Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions
Abstract
1. Introduction
2. Results
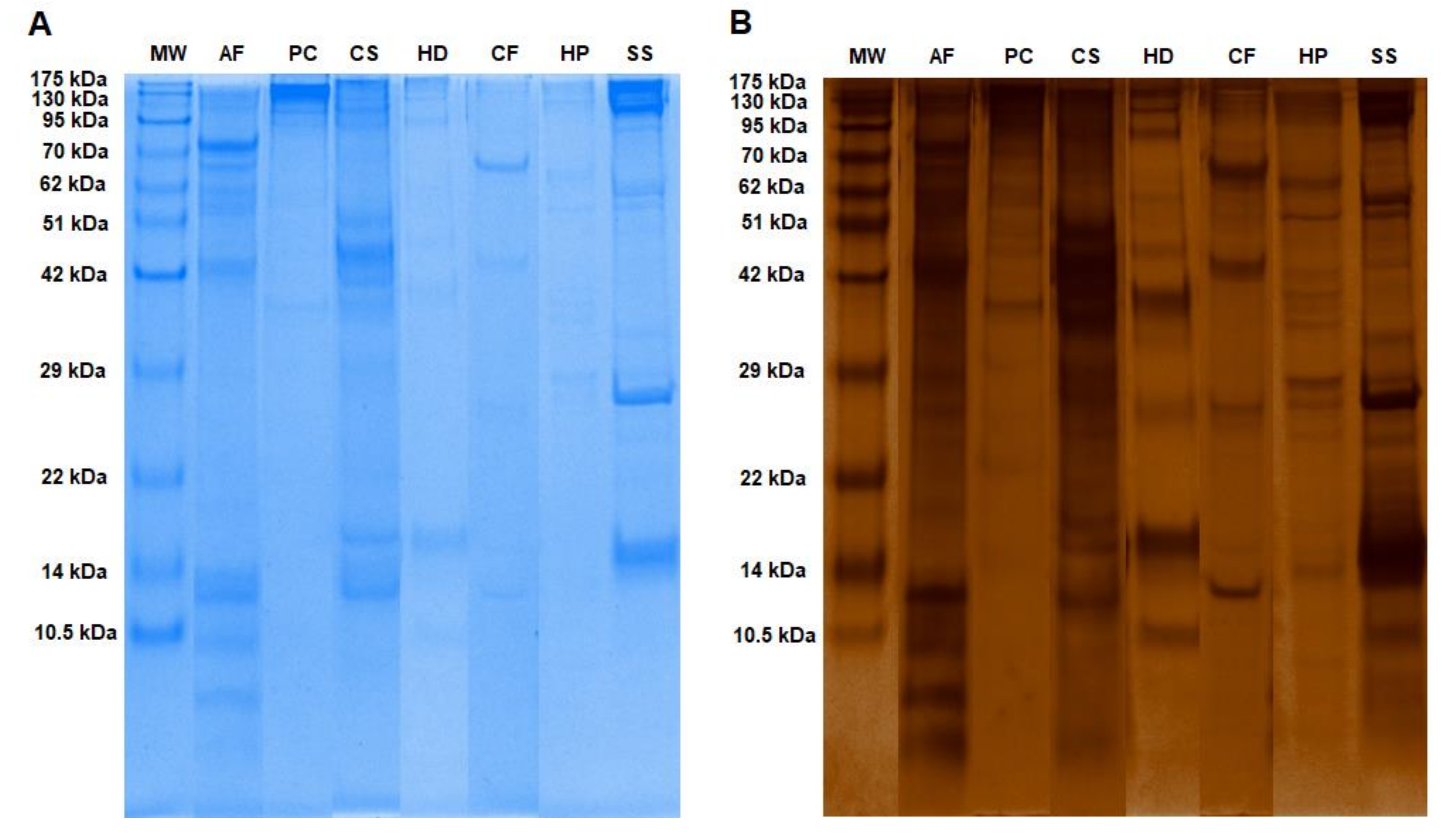
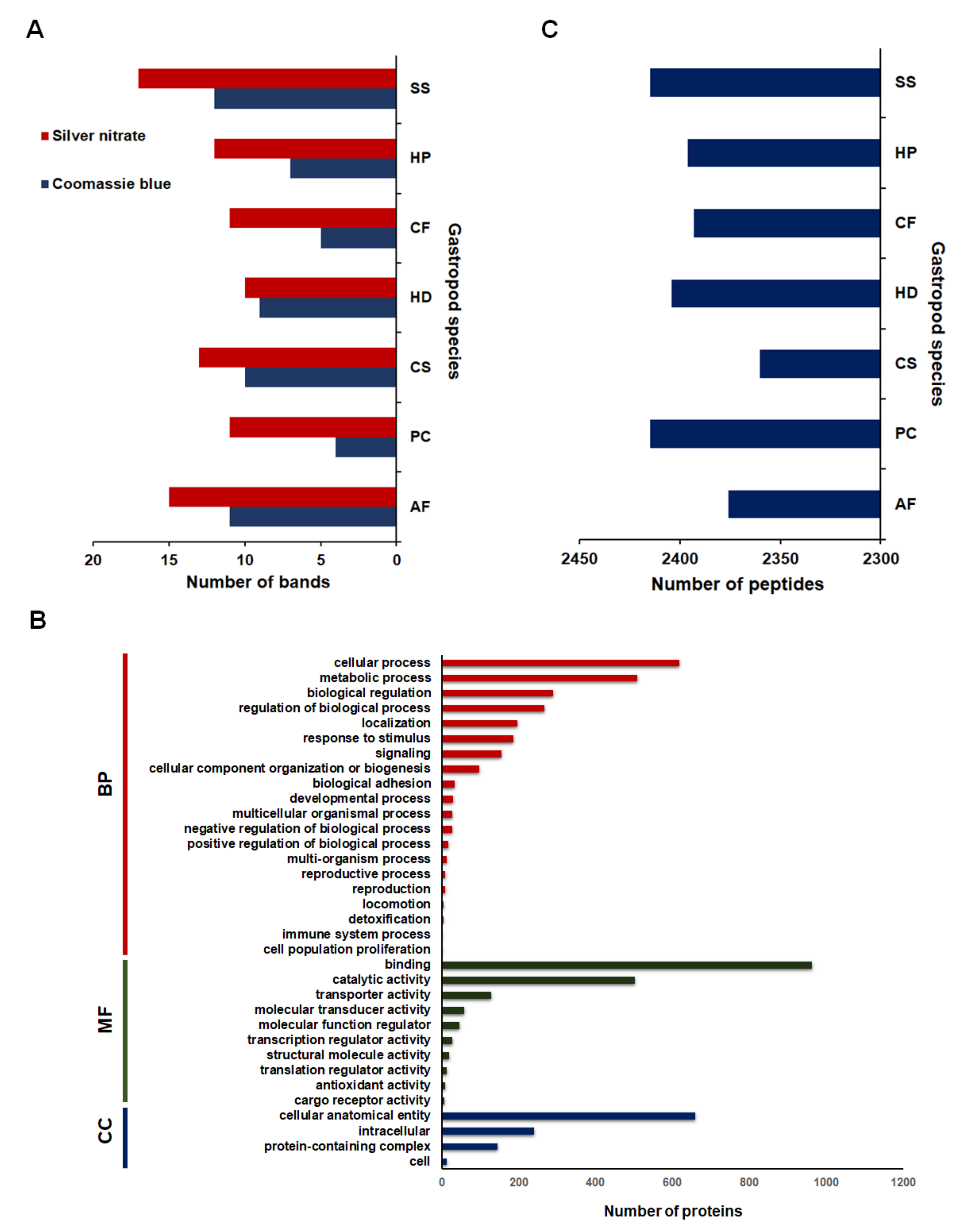
2.1. Mucous Proteins Separated on The 1D-SDS Polyacrylamide Gels and Identified by Mass Spectrometry
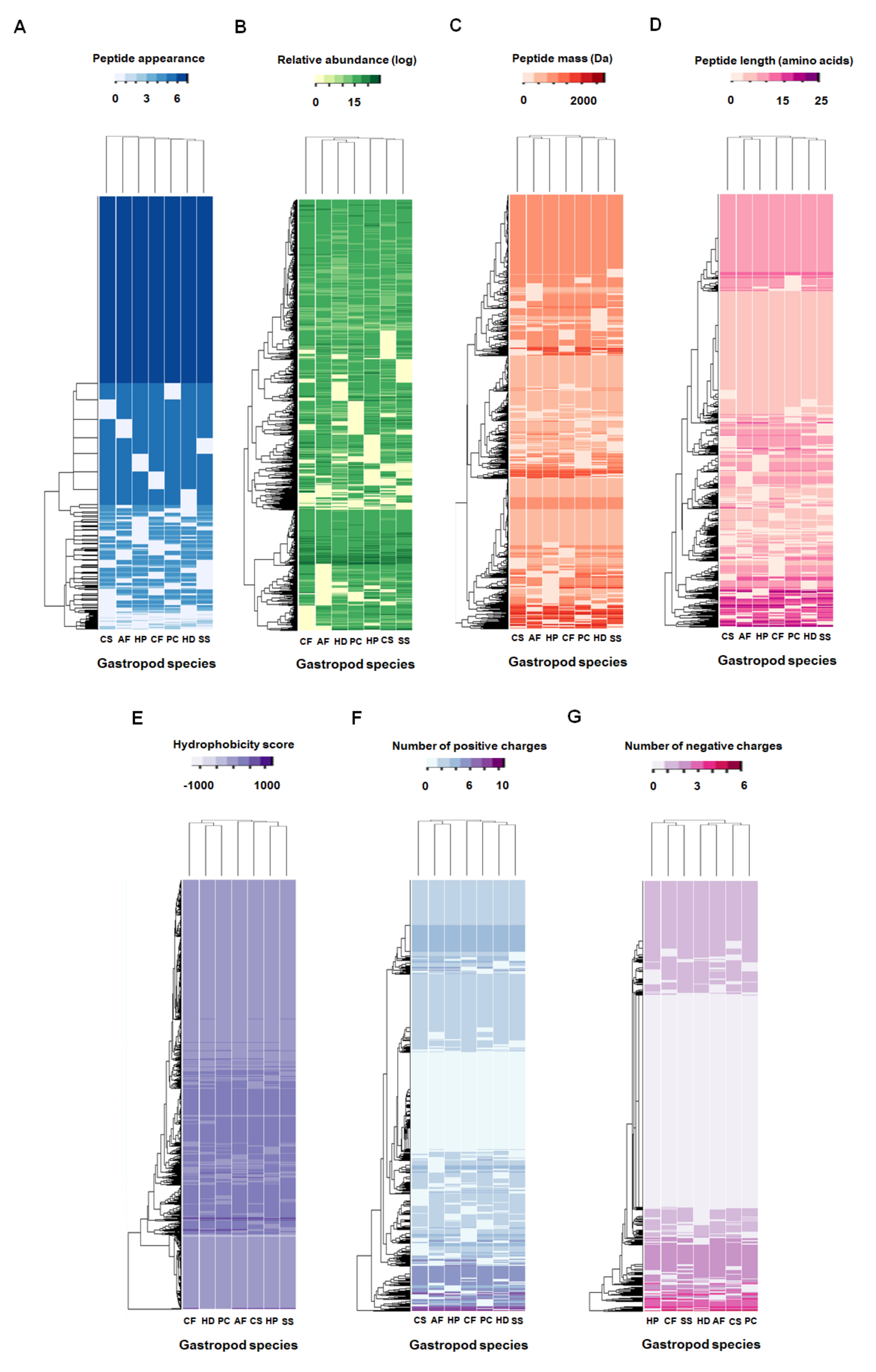
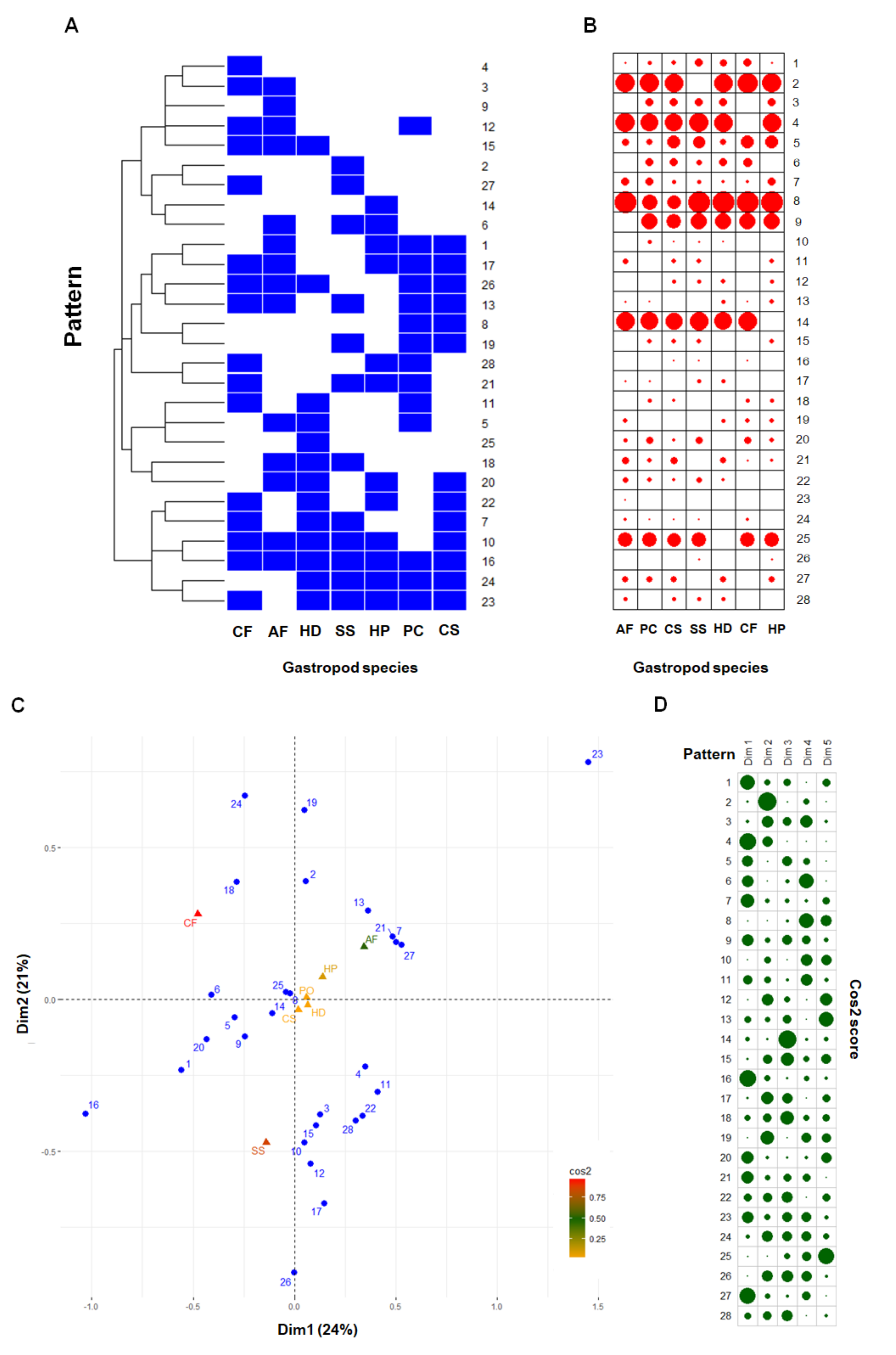
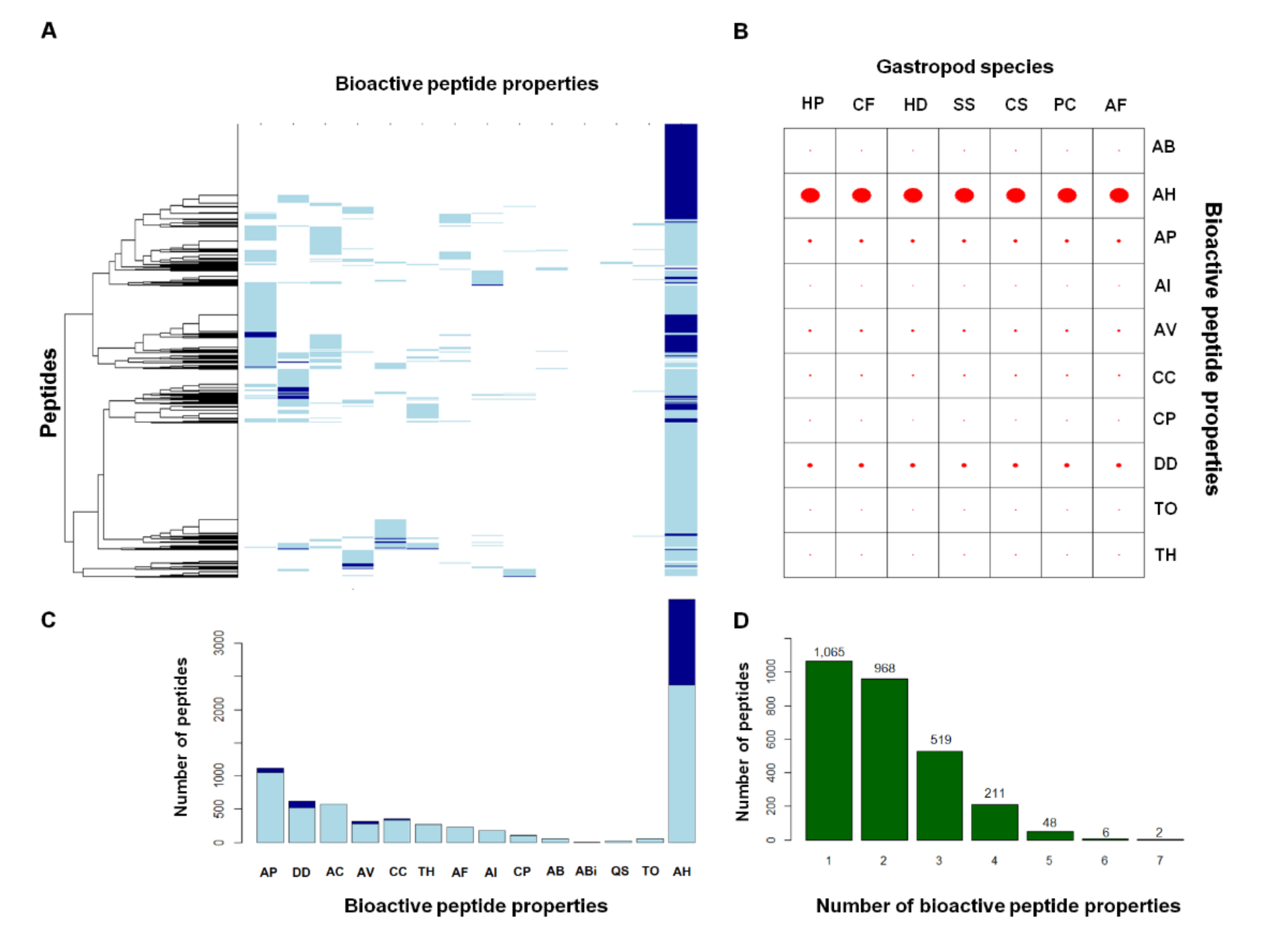
2.2. Comparative Mucus Peptides of Seven Gastropod Species
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. D-SDS-PAGE Experiment
4.3. LC-MS/MS
4.4. Proteomic Data Evaluation
4.5. Bioinformatics and Machine Learning
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Grande, C.; Templado, J.; Zardoya, R. Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 2008, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Vitta, A.; Polseela, R.; Nateeworanart, S.; Tattiyapong, M. Survey of Angiostrongylus cantonensis in rats and giant African land snails in Phitsanulok province, Thailand. Asian Pac. J. Trop. Med. 2011, 4, 597–599. [Google Scholar] [CrossRef]
- Martin-Alonso, A.; Abreu-Yanes, E.; Feliu, C.; Mas-Coma, S.; Bargues, M.D.; Valladares, B.; Foronda, P. Intermediate Hosts of Angiostrongylus cantonensis in Tenerife, Spain. PLoS ONE 2015, 10, e0120686. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M. Gastropod Secretory Glands and Adhesive Gels. In Biological Adhesive Systems: From Nature to Technical and Medical Application; von Byern, J., Grunwald, I., Eds.; Springer: Vienna, Austria, 2010; pp. 41–51. [Google Scholar]
- Seehabutr, V. Shell Repair by Glandular Cells at the Mantle Edge of Giant African Snail, Achatina fulica. Kamphaengsaen Acad. J. 2008, 6, 40–46. [Google Scholar]
- Chase, R. The function of dart shooting in helicid snails. AM Malacol. Bull. 2007, 23, 183–189. [Google Scholar] [CrossRef]
- Wagge, L.E. The Activity of Amoebocytes and of Alkaline Phosphatases during the Regeneration of the Shell in the Snail, Helix aspersa. Q. J. Micros. Sci. 1951, 3, 307–321. [Google Scholar]
- Denny, M. 10—Molecular Biomechanics of Molluscan Mucous Secretions A2—HOCHACHKA, PETER W. In Metabolic Biochemistry and Molecular Biomechanics; Academic Press: New York, NY, USA, 1983; pp. 431–465. [Google Scholar]
- Biswas, C.; Sinha, D.; Mandal, C. Investigation on interaction of Achatinin, a 9-O-acetyl sialic acid-binding lectin, with lipopolysaccharide in the innate immunity of Achatina fulica snails. Mol. Immunol. 2000, 37, 745–754. [Google Scholar] [CrossRef]
- Vieira, T.C.R.G.; Costa-Filho, A.; Salgado, N.C.; Allodi, S.; Valente, A.-P.; Nasciutti, L.E.; Silva, L.-C.F. Acharan sulfate, the new glycosaminoglycan from Achatina fulica Bowdich 1822. Eur. J. Biochem. 2004, 271, 845–854. [Google Scholar] [CrossRef]
- Pitt, S.J.; Graham, M.A.; Dedi, C.G.; Taylor-Harris, P.M.; Gunn, A. Antimicrobial properties of mucus from the brown garden snail Helix aspersa. Br. J. Biomed. Sci. 2015, 72, 174–181. [Google Scholar] [CrossRef]
- Otsuka-Fuchino, H.; Watanabe, Y.; Hirakawa, C.; Tamiya, T.; Matsumoto, J.J.; Tsuchiya, T. Bactericidal action of a glycoprotein from the body surface mucus of giant African snail. Comp. Biochem. Physiol. C 1992, 101, 607–613. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, W.; Yang, X.; Yan, X.; Liu, R. A novel cysteine-rich antimicrobial peptide from the mucus of the snail of Achatina fulica. Peptides 2013, 39, 1–5. [Google Scholar] [CrossRef]
- Smith, A.M.; Quick, T.J.; St Peter, R.L. Differences in the Composition of Adhesive and Non-Adhesive Mucus from the Limpet Lottia limatula. Biol. Bull. 1999, 196, 34–44. [Google Scholar] [CrossRef]
- Pawlicki, J.M.; Pease, L.B.; Pierce, C.M.; Startz, T.P.; Zhang, Y.; Smith, A.M. The effect of molluscan glue proteins on gel mechanics. J. Exp. Biol. 2004, 207, 1127–1135. [Google Scholar] [CrossRef]
- CAMPION, M. The Structure and Function of the Cutaneous Glands in Helix aspersa. Q. J. Microsc. Sci. 1961, 3, 195–216. [Google Scholar]
- Luchtel, D.L.; Martin, A.W.; Deyrup-Olsen, I. The channel cell of the terrestrial slug Ariolimax columbianus (Stylommatophora, Arionidae). Cell. Tissue. Res. 1984, 235, 143–151. [Google Scholar] [CrossRef]
- Cook, A.; Shirbhate, R. The mucus producing glands and the distribution of the cilia of the pulmonate slug Limax pseudoflavus. J. Zool. 1983, 201, 97–116. [Google Scholar] [CrossRef]
- Bonnemain, B. Helix and Drugs: Snails for Western Health Care from Antiquity to the Present. Evid. Based Complement. Altern. Med. 2005, 2, 25–28. [Google Scholar] [CrossRef]
- Brieva, A.; Philips, N.; Tejedor, R.; Guerrero, A.; Pivel, J.P.; Alonso-Lebrero, J.L.; Gonzalez, S. Molecular Basis for the Regenerative Properties of a Secretion of the Mollusk Cryptomphalus aspersa. Skin. Pharmacol. Physiol. 2008, 21, 15–22. [Google Scholar] [CrossRef]
- E-kobon, T.; Thongararm, P.; Roytrakul, S.; Meesuk, L.; Chumnanpuen, P. Prediction of anticancer peptides against MCF-7 breast cancer cells from the peptidomes of Achatina fulica mucus fractions. CSBJ 2016, 14, 49–57. [Google Scholar] [CrossRef]
- Etim, L.B.; Aleruchi, C.; Obande, G.A. Antibacterial properties of snail mucus on bacteria isolated from patients with wound infection. Br. Microbiol. Res. J. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Li, D.; Graham, L.D. Epidermal secretions of terrestrial flatworms and slugs: Lehmannia valentiana mucus contains matrilin-like proteins. Comp. Biochem. Phys. B Biochem. Mol. Biol. 2007, 148, 231–244. [Google Scholar] [CrossRef]
- Smith, A.M.; Morin, M.C. Biochemical differences between trail mucus and adhesive mucus from marsh periwinkle snails. Biol. Bull. 2002, 203, 338–346. [Google Scholar] [CrossRef]
- Li, D.; Graham, L.D. Epiphragmin, the major protein of epiphragm mucus from the vineyard snail, Cernuella virgata. Comp. Biochem. Phys. B Biochem. Mol. Biol. 2007, 148, 192–200. [Google Scholar] [CrossRef]
- Bulat, T.; Smidak, R.; Sialana, F.J.; Jung, G.; Rattei, T.; Bilban, M.; Sattmann, H.; Lubec, G.; Aradska, J. Transcriptomic and Proteomic Analysis of Arion vulgaris Proteins for Probably Successful Survival Strategies? PLoS ONE 2016, 11, e0150614. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Zhang, H.; Wang, H.; Heras, H.; Dreon, M.S.; Ituarte, S.; Ravasi, T.; Qian, P.-Y.; Qiu, J.-W. First proteome of the egg perivitelline fluid of a freshwater gastropod with aerial oviposition. J. Proteome Res. 2012, 11, 4240–4248. [Google Scholar] [CrossRef]
- Giusti, A.; Leprince, P.; Mazzucchelli, G.; Thomé, J.-P.; Lagadic, L.; Ducrot, V.; Joaquim-Justo, C. Proteomic analysis of the reproductive organs of the hermaphroditic gastropod Lymnaea stagnalis exposed to different endocrine disrupting chemicals. PLoS ONE 2013, 8, e81086. [Google Scholar] [CrossRef]
- Mann, K.; Cerveau, N.; Gummich, M.; Fritz, M.; Mann, M.; Jackson, D.J. In-depth proteomic analyses of Haliotis laevigata (greenlip abalone) nacre and prismatic organic shell matrix. Proteome. Sci. 2018, 16, 11. [Google Scholar] [CrossRef]
- Espinosa, E.P.; Koller, A.; Allam, B. Proteomic characterization of mucosal secretions in the eastern oyster, Crassostrea virginica. J. Proteomics 2016, 132, 63–76. [Google Scholar] [CrossRef]
- Eylers, J. Mucus and Slime: Structure and Rheology of Natural Polysaccharides; Elsevier: Amsterdam, The Netherlands, 2001. [Google Scholar]
- Greistorfer, S.; Klepal, W.; Cyran, N.; Gugumuck, A.; Rudoll, L.; Suppan, J.; von Byern, J. Snail mucus—Glandular origin and composition in Helix pomatia. Zoology 2017, 122, 126–138. [Google Scholar] [CrossRef]
- Joo, N.S.; Evans, I.A.T.; Cho, H.-J.; Park, I.-H.; Engelhardt, J.F.; Wine, J.J. Proteomic analysis of pure human airway gland mucus reveals a large component of protective proteins. PLoS ONE 2015, 10, e0116756. [Google Scholar] [CrossRef]
- Gauri, S.S.; Mandal, S.M.; Pati, B.R.; Dey, S. Purification and structural characterization of a novel antibacterial peptide from Bellamya bengalensis: Activity against ampicillin and chloramphenicol resistant Staphylococcus epidermidis. Peptides 2011, 32, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Sonthi, M.; Cantet, F.; Toubiana, M.; Trapani, M.-R.; Parisi, M.-G.; Cammarata, M.; Roch, P. Gene expression specificity of the mussel antifungal mytimycin (MytM). Fish Shellfish Immun. 2012, 32, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.-F.; Hu, F.-Y.; Wang, B.; Li, T.; Ding, G.-F. Antioxidant and anticancer peptides from the protein hydrolysate of blood clam (Tegillarca granosa) muscle. J. Funct. Foods 2015, 15, 301–313. [Google Scholar] [CrossRef]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Gutiérrez-López, G.F.; Dávila-Ortiz, G. Use of proteomics and peptidomics methods in food bioactive peptide science and engineering. Food Eng. Rev. 2012, 4, 224–243. [Google Scholar] [CrossRef]
- Fields, K.; Falla, T.; Rodan, K.; Bush, L. Bioactive peptides: Signaling the future. J. Cosmet. Dermatol. 2009, 8, 8–13. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic. Acids. Res. 2014, 43, D956–D962. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Lee, B.H.; Oh, D.H. Current perspectives on antihypertensive probiotics. Probiotics Antimicrob. Proteins 2017, 9, 91–101. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and characterization of angiotensin I-converting enzyme inhibitors from sour milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef]
- Abuine, R.; Rathnayake, A.U.; Byun, H.-G. Biological activity of peptides purified from fish skin hydrolysates. Fish Aquat. Sci. 2019, 22, 10. [Google Scholar] [CrossRef]
- Pärn, K.; Eriste, E.; Langel, Ü. The antimicrobial and antiviral applications of cell-penetrating peptides. In Cell-Penetrating Peptides; Springer: Berlin/Heidelberg, Germany, 2015; pp. 223–245. [Google Scholar]
- El-Andaloussi, S.; Holm, T.; Langel, U. Cell-penetrating peptides: Mechanisms and applications. Curr. Pharm. Des. 2005, 11, 3597–3611. [Google Scholar] [CrossRef]
- Lindgren, M.; Hällbrink, M.; Prochiantz, A.; Langel, Ü. Cell-penetrating peptides. Trends Pharmaco. Sci. 2000, 21, 99–103. [Google Scholar] [CrossRef]
- Wang, L.; Dong, C.; Li, X.; Han, W.; Su, X. Anticancer potential of bioactive peptides from animal sources. Oncol. Rep. 2017, 38, 637–651. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Pept. Sci. 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Gwadz, R.; Kaslow, D.; Lee, J.-Y.; Maloy, W.; Zasloff, M.; Miller, L. Effects of magainins and cecropins on the sporogonic development of malaria parasites in mosquitoes. Infect. Immun. 1989, 57, 2628–2633. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Saugar, J.M.; Dellisanti, M.; Barra, D.; Simmaco, M.; Rivas, L. Temporins, small antimicrobial peptides with leishmanicidal activity. J. Biol. Chem. 2005, 280, 984–990. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Lazar, I.; Zwecker-Lazar, I.; Lazar, R.H. Gel Analyzer 2010a: Freeware 1D Gel Electrophoresis Image Analysis Software; ScienceOpen, Inc.: Burlington, MA, USA, 2010. [Google Scholar]
- Johansson, C.; Samskog, J.; Sundström, L.; Wadensten, H.; Björkesten, L.; Flensburg, J. Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data. Proteomics 2006, 6, 4475–4485. [Google Scholar] [CrossRef]
- Thorsell, A.; Portelius, E.; Blennow, K.; Westman-Brinkmalm, A. Evaluation of sample fractionation using micro-scale liquid-phase isoelectric focusing on mass spectrometric identification and quantitation of proteins in a SILAC experiment. Rapid Commun. Mass Spectrom. Int. J. Devoted Rapid Dissem. Minute Res. Mass Spectrom. 2007, 21, 771–778. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophor. Int. J. Res. 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. Mev: Multiexperiment viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. [Google Scholar]
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. klaR analyzing German business cycles. In Data Analysis and Decision Support; Springer: Berlin/Heidelberg, Germany, 2005; pp. 335–343. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. R Package Version 2016, 1, 337–354. [Google Scholar]
- Zhang, L.; Zhang, C.; Gao, R.; Yang, R.; Song, Q. Sequence based prediction of antioxidant proteins using a classifier selection strategy. PLoS ONE 2016, 11, e0163274. [Google Scholar] [CrossRef]
- Wang, C.; Zolotarskaya, O.Y.; Nair, S.S.; Ehrhardt, C.J.; Ohman, D.E.; Wynne, K.J.; Yadavalli, V.K. Real-Time Observation of Antimicrobial Polycation Effects on Escherichia coli: Adapting the Carpet Model for Membrane Disruption to Quaternary Copolyoxetanes. Langmuir 2016, 32, 2975–2984. [Google Scholar] [CrossRef]
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Hamida, J.B.; Fliss, I. BACTIBASE: A new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Holton, T.A.; Pollastri, G.; Shields, D.C.; Mooney, C. CPPpred: Prediction of cell penetrating peptides. Bioinformatics 2013, 29, 3094–3096. [Google Scholar] [CrossRef]
- Novković, M.; Simunić, J.; Bojović, V.; Tossi, A.; Juretić, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. dPABBs: A novel in silico approach for predicting and designing anti-biofilm peptides. Sci. Rep. 2016, 6, 21839. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Qureshi, A.; Thakur, N.; Kumar, M. HIPdb: A database of experimentally validated HIV inhibiting peptides. PLoS ONE 2013, 8, e54908. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Wang, P.; Lin, W.-Z.; Jia, J.-H.; Chou, K.-C. iAMP-2L: A two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013, 436, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Madhu, M.K.; Sharma, A.K.; Sharma, V.K. ProInflam: A webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J. Transl. Med. 2016, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Gupta, A.K.; Kumar, M. Prediction and analysis of quorum sensing peptides based on sequence features. PLoS ONE 2015, 10, e0120066. [Google Scholar] [CrossRef]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2015, 44, D1119–D1126. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.; Consortium, O.S.D.D. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef]
| Sample | Length (cm) | Shell Diameter (cm) | Weight (g) | Location (Latitude, Longitude) |
|---|---|---|---|---|
| Achatina fulica (Heterobranchia) | 7–8 | 7–8 ** | 120–140 | A garden at Faculty of Science, Kasetsart University, Bangkok, Thailand (13.845416, 100572014) |
| Cryptozona siamensis (Heterobranchia) | 5–6 | 3–4 | 30–35 | Sakaerat Environmental Research Station Sakaerat Biosphere Reserves, Nakhon Ratchasima, Thailand (14.510218, 101.930995) |
| Semperula siamensis (Heterobranchia) | 5–6 | - | 15–20 | A tree shop, Patumthani, Thailand (14.918298, 100.9163500) |
| Hemiplecta distincta (Heterobranchia) * | 7–8 | 5–6 | 120–140 | Chet Khot-Pong Kon Sao Nature Study Centre, Saraburi, Thailand (14.493871, 101.161605) |
| Pomacea canaliculata (Caenogastropoda) * | 6–7 | 4–5 | 80–100 | A canal near Faculty of Science, Kasetsart University, Bangkok, Thailand (13.845505, 100.572566) |
| Cyclophorus fulguratus (Caenogastropoda) | 6–7 | 4–5 | 80–100 | Sakaerat Environmental Research Station Sakaerat Biosphere Reserves, Nakhon Ratchasima, Thailand (14.510218, 101.930995) |
| Helix pomatia (Heterobranchia) | 6–7 | 4–5 | 70–90 | A garden in Lisbon, Portugal (38.755233, −9.174415) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tachapuripunya, V.; Roytrakul, S.; Chumnanpuen, P.; E-kobon, T. Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions. Molecules 2021, 26, 3475. https://doi.org/10.3390/molecules26113475
Tachapuripunya V, Roytrakul S, Chumnanpuen P, E-kobon T. Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions. Molecules. 2021; 26(11):3475. https://doi.org/10.3390/molecules26113475
Chicago/Turabian StyleTachapuripunya, Viroj, Sittiruk Roytrakul, Pramote Chumnanpuen, and Teerasak E-kobon. 2021. "Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions" Molecules 26, no. 11: 3475. https://doi.org/10.3390/molecules26113475
APA StyleTachapuripunya, V., Roytrakul, S., Chumnanpuen, P., & E-kobon, T. (2021). Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions. Molecules, 26(11), 3475. https://doi.org/10.3390/molecules26113475








