Biological Evaluation of 3-Benzylidenechromanones and Their Spiropyrazolines-Based Analogues
Abstract
:1. Introduction
2. Results and Discussion
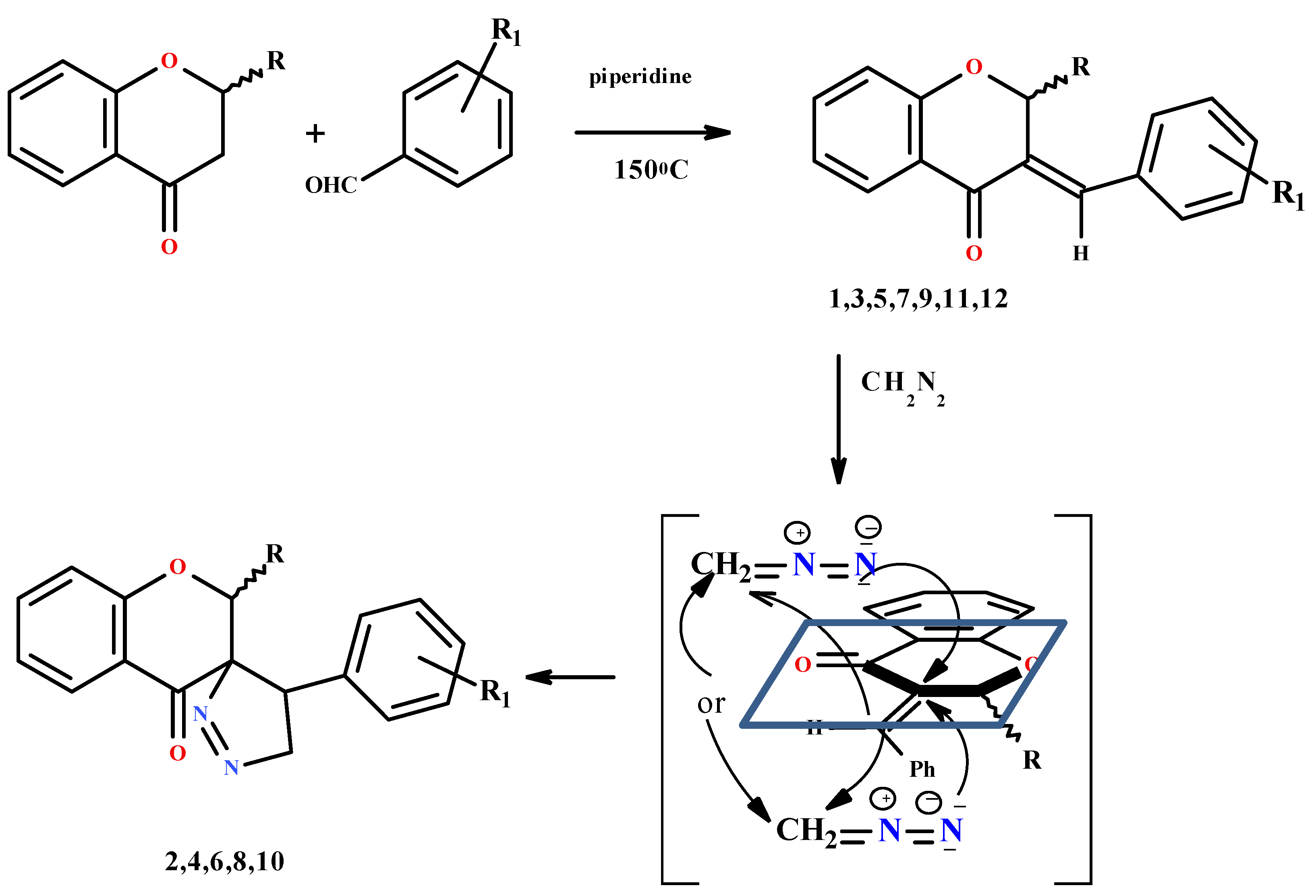
2.1. Chemistry
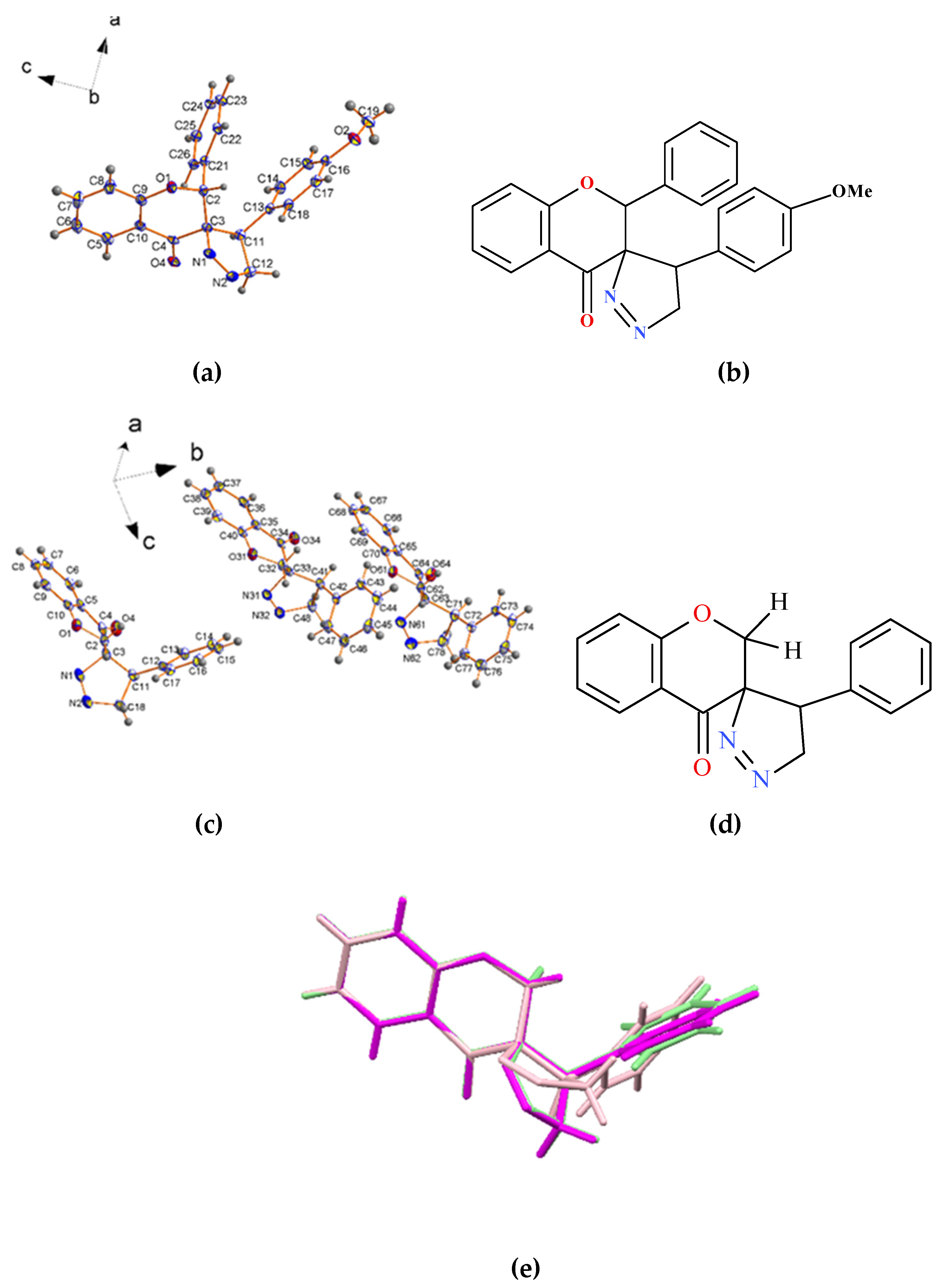
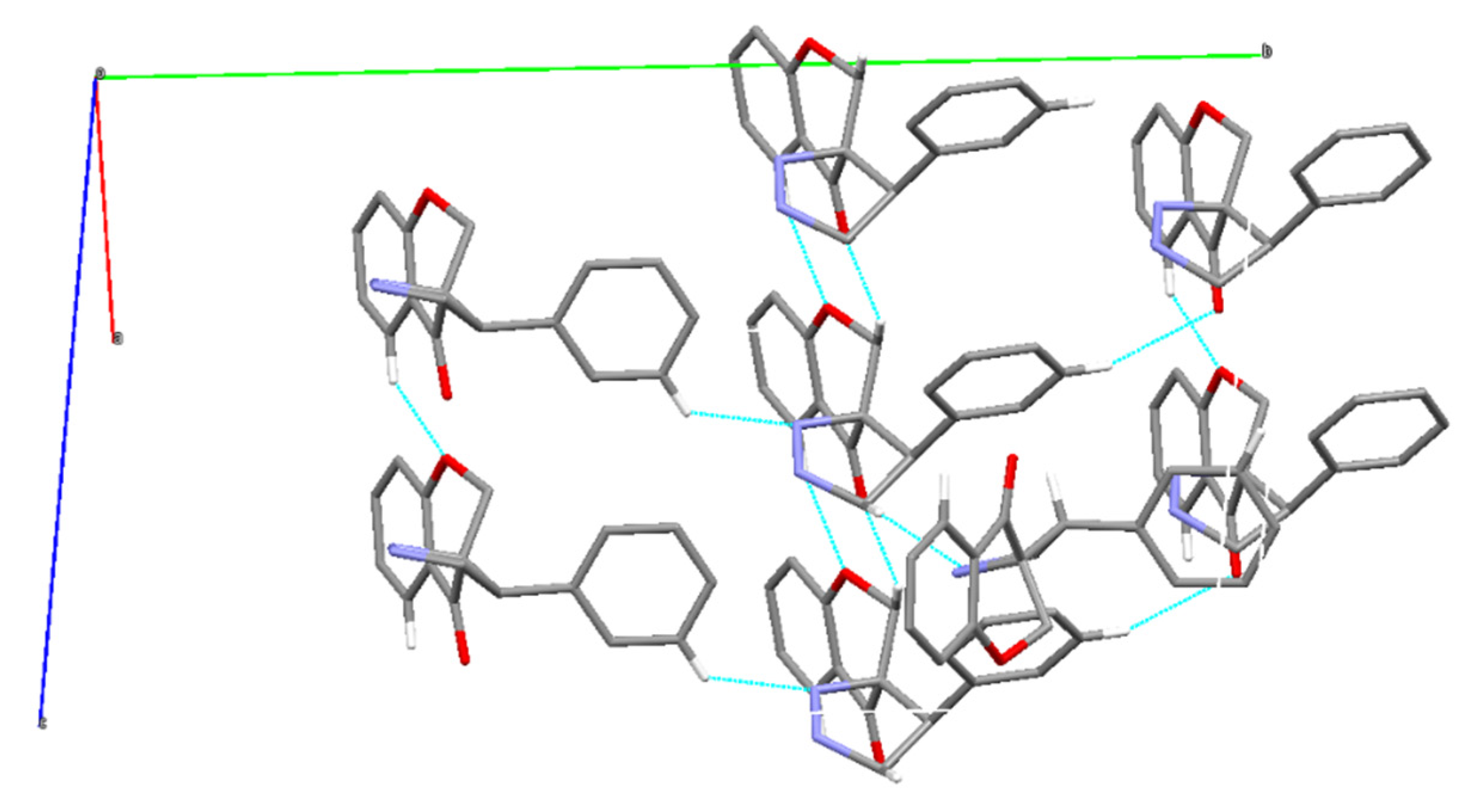
2.2. X-ray Single-Crystal Structure of 2 and 8
2.3. Experimental Lipophilicity of the Synthesized Flavone Analogues
2.4. Cytotoxicity toward Human Cancer Cells
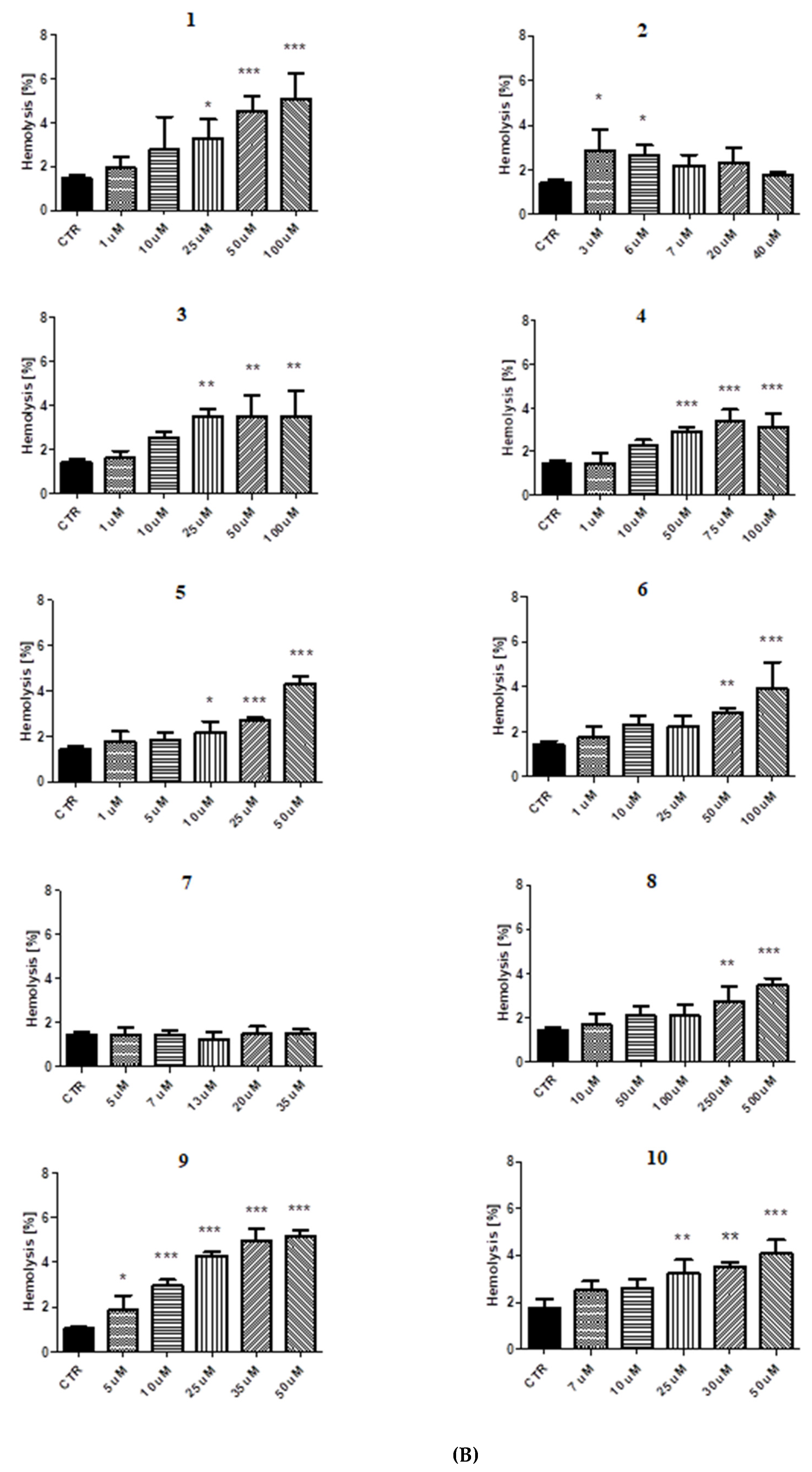
2.5. Red Blood Cell Lysis Assay
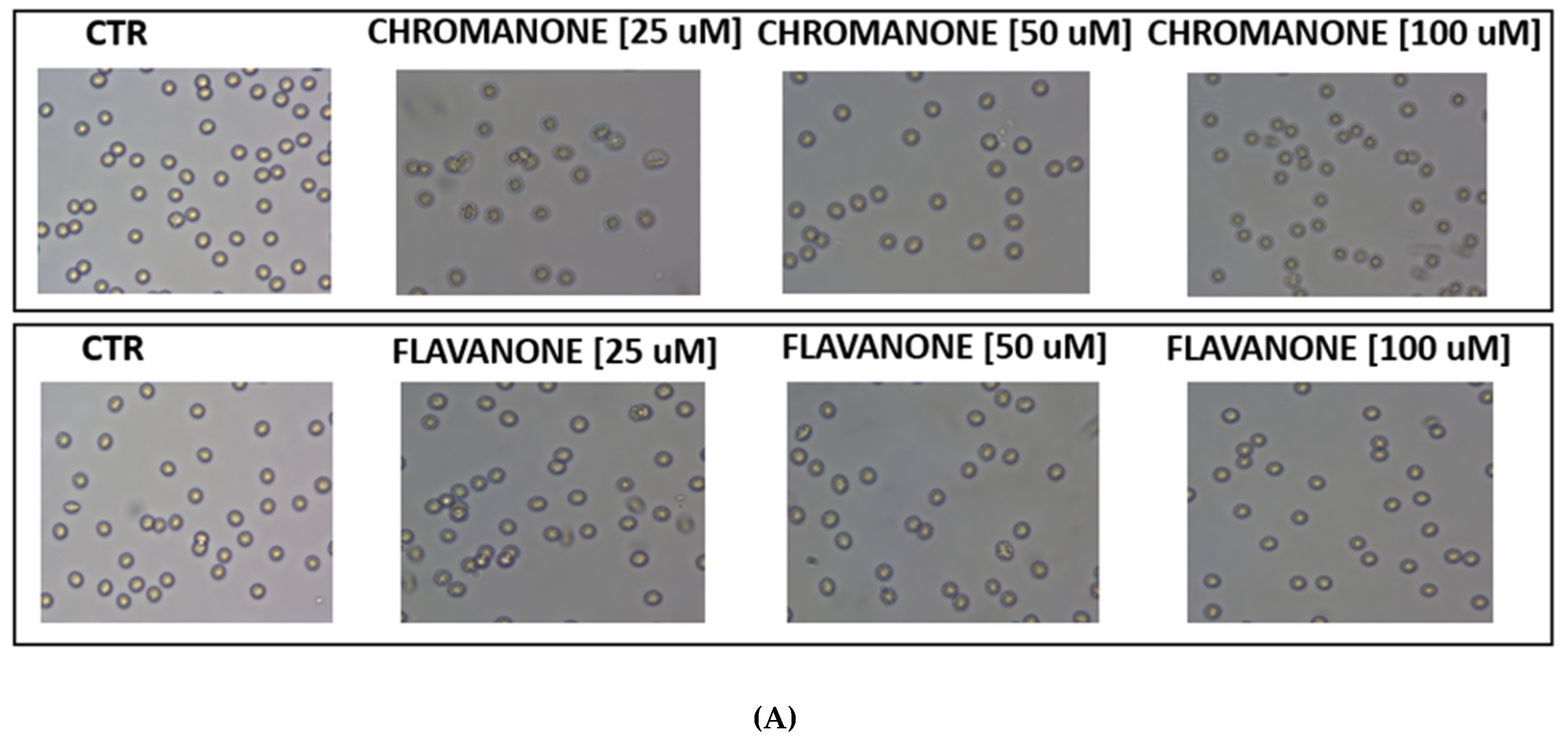
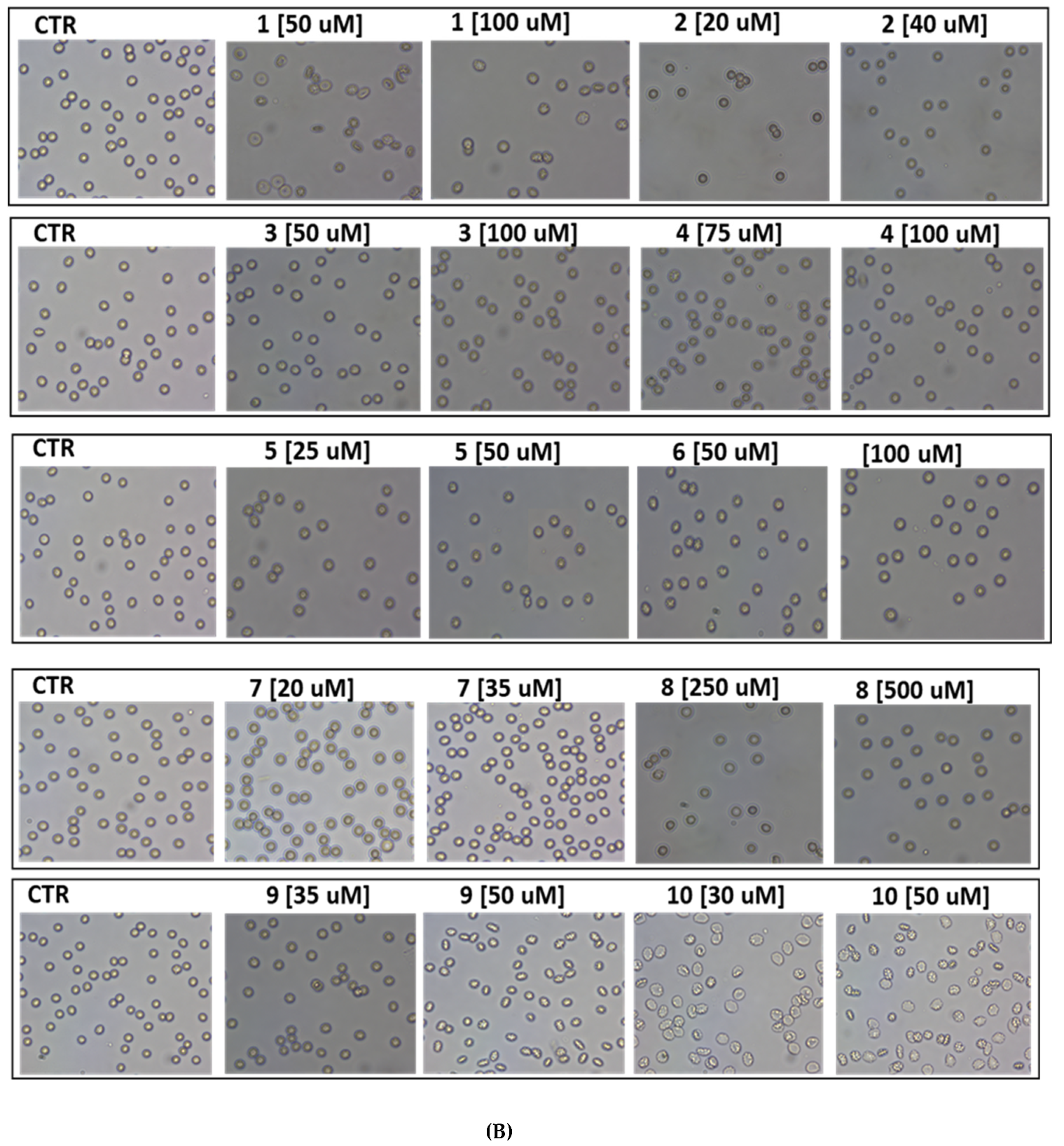
2.6. Red Blood Cell Morphology
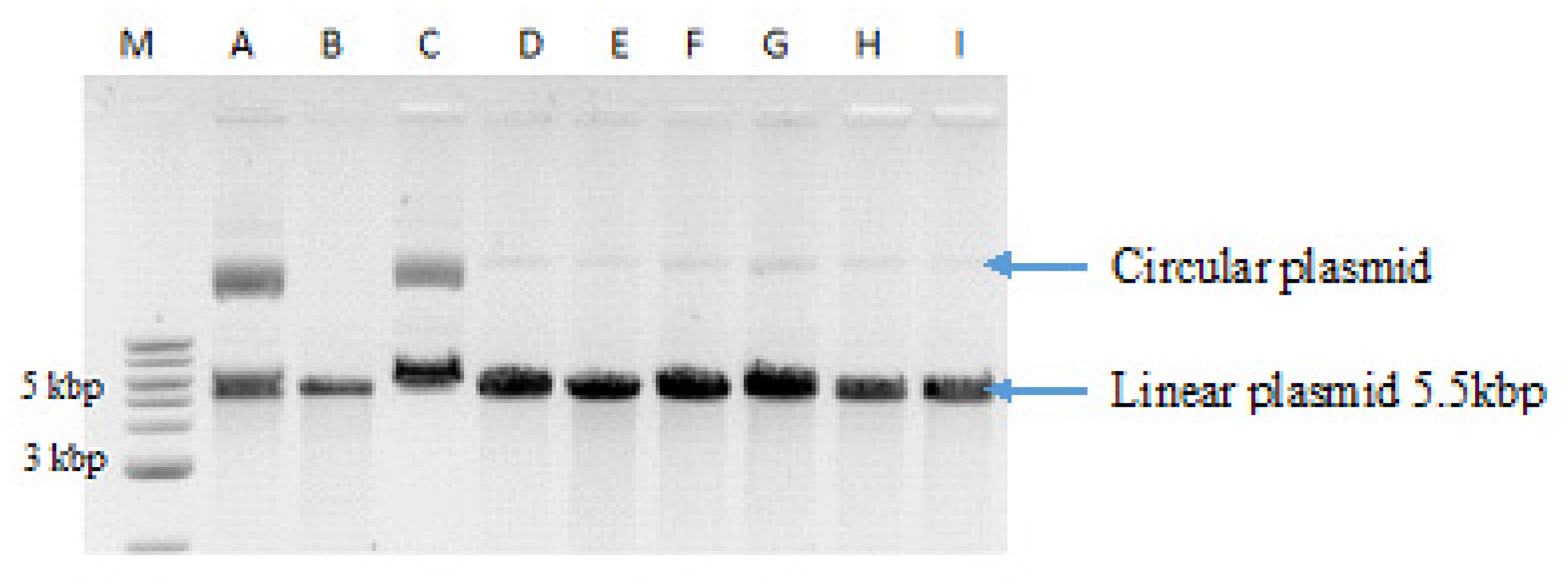
2.7. Digestion of Plasmid DNA with BamHI Restriction Nuclease
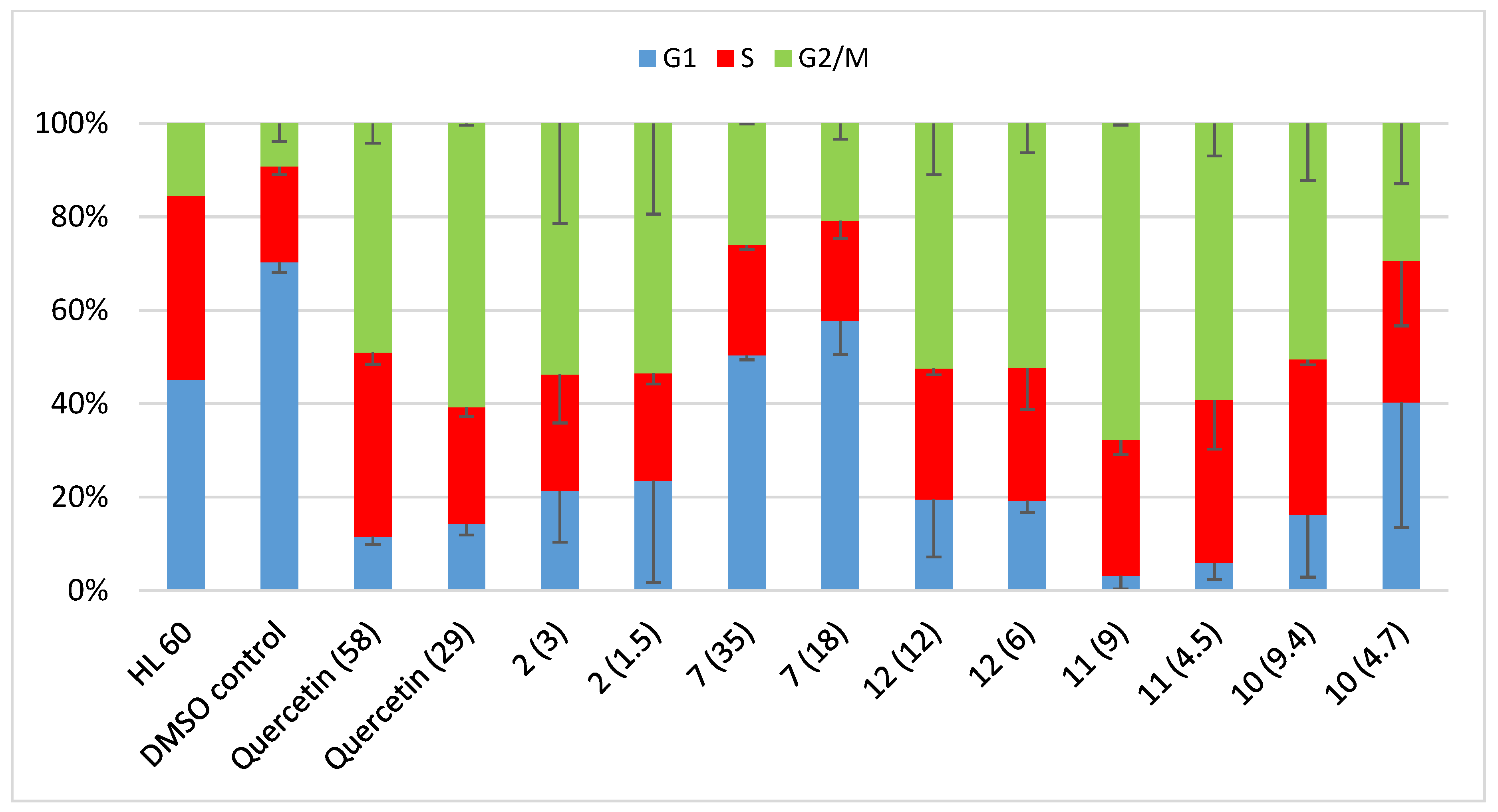
2.8. Cell Cycle Analysis
3. Materials and Methods
3.1. General Information
3.2. Synthesis of (E)-3-(4-methoxybenzylidene)-2-phenylchroman-4-one (1)
3.3. Synthesis of 5′-(4-methoxyphenyl)-2-phenylo -4′,5′-dihydro-4H-spiro [chromano-3,3′-pirazol]-4-one (2)
3.4. Synthesis of (E)-3-(4-methoxybenzylidene)chroman-4-one (3)
3.5. Synthesis of 4′-[(4-methoxy)phenyl]-4′,5′-dihydro-4H-spiro[chroman-3,3′-pirazol]-4-one (4)
3.6. Synthesis of (E)-3-benzylidene-2-phenylchroman-4-one (5)
3.7. Synthesis of 2′-phenylchromanone-3′-spiro-3,4-phenyl-1-pyrazoline (6)
3.8. Synthesis of (E)-3-benzylidene-chroman-4-one (7)
3.9. Synthesis of 4′-phenyl-4′,5′-dihydro-4H-spiro[chromano-3,3′-pirazol]-4-one (8)
3.10. Synthesis of (E)-3-(3-methoxybenzylidene)-2-phenylchroman-4-one (9)
3.11. Synthesis of 5′-(3-methoxyphenyl)-2-phenyl-4′,5′-dihydro-4H-spiro[chromano-3,3′-pyrazol]-4-one (10)
3.12. Determination of Lipophilicity of Flavone Analogues Using a RP-TLC Method
3.13. Cells Cultures and Cytotoxicity Assay by MTT
3.14. Red Blood Cells Lysis Assay
3.15. RBCs Morphology
3.16. Cell Cycle Analysis
3.17. Digestion of Plasmid DNA with BamHI Restriction Nuclease
3.18. X-ray Diffraction Experiment
3.19. Statistical Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marella, A.; Ali, R.; Alam, T.; Saha, R.; Tanwar, O.; Akhter, M.; Alam, M. Pyrazolines: A Biological Review. Mini-Rev. Med. Chem. 2013, 13, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Jain, P. Synthetic and biological studies of prrazolines and related heterocyclic compounds. Arab. J. Chem. 2014, 7, 553–596. [Google Scholar] [CrossRef] [Green Version]
- Amir, M.; Kumar, H.; Khan, S.A. Synthesis and pharmacological evaluation of pyrazoline derivatives as new anti-inflammatory and analgesic agents. Bio-org. Med. Chem. Lett. 2008, 18, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, M.S.; Holla, B.S.; Kumari, N.S. Synthesis and antimicrobial studies on novel chloro-fluorine containing hydroxypyrazolines. Eur. J. Med. Chem. 2007, 42, 30–36. [Google Scholar] [CrossRef]
- Prasad, Y.R.; Rao, A.L.; Prasoona, L.; Murali, K.; Kumar, P.R. Synthesis and antidepressant activity of some 1,3,5-triphenyl-2- pyrazolines and 3-(2”-hydroxynaphthalen-1”-yl)-1,5-diphenyl-2-pyrazolines. Bio-Org. Med. Chem. Lett. 2005, 15, 5030–5034. [Google Scholar] [CrossRef]
- Camacho, M.; Leon, J.; Entrena, A.; Velasco, G.; Carrion, M.; Escames, G.; Vivo, A.; Acuna-Castroviejo, D.; Gallo, M.; Espinosa, A. 4,5-Dihydro-1H-pyrazole Derivatives with Inhibitory nNOS Activity in Rat Brain: Synthesis and Structure−Activity Relationships. J. Med. Chem. 2004, 47, 5641–5650. [Google Scholar] [CrossRef]
- Carrión, M.; Lopez, C.; Tapias, V.; Escames, G.; Acuña-Castroviejo, D.; Espinosa, A.; Gallo, M.; Entrena, A. Pyrazoles and pyrazolines as neural and inducible nitric oxide synthase (nNOS and iNOS) potential inhibitors (III). Eur. J. Med. Chem. 2008, 43, 2579–2591. [Google Scholar] [CrossRef]
- Pijewska, L.; Kamiecki, J.; Perka-Karolczak, W. 3-Arylideneflavanones. Part 2 Reaction with diazomethane. Pharmazie 1993, 48, 254–257. [Google Scholar]
- Toth, G.; Szollosy, A.; Lévai, A.; Kotovych, G. Synthesis and stereochemistry of spiropyrazolines. J. Chem. Soc. Perkin. Trans. II 1986, 12, 1895–1898. [Google Scholar] [CrossRef]
- Adamus-Grabicka, A.; Markowicz-Piasecka, M.; Ponczek, M.; Kusz, J.; Malecka, M.; Krajewska, U.; Budzisz, E. Interaction of Arylidenechromanone/Flavanone Derivatives with Biological Macromolecules Studied as Human Serum Albumin Binding, Cytotoxic Effect, Biocompatibility Towards Red Blood Cells. Molecules 2018, 23, 3172. [Google Scholar] [CrossRef] [Green Version]
- Cremer, D.; Pople, J.A. General definition of ring puckering coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Duax, W.L.; Norton, D.A. Atlas of Steroid Structure; Plenum Press: London, UK, 1975; pp. 3273–3274. [Google Scholar]
- Suchojad, K.; Dołega, A.; Adamus-Grabicka, A.; Budzisz, E.; Małecka, M. Crystal structures of (E)-3-(4-hydroxybenzylidene)chroman-4-one and (E)-3-(3-hydroxybenzylidene)-2-phenylchroman-4-one. Acta Cryst. E. 2019, E75, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Pastuszko, A.; Majchrzak, K.; Czyz, M.; Kupcewicz, B.; Budzisz, E. The synthesis, lipophilicity and cytotoxic effects of new ruthenium(II) arene complexes with chromone derivatives. J. Innorg. Biochem. 2016, 159, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jóźwiak, K.; Szumiło, H.; Soczewiński, E. Lipofilowość, metody wyznaczania i rola w działaniu biologicznym substancji chemicznych. Wiad. Chem. 2001, 55, 1047–1074. [Google Scholar]
- Gao, S.; Cao, C. A New Approach on Estimation of Solubility and n-Octanol/Water Partition Coefficient for Organohalogen Compounds. Int. J. Mol. Sci. 2008, 6, 962–977. [Google Scholar]
- Dearden, J.C. The History and Development of Quantitative Structure-Activity Relationships (QSARs). IJQSPR 2016, 1, 1–44. [Google Scholar] [CrossRef] [Green Version]
- Hoekman, D. Exploring QSAR Fundamentals and Applications in Chemistry and Biology, Volume 1. Hydrophobic, Electronic and Steric Constants. J. Am. Chem. Soc. 1996, 118, 10678. [Google Scholar] [CrossRef] [Green Version]
- Soskic, M.; Magnus, V. Binding of ring-substituted indole-3-acetic acids to human serum albumin. Bioorg. Med. Chem. 2007, 15, 4595–5000. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Gallagher, P.G. Red Blood Cell Membrane Disorders. In Hematology, 7th ed.; Hoffman, R., Benz, E., Heslop, H., Weitz, J., Eds.; Elsevier: Philadelphia, PA, USA, 2018. [Google Scholar]
- Stasiuk, M.; Kijanka, G.; Kozubek, A. Changes in the shape of erythrocytes and the factors causing them. Post. Biochem. 2009, 55, 425–433. [Google Scholar]
- Levai, A.; Schag, J.B. Synthesis of 3-benzylidenechroman-4-ones and -1-thiochroman-4-ones. Pharmazie 1979, 34, 747–749. [Google Scholar]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef]
- CrysAlis PRO; Agilent Technologies: Yarnton, Oxfordshire, UK, 2013.
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Cryst. D. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Van de Streek, J.; Wood, P.A. New Features for the Visualization and Investigation of Crystal Structures. J. Appl. Cryst. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Kupcewicz, B.; Balcerowska-Czerniak, G.; Małecka, M.; Paneth, P.; Krajewska, U.; Rozalski, M. Structure-cytotoxic activity relationship of 3-arylideneflavanone and chromanone (E, Z isomers) and 3-arylflavones. Bioorg. Med. Chem. Lett. 2012, 23, 4102–4106. [Google Scholar] [CrossRef]
- Kupcewicz, B.; Małecka, M.; Zapadka, M.; Krajewska, U.; Rozalski, M.; Budzisz, E. Quantitative relationships between structure and cytotoxic activity of flavonoid derivatives. An application of Hirshfeld surface derived descriptors. Bioorg. Med. Chem. Lett. 2016, 26, 3336–3341. [Google Scholar] [CrossRef]
- Velma, V.; Dasari, S.R.; Tchounwou, P.B. Low Doses of Cisplatin Induce Gene Alterations, Cell Cycle Arrest, and Apoptosis in Human Promyelocytic Leukemia Cells. Biomark. Insights 2016, 24, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Żuryń, A.; Krajewski, A.; Szulc, D.; Litwiniec, .A.; Grzanka, A. Activity of cyclin B1 in HL-60 cells treated with etoposide. Acta Histochem. 2016, 118, 537–543. [Google Scholar] [CrossRef]
- Ghelli, A.; di Rora’, L.; Iacobucci, I.; Martinelli, G. The cell cycle checkpoint inhibitors in thetreatment of leukemia. J. Hematol. Oncol. 2017, 10, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasari, S.R.; Velma, V.; Yedjou, C.G.; Tchounwou, P.B. Preclinical assessment of low doses of cisplatin in the management of acute promyelocytic leukemia. Int. J. Cancer Res. Mol. Mech. 2015, 1, 1–6. [Google Scholar]
- Srivastava, S.; Somasagara, R.; Hedge, M. Quercetin, a Natural Flavonoid Interacts with DNA, Arrests Cell Cycle and Causes Tumor Regression by Activating Mitochondrial Pathway of Apoptosis. Sci. Rep. 2016, 6, 240–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 are available from the author Angelika A. Adamus-Grabicka. |


| Compound/Molecule 5-Membered Ring 6-Membered Ring | Q (Å) | θ (°) | Φ (°) | Asymmetry Parameter (°) | Conformation |
|---|---|---|---|---|---|
| 2 | 0.211(2) | - | 108.9(3) | ΔCs(C11) = 0.2(2) | E |
| 0.411(2) | 51.2(2) | 73.2(2) | ΔCs(C10) = 5.9(2) | E | |
| 8/molecule 1 | 0.307(2) | - | 288.45(4) | ΔCs(C11) = 0.4(2) | E |
| 0.469(2) | 54.0(2) | 83.8(3) | ΔC2(C2-C3) = 9.5(2) | distorted H | |
| molecule 2 | 0.236(2) | - | 108.8(5) | ΔCs(C41) = 0.2(2) | E |
| 0.431(2) | 53.9(2) | 78.6(3) | ΔCs(C32) = 10.5(2) | distorted E | |
| molecule 3 | 0.240(2) | - | 108.8(5) | ΔCs(C61) = 0.1(2) | E |
| 0.414(2) | 53.9(3) | 74.7(3) | ΔCs(C62) = 7.4(2) | distorted E |
| D-H | H…A | D…A | <D-H…A | |
|---|---|---|---|---|
| 2 | ||||
| C12-H12A…O1i | 0.99 | 2.56 | 3.392(2) | 136 |
| C2-H2…O4ii | 1.00 | 2.61 | 3.388(2) | 135 |
| C24-H24…O4iii | 0.95 | 2.53 | 3.239(2) | 131 |
| C26-H26…N2i | 0.95 | 2.74 | 3.416(2) | 129 |
| C11-H11…N1i | 0.85 | 2.67 | 3.653(2) | 180 |
| C11-H11…O4intra | 0.93 | 2.68 | 3.497(2) | 147 |
| 8 | ||||
| Molecule 1 | ||||
| C6-H6…O1iv | 0.95 | 2.59 | 3.304(2) | 132 |
| C14-H14…N31 | 0.95 | 2.60 | 3.316(3) | 133 |
| C11-H11…O4intra | 1.00 | 2.45 | 2.863(2) | 104 |
| Molecule 2 | ||||
| C32-H32A…O34v | 0.99 | 2.47 | 3.373(2) | 152 |
| C36-H36…O31iii | 0.99 | 2.45 | 3.361(2) | 161 |
| C48-H48A…N2vi | 0.99 | 2.61 | 3.473(2) | 146 |
| C44-H44…O64 | 0.95 | 2.53 | 3.418(2) | 157 |
| C41-H41…O34intra | 1.00 | 2.45 | 2.799(2) | 100 |
| Molecule 3 | ||||
| C66-H66…O61iii | 0.95 | 2.57 | 3.281(2) | 132 |
| C71-H71…O64intra | 1.00 | 2.43 | 2.783(2) | 100 |
| Number of Compound | Chemical Structure | IC50 (µM) | ||||
|---|---|---|---|---|---|---|
| HL-60 | NALM-6 | WM-115 | COLO-205 | logP | ||
| 1 | 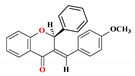 | 53.38 ± 2.49 | 49.81 ± 2.35 | 58.13 ± 4.93 | 51.0 ± 2.3 | 4.79 |
| 2 | 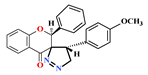 | 3.0 ± 0.3 | 6.8 ± 0.4 | 5.5 ± 0.4 | 29.3 ± 2.35 | 5.42 |
| 3 | 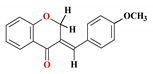 | 45.36 ± 4.77 | 30.07 ± 3.15 | 23.53 ± 1.71 | 193.1 ± 2.9 | 4.08 |
| 4 | 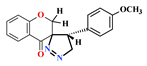 | 74.05 ± 1.85 | 72.60 ± 4.56 | 75.27 ± 4.89 | 26.5 ± 1.8 | 4.68 |
| 5 | 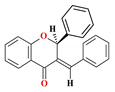 | 33.74 ± 3.87 | 27.02 ± 3.10 | 47.41 ± 4.62 | 77.1 ± 1.9 | 4.90 |
| 6 | 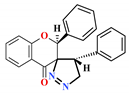 | >1000 | 44.4 ± 6.3 | 57.4 ± 5.8 | 53.6 ± 1.6 | 5.88 |
| 7 |  | 34.78 ± 2.87 | 6.83 ± 0.18 | 12.75 ± 1.83 | 31 ± 1.4 | 4.23 |
| 8 |  | 473.9 ± 29.0 | 281.6 ± 49.4 | 415.0 ± 75.4 | >1000 | 2.35 |
| 9 | 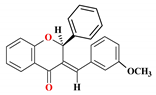 | 35.89 ± 2.96 | 15.7 ± 6.23 | 16.89 ± 3.78 | 169.94 ± 3.81 | 5.83 |
| 10 |  | 9.4 ± 0.4 | 25.0 ± 3.3 | 49.2 ± 2.7 | 487.1 ± 4.1 | 5.42 |
| 11 | 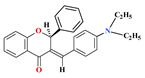 | 8.36 ± 0.63 * | 9.08 ± 0.30 * | 6.45 ± 0.69 * | - | 5.69 |
| 12 | 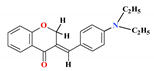 | 11.76 ± 1.97 * | 8.69 ± 0.40 * | 18.09 ± 3.14 * | - | 3.43 |
| 4-Chromanone ** | 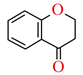 | 676.7 ± 32.6 | 673.7 ± 22.5 | >1000 | 721.5 ± 6.4 | 2.19 |
| Quercetin ** |  | 58.0 ±4.0 | 77.1 ± 7.8 | 177.5 ± 21.5 | - | 2.16 |
| Cisplatin ** | 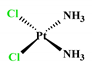 | 0.8 ± 0.1 | 0.7 ± 0.3 | 18.2 ± 4.3 | 61.45 ± 3.2 | −2.21 |
| Carboplatin ** | 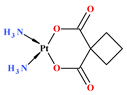 | 4.3 ± 1.3 | 0.7 ± 0.2 | 422.2 ± 50.2 | 354.6 ± 73.7 | −2.30 |
| HL 60 | DMSO Control | Quercetin (58) | Quercetin (29) | 2 (3) | 2 (1.5) | 7 (35) | 7 (18) | 12 (12) | 12 (6) | 11 (4.5) | 11 (9) | 10 (9.4) | 10 (4.7) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 45.2 | 70.3 ± 2.2 | 11.6 ± 1.7 | 14.3 ± 2.4 | 21.3 ± 10.9 | 23.5 ± 21.7 | 50.4 ± 1.0 | 57.7 ± 7.2 | 19.5 ± 12.3 | 19.3 ± 2.6 | 3.2 ± 2.8 | 6 ± 3.5 | 16.2 ± 13.4 | 40.3 ± 26.8 |
| G2 | 39.2 | 20.5 ± 1.7 | 39.4 ± 2.5 | 25.0 ± 2.0 | 25.0 ± 10.4 | 23 ± 2.3 | 23.5 ± 0.9 | 21.5 ± 3.8 | 28.0 ± 1.4 | 28.4 ± 8.8 | 29.1 ± 3.1 | 34.8 ± 10.5 | 33.3 ± 1.1 | 30.2 ± 13.9 |
| G2/M | 15.6 | 9.2 ± 3.9 | 49.0 ± 4.2 | 60.8 ± 0.4 | 53.7 ± 21.4 | 53.5 ± 19.4 | 26.1 ± 0.1 | 20.8 ± 3.4 | 52.4 ± 10.9 | 52.4 ± 6.2 | 67.8 ± 0.3 | 59.2 ± 6.9 | 50.5 ± 12.2 | 29.4 ± 12.9 |
| 2 | 8 | |
|---|---|---|
| Chemical formula | C24H20N2O3 | C17H14N2O2 |
| Formula weight | 384.42 | 278.30 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P21/n | P21/n |
| Temperature (K) | 100.0(1) | 100.0(1) |
| a (Å) | 9.3885(2) | 6.7189(1) |
| b (Å) | 10.6241(2) | 23.6127(5) |
| c (Å) | 19.2202(5) | 25.0637(7) |
| α (°) | 90 | 90 |
| β (°) | 90.768(2) | 95.700(2) |
| γ (°) | 90 | 90 |
| V (Å3) | 1916.93(7) | 3956.73(15) |
| Z | 4 | 12 |
| Radiationtype | Mo Kα | Cu Kα |
| Measuredreflections | 15,186 | 25,640 |
| Uniquereflections | 3965 | 7120 |
| Observedreflections | 3578 | 5196 |
| R[F2 > 2σ(F2)] | 0.0334 | 0.0466 |
| wR(F2) | 0.0874 | 0.1228 |
| S | 1.02 | 1.00 |
| No. of reflections | 2 3965 | 7120 |
| No. of parameters | 263 | 568 |
| No. of restraints | 0 | 0 |
| Δρmax (e Å−3) | 0.33 | 0.22 |
| Δρmin (e Å−3) | −0.20 | −0.25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamus-Grabicka, A.A.; Markowicz-Piasecka, M.; Cieślak, M.; Królewska-Golińska, K.; Hikisz, P.; Kusz, J.; Małecka, M.; Budzisz, E. Biological Evaluation of 3-Benzylidenechromanones and Their Spiropyrazolines-Based Analogues. Molecules 2020, 25, 1613. https://doi.org/10.3390/molecules25071613
Adamus-Grabicka AA, Markowicz-Piasecka M, Cieślak M, Królewska-Golińska K, Hikisz P, Kusz J, Małecka M, Budzisz E. Biological Evaluation of 3-Benzylidenechromanones and Their Spiropyrazolines-Based Analogues. Molecules. 2020; 25(7):1613. https://doi.org/10.3390/molecules25071613
Chicago/Turabian StyleAdamus-Grabicka, Angelika A., Magdalena Markowicz-Piasecka, Marcin Cieślak, Karolina Królewska-Golińska, Paweł Hikisz, Joachim Kusz, Magdalena Małecka, and Elzbieta Budzisz. 2020. "Biological Evaluation of 3-Benzylidenechromanones and Their Spiropyrazolines-Based Analogues" Molecules 25, no. 7: 1613. https://doi.org/10.3390/molecules25071613






