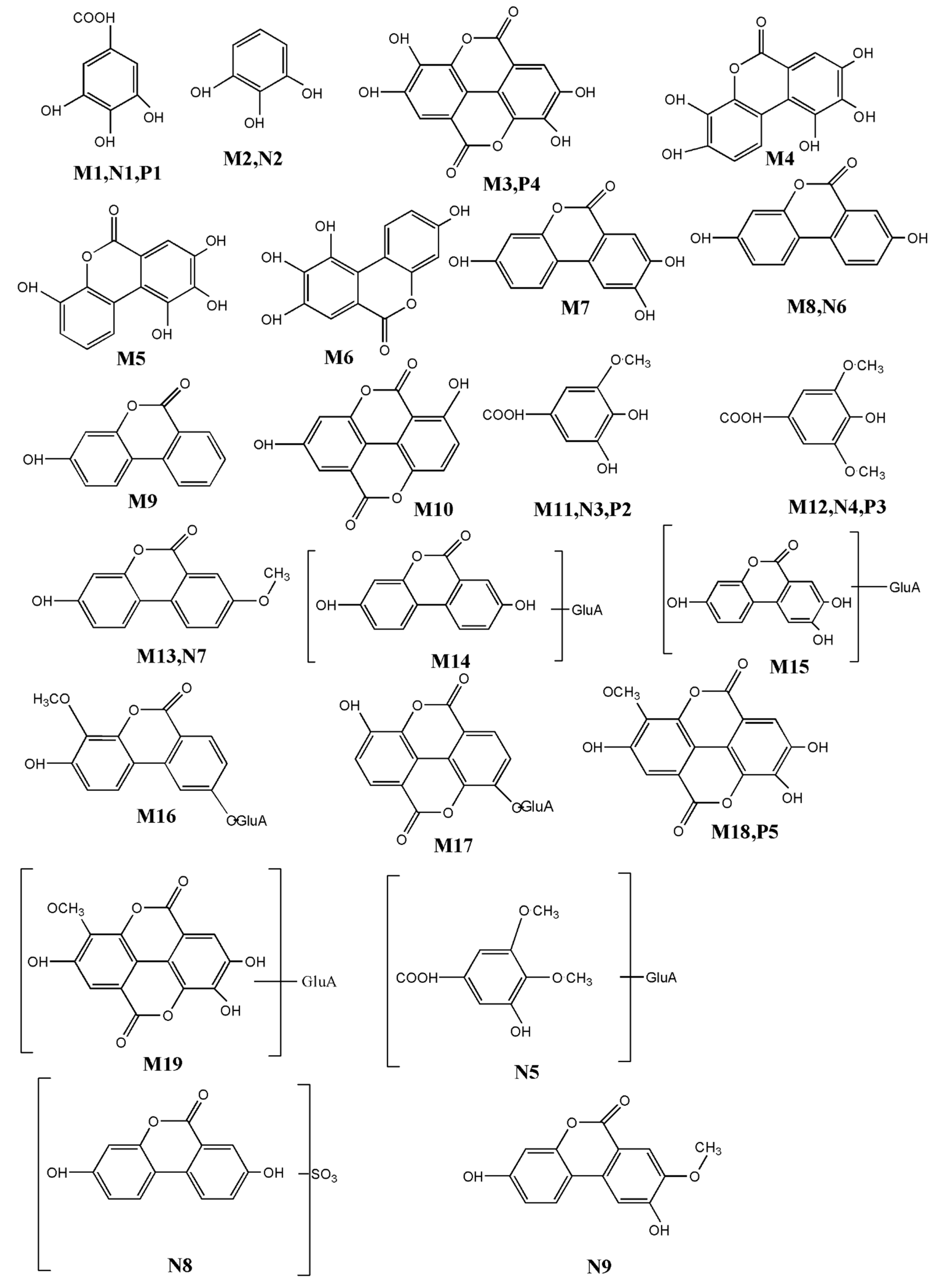
| M1 I | 3.1 | 169.0148 | 3.54 | 125.0241 | C7H6O5 | Gallic acid | Urine Feces Bile plasma |
| M2 I | 6.7 | 125.0257 | 2.05 | 81.0345 | C6H6O3 | Pyrogallol | Urine Feces Bile Plasma |
| M3 I | 28.1 | 300.9996 | 2.32 | 283.1526 257.0825 | C15H6O8 | Ellagic acid | Feces |
| M4 I | 24.1 | 275.0204 | 6.08 | 257.0106 229.0167 | C13H8O7 | Urolithin M5 (3,4,8,9,10-pentahydroxy-urolithin) | Urine Feces |
| M5 I | 25.2 | 259.0249 | 0.38 | 241.0176 213.0223 | C13H8O6 | Urolithin M6 | Feces |
| M6 I | 24.4 | 259.0251 | 1.16 | 241.0171 213.0225 | C13H8O6 | Urolithin D | Feces |
| M7 I | 28.4 | 243.0299 | 0.82 | 215.0367 187.0426 | C13H8O5 | Urolithin C | Urine Bile |
| M8 I | 30.4 | 227.0358 | 4.40 | 199.0346 | C13H8O4 | Urolithin A | Urine |
| M9 I | 38.1 | 211.0408 | 3.79 | 167.0502 139.0556 | C13H8O3 | Urolithin B | Urine |
| M10 I | 36.7 | 269.0424 | −1.12 | 224.0454 197.0649 | C14H6O6 | Nasutin A (4,9-dehydroxy-ellagic acid) | Urine |
| M11 II | 8.9 | 183.0294 | 2.73 | 169.0049 125.0126 | C8H8O5 | Gallic acid methyl | Urine Bile |
| M12 II | 4.2 | 197.0460 | 0.51 | 183.0854 169.0146 125.0196 | C9H10O5 | Gallic acid dimethyl | Bile |
| M13 II | 34.3 | 241.0513 | −2.9 | 210.9041 196. 9863 | C14H10O4 | Urolithin A methyl | Bile |
| M14 II | 26.6 | 402.0678 | 7.89 | 227.0305 227.1005 175.2006 | C19H16O10 | Urolithin A glucuronide | Urine |
| M15 II | 26.8 | 419.0645 | 1.53 | 243.0314 214.9997 187.0426 | C19H16O11 | Urolithin C glucuronide | Urine |
| M16 II | 26.5 | 433.0787 | 5.07 | 257.0485 243.0256 | C20H18O11 | Urolithin C methyl ether glucuronide | Urine |
| M17 II | 29.4 | 445.0792 | 2.47 | 269.0463 | C21H18O11 | Nasutin A glucuronide | Urine |
| M18 II | 31.8 | 315.0140 | −9.52 | 300.9991 | C15H8O8 | Ellagic acid methyl ether | Bile |
| M19 II | 25.8 | 491.0013 | 0. 26 | 315.0128 299.7270 | C21H16O14 | Ellagic acid methyl ether glucuronide | Bile Kidney |
| NO. | tR (min) | MS1 | Diff(ppm) | MS2 | Formula | Compound |
|---|---|---|---|---|---|---|
| N1 I | 2.2 | 169.0115 | 15.97 | 125.0225 | C7H6O5 | Gallic acid |
| N2 I | 5.8 | 125.3446 | 2.75 | 81.1342 | C6H6O3 | Pyrogallol |
| N3 II | 8.4 | 183.0298 | 0.54 | 169.0052 125.0177 | C8H8O5 | Gallic acid methyl |
| N4 II | 4.1 | 197.0466 | 0.54 | 169.0118 125.0232 | C9H10O5 | Gallic acid dimethyl |
| N5 II | 9.6 | 359.0264 | 1.86 | 183.0205 169.1322 | C14H16O11 | Gallic acid methyl ether glucuronide |
| N6 I | 32.3 | 227.0312 | 1.58 | 199.0344 | C13H8O4 | Urolithin A |
| N7 II | 36.6 | 241.0531 | 10.4 | 210.9047 196.9068 | C14H10O4 | Urolithin A methyl |
| N8 II | 8.5 | 309.9918 | 2.83 | 227.0374 | C13H7O7S | Urolithin A sulfate |
| N9 II | 37.8 | 257.0458 | 0.78 | 243.0276 215.0385 | C14H10O5 | Urolithin C methyl |
| NO. | tR(min) | MS1 | Diff(ppm) | MS2 | Formula | Compound |
|---|---|---|---|---|---|---|
| P1 I | 2.7 | 169.0115 | 2.96 | 125.4925 | C7H6O5 | Gallic acid |
| P2 II | 8.4 | 183.0292 | −3.82 | 125.0236 | C8H8O5 | Gallic acid methyl |
| P3 II | 5.3 | 197.0414 | 0.51 | 125.0788 | C9H10O5 | Gallic acid dimethyl |
| P4 I | 27.8 | 300.9996 | 2.32 | 283.9996 257.0067 229.0102 185.0274 | C15H6O8 | Ellagic acid |
| P5 II | 30.8 | 315.0148 | 3.25 | 299.9936 | C15H8O8 | Ellagic acid methyl ether |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Li, Q.; Zhou, B. The Tannins from Sanguisorba officinalis L. (Rosaceae): A Systematic Study on the Metabolites of Rats Based on HPLC–LTQ–Orbitrap MS2 Analysis. Molecules 2021, 26, 4053. https://doi.org/10.3390/molecules26134053
Tu J, Li Q, Zhou B. The Tannins from Sanguisorba officinalis L. (Rosaceae): A Systematic Study on the Metabolites of Rats Based on HPLC–LTQ–Orbitrap MS2 Analysis. Molecules. 2021; 26(13):4053. https://doi.org/10.3390/molecules26134053
Chicago/Turabian StyleTu, Jingjing, Qiaoling Li, and Benhong Zhou. 2021. "The Tannins from Sanguisorba officinalis L. (Rosaceae): A Systematic Study on the Metabolites of Rats Based on HPLC–LTQ–Orbitrap MS2 Analysis" Molecules 26, no. 13: 4053. https://doi.org/10.3390/molecules26134053
APA StyleTu, J., Li, Q., & Zhou, B. (2021). The Tannins from Sanguisorba officinalis L. (Rosaceae): A Systematic Study on the Metabolites of Rats Based on HPLC–LTQ–Orbitrap MS2 Analysis. Molecules, 26(13), 4053. https://doi.org/10.3390/molecules26134053






