Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Cytotoxicity Assay
2.3. Aromatase Inhibition Assay
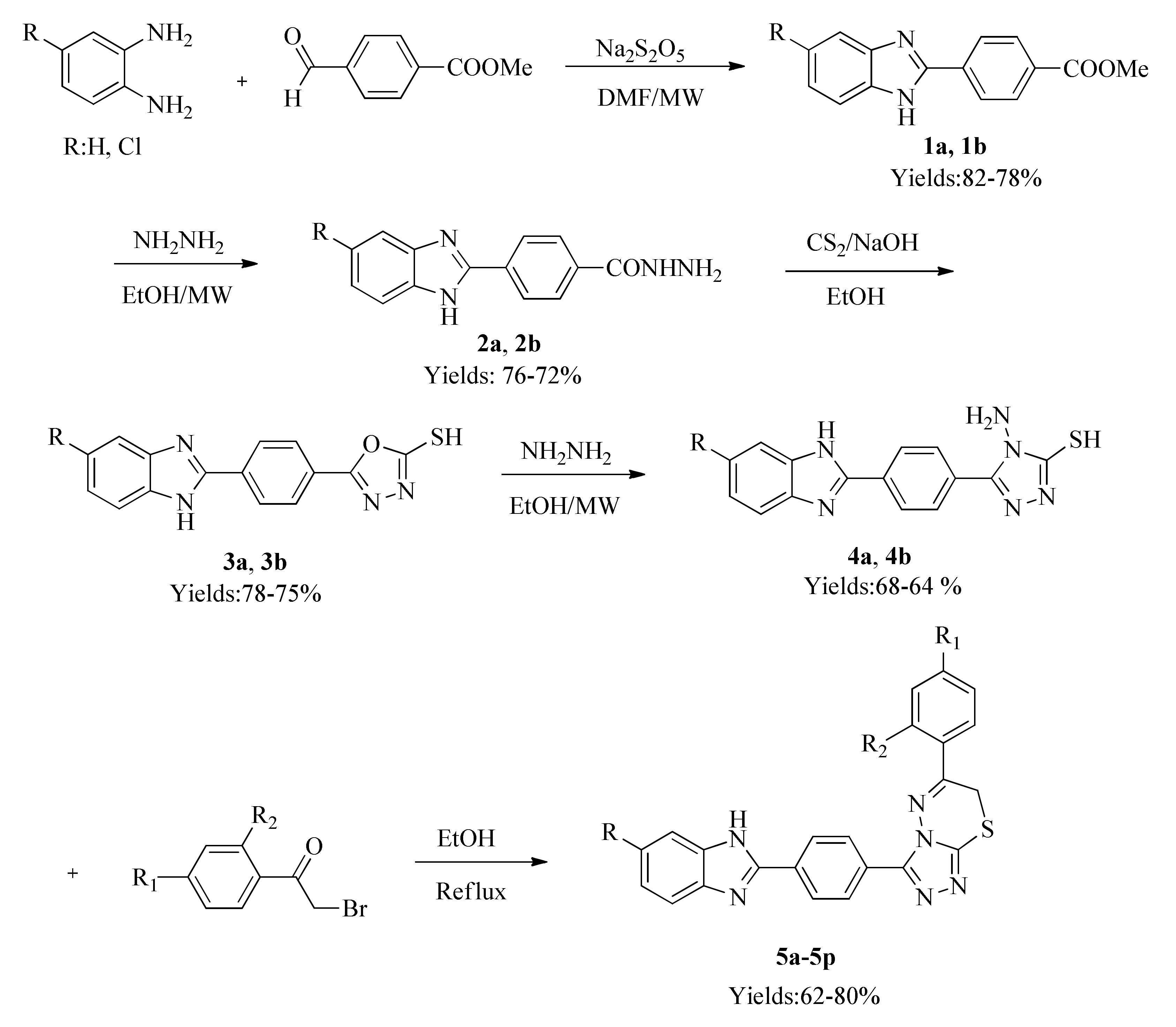
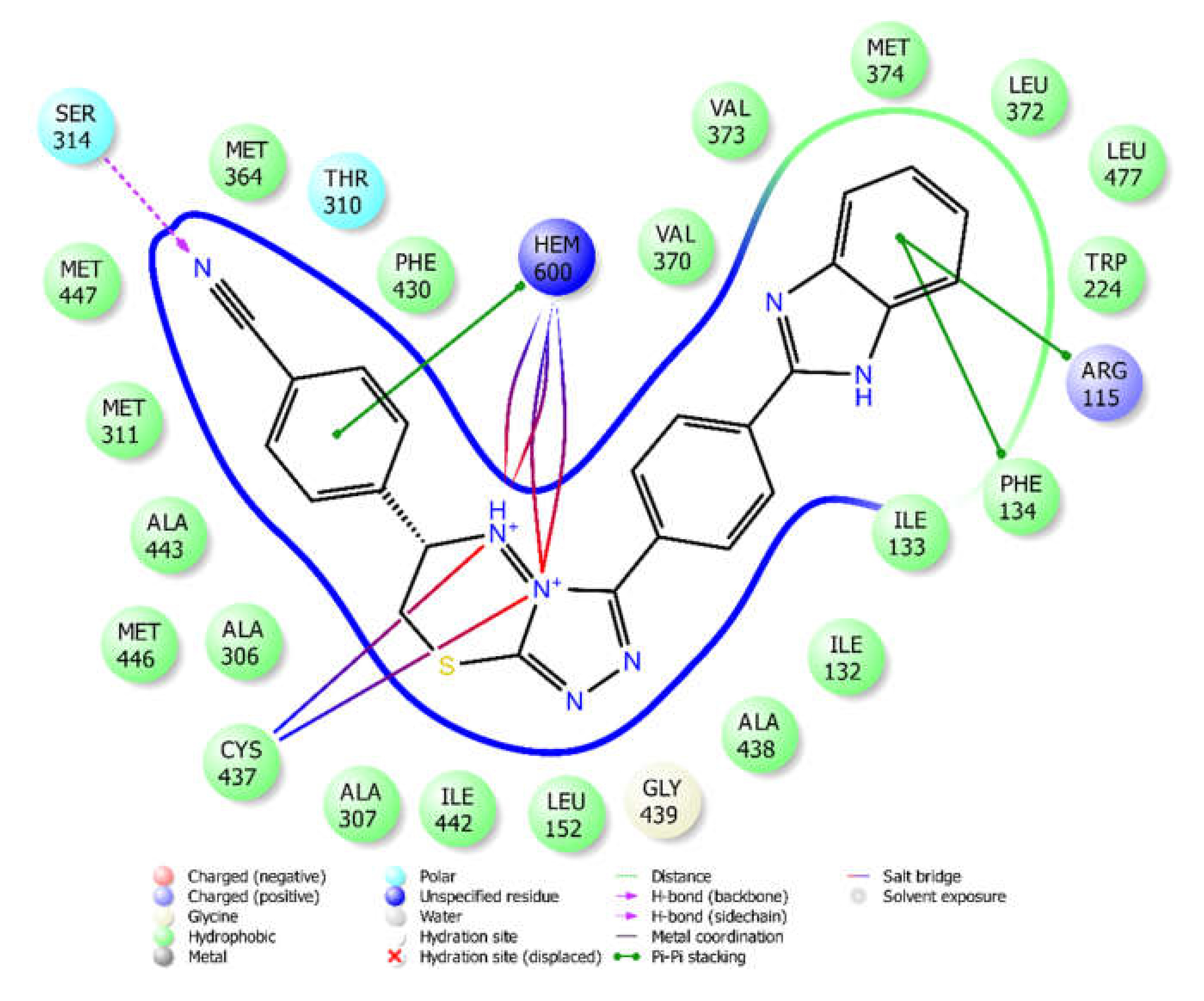
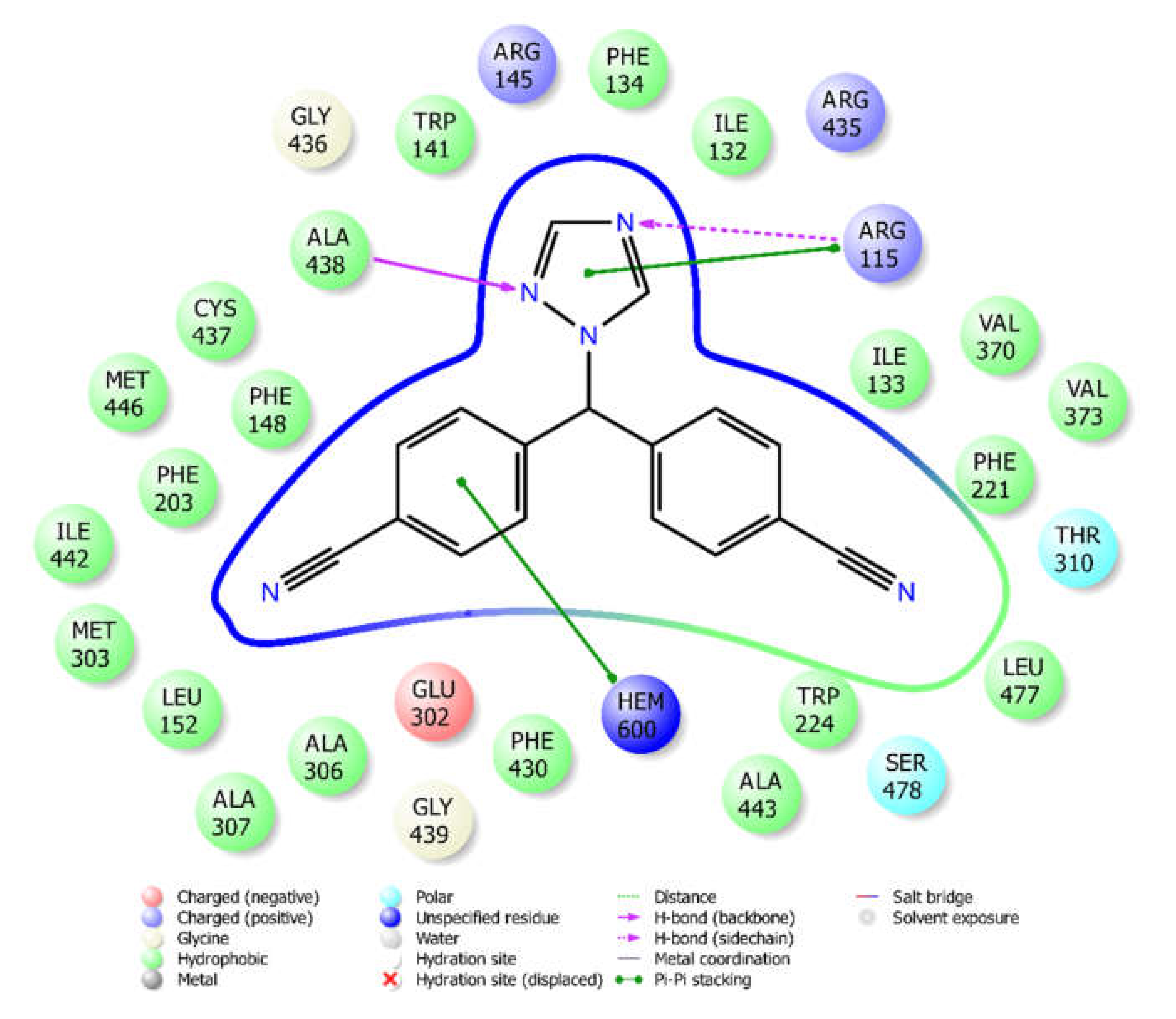
2.4. Molecular Docking
2.5. Theoretical Determination of ADME Properties
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of 4-(5-substitüe-1H-benz[d]imidazol-2-yl)benzoic Acid Methyl Ester (1a,1b)
3.1.2. 4-Amino-5-(4-(5-substitüe-1H-benzo[d]imidazol-2-yl)phenyl)-4H-1,2,4-triazole-3-thiol (4a,4b)
3.1.3. 3-(4-(5-substitüe-1H-benzo[d]imidazol-2-yl)phenyl)-6-(substitüephenyl)-7H-[1,2,4]triazole [3,4-b][1,3,4]thiadiazine (5a–5p)
3.2. Cytotoxicity Assay
3.3. Aromatase Inhibition Assay
3.4. Molecular Docking
3.5. Theoretical Determination of ADME Properties
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chamduang, C.; Pingaew, R.; Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Novel triazole-tetrahydroisoquinoline hybrids as human aromatase inhibitors. Bioorg. Chem. 2019, 93, 103327. [Google Scholar] [CrossRef] [PubMed]
- Doiron, J.; Richard, R.; Touré, M.M.; Picot, N.; Richard, R.; Čuperlović-Culf, M.; Robichaud, G.A.; Touaibia, M. Synthesis and structure–activity relationship of 1-and 2-substituted-1, 2, 3-triazole letrozole-based analogues as aromatase inhibitors. Eur. J. Med. Chem. 2011, 46, 4010–4024. [Google Scholar] [CrossRef] [PubMed]
- Henneberta, O.; Montes, M.; Favre-Reguillon, A.; Chermetted, H.; Ferroudc, C.; Mortin, R. Epimerase activity of the human 11β-hydroxysteroid dehydrogenase type 1 on 7-hydroxylated C19-steroids. J. Steroid Biochem. Mol. Biol. 2009, 114, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Leechaisit, R.; Pingaew, R.; Prachayasittikul, V.; Worachartcheewan, A.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Synthesis, molecular docking, and QSAR study of bis-sulfonamide derivatives as potential aromatase inhibitors. Bioorg. Med. Chem. 2019, 27, 115040. [Google Scholar] [CrossRef]
- Brueggemeier, R.W.; Hackett, J.C.; Diaz-Cruz, E.S. Aromatase inhibitors in the treatment of breast cancer. Endocr. Rev. 2005, 26, 331–345. [Google Scholar] [CrossRef]
- Cepa, M.M.; da Silva, E.J.T.; Correia-da-Silva, G.; Roleira, F.M.; Teixeira, N.A. Synthesis and biochemical studies of 17-substituted androst-3-enes and 3,4-epoxyandrostanes as aromatase inhibitors. Steroids 2008, 73, 1409–1415. [Google Scholar] [CrossRef]
- Bonfield, K.; Amato, E.; Bankemper, T.; Agard, H.; Steller, J.; Keeler, J.M.; Roy, D.; McCallum, A.; Paula, S.; Ma, L. Development of a new class of aromatase inhibitors: Design, synthesis and inhibitory activity of 3-phenylchroman-4-one (isoflavanone) derivatives. Bioorg. Med. Chem. 2012, 20, 2603–2613. [Google Scholar] [CrossRef]
- Rampogu, S.; Baek, A.; Bavi, R.S.; Son, M.; Cao, G.P.; Kumar, R.; Park, C.; Zeb, A.; Rana, R.M.; Park, S.J.; et al. Identification of Novel Scaffolds with Dual Role as Antiepileptic and Anti-Breast Cancer. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 1663–1674. [Google Scholar] [CrossRef]
- Trunet, P.F.; Vreeland, F.; Royce, C.; Chaudri, H.A.; Cooper, J.; Bhatnagar, A.S. Clinical use of aromatase inhibitors in the treatment of advanced breast cancer. J. Steroid. Biochem. Mol. Biol. 1997, 61, 241–245. [Google Scholar] [CrossRef]
- Steele, R.E.; Mellor, L.B.; Sawyer, W.K.; Wasvary, J.M.; Browne, L.J. In vitro and in vivo studies demonstrating potent and selective estrogen inhibition with the nonsteroidal aromatase inhibitor CGS 16949A. Steroids 1987, 50, 147161. [Google Scholar] [CrossRef]
- Kato, S.; Endoh, H.; Masuhiro, Y.; Kitamoto, T.; Uchiyama, S.; Sasaki, H.; Masushige, S.; Gotoh, Y.; Nishida, E.; Kawashima, H.; et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 1995, 270, 1491–1494. [Google Scholar] [CrossRef] [PubMed]
- Sable, P.M.; Potey, L.C. Synthesis and antiproliferative activity of imidazole and triazole derivatives of flavonoids. Pharm. Chem. J. 2018, 52, 438–443. [Google Scholar] [CrossRef]
- Gilardi, G.; Di Nardo, G. Heme iron centers in cytochrome P450: Structure and catalytic activity. Rend. Lincei 2017, 28, 159–167. [Google Scholar] [CrossRef]
- Asadi, P.; Khodarahmi, G.; Farrokhpour, H.; Hassanzadeh, F.; Saghaei, L. Quantum mechanical/molecular mechanical and docking study of the novel analogues based on hybridization of common pharmacophores as potential anti-breast cancer agents. Res. Pharm. Sci. 2017, 12, 233. [Google Scholar]
- Mojaddami, A.; Sakhteman, A.; Fereidoonnezhad, M.; Faghih, Z.; Najdian, A.; Khabnadideh, S.; Rezaei, Z. Binding mode of triazole derivatives as aromatase inhibitors based on docking, protein ligand interaction fingerprinting, and molecular dynamics simulation studies. Res. Pharm. Sci. 2017, 12, 21. [Google Scholar] [CrossRef]
- Song, Z.; Liu, Y.; Dai, Z.; Liu, W.; Zhao, K.; Zhang, T.; Dai, Y. Synthesis and aromatase inhibitory evaluation of 4-N-nitrophenyl substituted amino-4H-1,2,4-triazole derivatives. Bioorg. Med. Chem. 2016, 24, 4723–4730. [Google Scholar] [CrossRef]
- Adhikari, N.; Amin, S.A.; Jha, T.; Gayen, S. Integrating regression and classification-based QSARs with molecular docking analyses to explore the structure-antiaromatase activity relationships of letrozole-based analogs. Can. J. Chem. 2017, 95, 1285–1295. [Google Scholar] [CrossRef]
- Prachayasittikul, V.; Pingaew, R.; Worachartcheewan, A.; Sitthimonchai, S.; Nantasenamat, C.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Aromatase inhibitory activity of 1, 4-naphthoquinone derivatives and QSAR study. EXCLI J. 2017, 16, 714. [Google Scholar]
- Augusto, T.V.; Amaral, C.; Varela, C.L.; Bernardo, F.; da Silva, E.T.; Roleira, F.F.; Costa, S.; Teixeria, N.; Correia-da-Silva, G. Effects of new C6-substituted steroidal aromatase inhibitors in hormone-sensitive breast cancer cells: Cell death mechanisms and modulation of estrogen and androgen receptors. J. Steroid. Biochem. Mol. Biol. 2019, 195, 105486. [Google Scholar] [CrossRef]
- Shoombuatong, W.; Schaduangrat, N.; Nantasenamat, C. Towards understanding aromatase inhibitory activity via QSAR modeling. EXCLI J. 2018, 17, 688. [Google Scholar]
- Prior, A.M.; Yu, X.; Park, E.J.; Kondratyuk, T.P.; Lin, Y.; Pezzuto, J.M.; Sun, D. Structure-activity relationships and docking studies of synthetic 2-arylindole derivatives determined with aromatase and quinone reductase 1. Bioorg. Med. Chem. Lett. 2017, 27, 5393–5399. [Google Scholar] [CrossRef] [PubMed]
- Acar Çevik, U.; Sağlık, B.N.; Osmaniye, D.; Levent, S.; Kaya Çavuşoğlu, B.; Karaduman, A.B.; Ozkay, Y.; Kaplancıklı, Z.A. Synthesis and docking study of benzimidazole–triazolothiadiazine hybrids as aromatase inhibitors. Arch. Pharm. 2020, e2000008. [Google Scholar] [CrossRef] [PubMed]
- Çevik, U.A.; Osmaniye, D.; Çavuşoğlu, B.K.; Sağlik, B.N.; Levent, S.; Ilgin, S.; Ozkay, Y.; Kaplancikli, Z.A. Synthesis of novel benzimidazole–oxadiazole derivatives as potent anticancer activity. Med. Chem. Res. 2019, 28, 2252–2261. [Google Scholar] [CrossRef]
- Evren, A.E.; Yurttaş, L.; Eksellı, B.; Akalın-Cıftcı, G. Novel tri-substituted thiazoles bearing piperazine ring: Synthesis and evaluation of their anticancer activity. Lett. Drug Des. Discov. 2019, 16, 547–555. [Google Scholar] [CrossRef]
- Çevik, U.A.; Sağlık, B.N.; Ardıç, C.M.; Özkay, Y.; Atlı, Ö. Synthesis and evaluation of new benzimidazole derivatives with hydrazone moiety as anticancer agents. Turk. J. Biochem. 2018, 43, 151–158. [Google Scholar] [CrossRef]
- Ghosh, D.; Griswold, J.; Erman, M.; Pangborn, W. Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 2009, 457, 219. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Duffy, E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Maestro, version 10.6; Schrödinger, LLC: New York, NY, USA, 2016.
- The Schrödinger Suite 2016 Update 2; Schrödinger, LLC: New York, NY, USA, 2016.
- LigPrep, version 3.8; Schrödinger, LLC: New York, NY, USA, 2016.
- Glide, version 7.1; Schrödinger, LLC: New York, NY, USA, 2016.
- QikProp, version 4.8; Schrödinger, LLC: New York, NY, USA, 2016.
Sample Availability: Samples of the compounds 5a–5p are available from the authors. |

| Comp. | R | R1 | R2 |
|---|---|---|---|
| 5a | -H | -H | -H |
| 5b | -H | -Cl | -H |
| 5c | -H | -F | -H |
| 5d | -H | -CH3 | -H |
| 5e | -H | -CN | -H |
| 5f | -H | -Br | -H |
| 5g | -H | -F | -F |
| 5h | -H | -Cl | -Cl |
| 5ı | -Cl | -H | -H |
| 5j | -Cl | -Cl | -H |
| 5k | -Cl | -F | -H |
| 5l | -Cl | -CH3 | -H |
| 5m | -Cl | -CN | -H |
| 5n | -Cl | -Br | -H |
| 5o | -Cl | -F | -F |
| 5p | -Cl | -Cl | -Cl |
| Comp. | MCF-7 | NIH3T3 |
|---|---|---|
| 5a | 0.142 ± 0.007 | - |
| 5b | 0.0414 ± 0.001 | - |
| 5c | 0.119 ± 0.005 | 81.8 ± 2.4 |
| 5d | 0.194 ± 0.006 | - |
| 5e | 0.016 ± 0.001 | ≥1000 |
| 5f | 0.318 ± 0.019 | - |
| 5g | 0.245 ± 0.004 | - |
| 5h | 0.342 ± 0.017 | - |
| 5ı | 0.205 ± 0.005 | - |
| 5j | 0.258 ± 0.011 | - |
| 5k | 0.110 ± 0.005 | 77.6 ± 2.1 |
| 5l | 0.352 ± 0.019 | - |
| 5m | 0.018 ± 0.001 | ≥1000 |
| 5n | 0.264 ± 0.011 | - |
| 5o | 0.373 ± 0.011 | - |
| 5p | 0.147 ± 0.001 | - |
| Cisplatin | 0.020 ± 0.009 | ≥1000 |
| Comp. | Aromatase Inhibition |
|---|---|
| 5c | 1.716 ± 0.042 |
| 5e | 0.032 ± 0.001 |
| 5k | 2.276 ± 0.106 |
| 5m | 1.562 ± 0.064 |
| Letrozole | 0.024 ± 0.001 |
| Comp. | MW | RB | DM | MV | DHB | AHB | PSA | logP | logS | PCaco | logBB | PMDCK | PM | CNS | %HOA | VRF | VRT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 408.48 | 0 | 13.539 | 1230.58 | 1 | 4.5 | 66.81 | 5.226 | −7.577 | 1145.907 | −0.361 | 1066.282 | 2 | 0 | 100 | 1 | 1 |
| 5b | 442.925 | 0 | 11.536 | 1274.691 | 1 | 4.5 | 66.813 | 5.722 | −8.323 | 1145.871 | −0.207 | 2630.568 | 2 | 0 | 100 | 1 | 1 |
| 5c | 426.47 | 0 | 11.294 | 1246.689 | 1 | 4.5 | 66.815 | 5.463 | −7.944 | 1145.883 | −0.255 | 1928.038 | 2 | 0 | 100 | 1 | 1 |
| 5d | 422.506 | 0 | 14.175 | 1289.517 | 1 | 4.5 | 66.81 | 5.533 | −8.145 | 1145.907 | −0.388 | 1066.282 | 3 | 0 | 100 | 1 | 1 |
| 5e | 433.489 | 1 | 7.968 | 1297.278 | 1 | 6 | 92.606 | 4.468 | −8.534 | 236.976 | −1.255 | 194.119 | 2 | −2 | 95.607 | 0 | 1 |
| 5f | 487.376 | 0 | 11.913 | 1283.596 | 1 | 4.5 | 66.813 | 5.799 | −8.439 | 1145.858 | −0.198 | 2828.358 | 2 | 0 | 100 | 1 | 1 |
| 5g | 444.461 | 0 | 12.598 | 1251.337 | 1 | 4.5 | 65.595 | 5.544 | −8.02 | 1167.271 | −0.2 | 2431.401 | 2 | 0 | 100 | 1 | 1 |
| 5h | 477.37 | 0 | 12.709 | 1287.743 | 1 | 4.5 | 64.6 | 5.897 | −8.422 | 1199.257 | −0.099 | 3927.141 | 2 | 0 | 100 | 1 | 1 |
| 5i | 442.925 | 0 | 12.609 | 1274.528 | 1 | 4.5 | 66.806 | 5.721 | −8.321 | 1145.207 | −0.208 | 2625.13 | 2 | 0 | 100 | 1 | 1 |
| 5j | 477.37 | 0 | 10.697 | 1318.639 | 1 | 4.5 | 66.809 | 6.217 | −9.068 | 1145.171 | −0.054 | 6476.318 | 2 | 0 | 100 | 1 | 1 |
| 5k | 460.915 | 0 | 10.455 | 1290.637 | 1 | 4.5 | 66.811 | 5.957 | −8.688 | 1145.183 | −0.101 | 4746.726 | 2 | 0 | 100 | 1 | 1 |
| 5l | 456.951 | 0 | 13.229 | 1333.465 | 1 | 4.5 | 66.806 | 6.028 | −8.891 | 1145.207 | −0.235 | 2625.13 | 3 | 0 | 100 | 1 | 1 |
| 5m | 467.934 | 1 | 7.409 | 1341.226 | 1 | 6 | 92.602 | 4.96 | −9.274 | 236.832 | −1.117 | 477.911 | 2 | −2 | 100 | 0 | 1 |
| 5n | 521.821 | 0 | 11.046 | 1327.544 | 1 | 4.5 | 66.809 | 6.294 | −9.184 | 1145.158 | −0.044 | 6963.267 | 2 | 0 | 92.631 | 2 | 1 |
| 5o | 478.906 | 0 | 11.99 | 1295.266 | 1 | 4.5 | 65.591 | 6.038 | −8.764 | 1166.535 | −0.046 | 5985.589 | 2 | 0 | 100 | 1 | 1 |
| 5p | 511.815 | 0 | 12.056 | 1331.676 | 1 | 4.5 | 64.597 | 6.391 | −9.166 | 1198.487 | 0.055 | 9667.877 | 2 | 1 | 93.553 | 2 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acar Çevik, U.; Kaya Çavuşoğlu, B.; Sağlık, B.N.; Osmaniye, D.; Levent, S.; Ilgın, S.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor. Molecules 2020, 25, 1642. https://doi.org/10.3390/molecules25071642
Acar Çevik U, Kaya Çavuşoğlu B, Sağlık BN, Osmaniye D, Levent S, Ilgın S, Özkay Y, Kaplancıklı ZA. Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor. Molecules. 2020; 25(7):1642. https://doi.org/10.3390/molecules25071642
Chicago/Turabian StyleAcar Çevik, Ulviye, Betül Kaya Çavuşoğlu, Begüm Nurpelin Sağlık, Derya Osmaniye, Serkan Levent, Sinem Ilgın, Yusuf Özkay, and Zafer Asım Kaplancıklı. 2020. "Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor" Molecules 25, no. 7: 1642. https://doi.org/10.3390/molecules25071642
APA StyleAcar Çevik, U., Kaya Çavuşoğlu, B., Sağlık, B. N., Osmaniye, D., Levent, S., Ilgın, S., Özkay, Y., & Kaplancıklı, Z. A. (2020). Synthesis, Docking Studies and Biological Activity of New Benzimidazole- Triazolothiadiazine Derivatives as Aromatase Inhibitor. Molecules, 25(7), 1642. https://doi.org/10.3390/molecules25071642






