Protein Conformational Dynamics upon Association with the Surfaces of Lipid Membranes and Engineered Nanoparticles: Insights from Electron Paramagnetic Resonance Spectroscopy
Abstract
1. Introduction
2. EPR Spectroscopy of Spin-Labeled Proteins
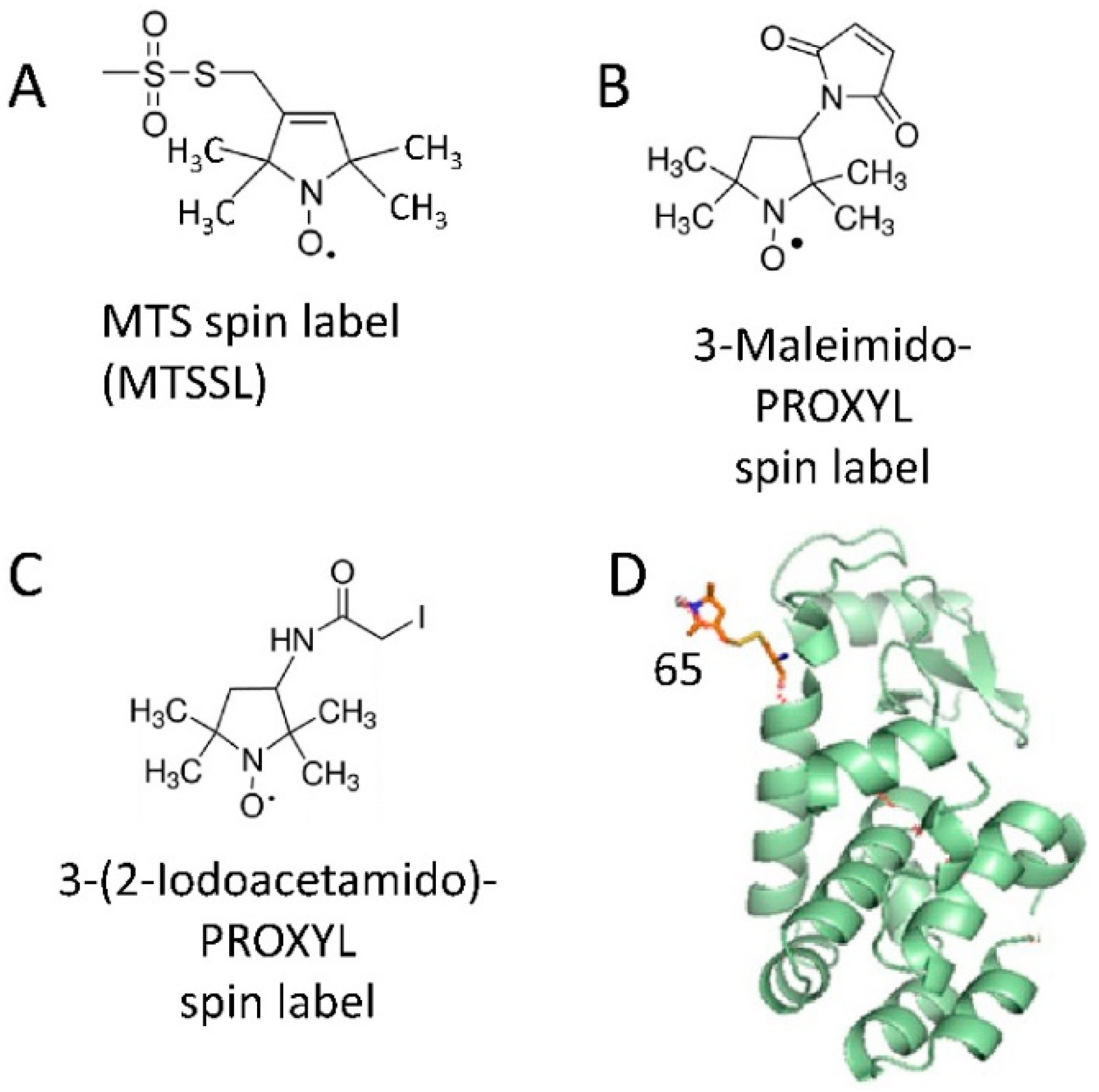
2.1. Spin Labeling of Protein Molecules with Nitroxides
2.2. CW and Pulse EPR Spectroscopy Applied to Spin-Labeled Proteins
3. EPR Studies Explore the Conformational Dynamics upon Interactions of Proteins with Biological and Engineered Surfaces
3.1. Conformational Dynamics Underlying Protein-Lipid Membrane Association
3.2. Conformational Dynamics and Structural Stability Underlying the Immobilization of Proteins on Engineered Surfaces
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Campitelli, P.; Modi, T.; Kumar, S.; Ozkan, S.B. The Role of Conformational Dynamics and Allostery in Modulating Protein Evolution. Annu. Rev. Biophys. 2020, 49, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.H.; Park, J.; Kim, E.; Hohng, S.; Kim, H.S. Protein conformational dynamics dictate the binding affinity for a ligand. Nat. Commun. 2014, 5, 3724. [Google Scholar] [CrossRef] [PubMed]
- Kovermann, M.; Grundstrom, C.; Sauer-Eriksson, A.E.; Sauer, U.H.; Wolf-Watz, M. Structural basis for ligand binding to an enzyme by a conformational selection pathway. Proc. Natl. Acad. Sci. USA 2017, 114, 6298–6303. [Google Scholar] [CrossRef]
- Chakrabarti, K.S.; Agafonov, R.V.; Pontiggia, F.; Otten, R.; Higgins, M.K.; Schertler, G.F.X.; Oprian, D.D.; Kern, D. Conformational Selection in a Protein-Protein Interaction Revealed by Dynamic Pathway Analysis. Cell Rep. 2016, 14, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cano, L.; Eliahoo, E.; Lasker, K.; Wolfson, H.J.; Glaser, F.; Manor, H.; Bernado, P.; Fernandez-Recio, J. Conformational transitions in human translin enable nucleic acid binding. Nucleic. Acids. Res. 2013, 41, 9956–9966. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Ramlall, T.F.; Borbat, P.P.; Freed, J.H.; Eliezer, D. The lipid-binding domain of wild type and mutant alpha-synuclein: Compactness and interconversion between the broken and extended helix forms. J. Biol. Chem. 2010, 285, 28261–28274. [Google Scholar] [CrossRef]
- Jao, C.C.; Der-Sarkissian, A.; Chen, J.; Langen, R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. USA 2004, 101, 8331–8336. [Google Scholar] [CrossRef]
- Tang, S.; Henne, W.M.; Borbat, P.P.; Buchkovich, N.J.; Freed, J.H.; Mao, Y.; Fromme, J.C.; Emr, S.D. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife 2015, 4, e12548. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Ramlall, T.F.; Borbat, P.P.; Freed, J.H.; Eliezer, D. Membrane-bound alpha-synuclein forms an extended helix: Long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 2008, 130, 12856–12857. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Hlady, V.V.; Buijs, J. Protein adsorption on solid surfaces. Curr. Opin. Biotechnol. 1996, 7, 72–77. [Google Scholar] [CrossRef]
- Gutierrez, R.; Valle, E.M.M.d.; Galan, M.A. Immobilized Metal-Ion Affinity Chromatography: Status and Trends. Sep. Purif. Rev. 2007, 36, 71–111. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Shukla, R.J.; Singh, S.P. Structural and catalytic properties of immobilized alpha-amylase from Laceyella sacchari TSI−2. Int. J. Biol. Macromol. 2016, 85, 208–216. [Google Scholar] [CrossRef]
- Kienle, D.F.; Falatach, R.M.; Kaar, J.L.; Schwartz, D.K. Correlating Structural and Functional Heterogeneity of Immobilized Enzymes. ACS Nano 2018, 12, 8091–8103. [Google Scholar] [CrossRef]
- Marcuello, C.; de Miguel, R.; Gomez-Moreno, C.; Martinez-Julvez, M.; Lostao, A. An efficient method for enzyme immobilization evidenced by atomic force microscopy. Protein Eng. Des. Sel. 2012, 25, 715–723. [Google Scholar] [CrossRef]
- Eschmann, N.A.; Georgieva, E.R.; Ganguly, P.; Borbat, P.P.; Rappaport, M.D.; Akdogan, Y.; Freed, J.H.; Shea, J.E.; Han, S. Signature of an aggregation-prone conformation of tau. Sci. Rep. 2017, 7, 44739. [Google Scholar] [CrossRef]
- Hubbell, W.L.; Cafiso, D.S.; Altenbach, C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000, 7, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. [Google Scholar] [CrossRef] [PubMed]
- Klare, J.P.; Steinhoff, H.J. Spin labeling EPR. Photosynth. Res. 2009, 102, 377–390. [Google Scholar] [CrossRef] [PubMed]
- McHaourab, H.S.; Lietzow, M.A.; Hideg, K.; Hubbell, W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry 1996, 35, 7692–7704. [Google Scholar] [CrossRef] [PubMed]
- McHaourab, H.S.; Steed, P.R.; Kazmier, K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure 2011, 19, 1549–1561. [Google Scholar] [CrossRef]
- More, C.; Belle, V.; Asso, M.; Fournel, A.; Roger, G.; Guigliarelli, B.; Bertrand, P. EPR spectroscopy: A powerful technique for the structural and functional investigation of metalloproteins. Biospectroscopy 1999, 5, S3–S18. [Google Scholar] [CrossRef]
- Horitani, M.; Kusubayashi, K.; Oshima, K.; Yato, A.; Sugimoto, H.; Watanabe, K. X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase. Sci. Rep. 2020, 10, 4368. [Google Scholar] [CrossRef]
- Orlando, B.J.; Borbat, P.P.; Georgieva, E.R.; Freed, J.H.; Malkowski, M.G. Pulsed Dipolar Spectroscopy Reveals That Tyrosyl Radicals Are Generated in Both Monomers of the Cyclooxygenase-2 Dimer. Biochemistry 2015, 54, 7309–7312. [Google Scholar] [CrossRef]
- Bennati, M.; Robblee, J.H.; Mugnaini, V.; Stubbe, J.; Freed, J.H.; Borbat, P. EPR distance measurements support a model for long-range radical initiation in E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2005, 127, 15014–15015. [Google Scholar] [CrossRef]
- Yee, E.F.; Diensthuber, R.P.; Vaidya, A.T.; Borbat, P.P.; Engelhard, C.; Freed, J.H.; Bittl, R.; Moglich, A.; Crane, B.R. Signal transduction in light-oxygen-voltage receptors lacking the adduct-forming cysteine residue. Nat. Commun. 2015, 6, 10079. [Google Scholar] [CrossRef]
- Jassoy, J.J.; Heubach, C.A.; Hett, T.; Bernhard, F.; Haege, F.R.; Hagelueken, G.; Schiemann, O. Site Selective and Efficient Spin Labeling of Proteins with a Maleimide-Functionalized Trityl Radical for Pulsed Dipolar EPR Spectroscopy. Molecules 2019, 24, 2735. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; McHaourab, H.S.; Altenbach, C.; Lietzow, M.A. Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [Google Scholar] [CrossRef]
- Klug, C.S.; Feix, J.B. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008, 84, 617–658. [Google Scholar]
- Giannoulis, A.; Yang, Y.; Gong, Y.J.; Tan, X.; Feintuch, A.; Carmieli, R.; Bahrenberg, T.; Liu, Y.; Su, X.C.; Goldfarb, D. DEER distance measurements on trityl/trityl and Gd(iii)/trityl labelled proteins. Phys. Chem. Chem. Phys. 2019, 21, 10217–10227. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Y.; Borbat, P.; Zweier, J.L.; Freed, J.H.; Hubbell, W.L. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J. Am. Chem. Soc. 2012, 134, 9950–9952. [Google Scholar] [CrossRef]
- Yang, Y.; Gong, Y.J.; Litvinov, A.; Liu, H.K.; Yang, F.; Su, X.C.; Goldfarb, D. Generic tags for Mn(ii) and Gd(iii) spin labels for distance measurements in proteins. Phys. Chem. Chem. Phys. 2017, 19, 26944–26956. [Google Scholar] [CrossRef]
- Merz, G.E.; Borbat, P.P.; Pratt, A.J.; Getzoff, E.D.; Freed, J.H.; Crane, B.R. Copper-based pulsed dipolar ESR spectroscopy as a probe of protein conformation linked to disease states. Biophys. J. 2014, 107, 1669–1674. [Google Scholar] [CrossRef]
- Cunningham, T.F.; Putterman, M.R.; Desai, A.; Horne, W.S.; Saxena, S. The double-histidine Cu(2)(+)-binding motif: A highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angew. Chem. Int. Ed. Engl. 2015, 54, 6330–6334. [Google Scholar] [CrossRef]
- Yardeni, E.H.; Bahrenberg, T.; Stein, R.A.; Mishra, S.; Zomot, E.; Graham, B.; Tuck, K.L.; Huber, T.; Bibi, E.; McHaourab, H.S.; et al. Probing the solution structure of the E. coli multidrug transporter MdfA using DEER distance measurements with nitroxide and Gd(III) spin labels. Sci. Rep. 2019, 9, 12528. [Google Scholar] [CrossRef]
- Galazzo, L.; Meier, G.; Timachi, M.H.; Hutter, C.A.J.; Seeger, M.A.; Bordignon, E. Spin-labeled nanobodies as protein conformational reporters for electron paramagnetic resonance in cellular membranes. Proc. Natl. Acad. Sci. USA 2020, 117, 2441–2448. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Borbat, P.P.; Wang, J.; Freed, J.H.; Edmondson, D.E. Determination of the oligomeric states of human and rat monoamine oxidases in the outer mitochondrial membrane and octyl beta-D-glucopyranoside micelles using pulsed dipolar electron spin resonance spectroscopy. Biochemistry 2008, 47, 1554–1566. [Google Scholar] [CrossRef] [PubMed]
- Polyhach, Y.; Bordignon, E.; Jeschke, G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011, 13, 2356–2366. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, C.; Marti, T.; Khorana, H.G.; Hubbell, W.L. Transmembrane protein structure: Spin labeling of bacteriorhodopsin mutants. Science 1990, 248, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, A.; Walko, M.; Hashemi Shabestari, M.; Kumar, P.; Huber, M.; Kocer, A. In situ, Reversible Gating of a Mechanosensitive Ion Channel through Protein-Lipid Interactions. Front. Physiol. 2016, 7, 409. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Xiao, S.; Borbat, P.P.; Freed, J.H.; Eliezer, D. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys. J. 2014, 107, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.S.; Georgieva, E.R.; Borbat, P.P.; Freed, J.H.; Heldwein, E.E. Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat. Struct. Mol. Biol. 2018, 25, 416–424. [Google Scholar] [CrossRef]
- Altenbach, C.; Greenhalgh, D.A.; Khorana, H.G.; Hubbell, W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 1994, 91, 1667–1671. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Borbat, P.P.; Ginter, C.; Freed, J.H.; Boudker, O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 2013, 20, 215–221. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Borbat, P.P.; Norman, H.D.; Freed, J.H. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci. Rep. 2015, 5, 11757. [Google Scholar] [CrossRef]
- Lai, A.L.; Clerico, E.M.; Blackburn, M.E.; Patel, N.A.; Robinson, C.V.; Borbat, P.P.; Freed, J.H.; Gierasch, L.M. Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J. Biol. Chem. 2017, 292, 8773–8785. [Google Scholar] [CrossRef]
- Puljung, M.C.; DeBerg, H.A.; Zagotta, W.N.; Stoll, S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. USA 2014, 111, 9816–9821. [Google Scholar] [CrossRef] [PubMed]
- Freed, J.H. Theory of Slow Tumbling ESR Spectra for Nitroxides. In Spin Labeling: Theory and Applications; Berliner, L.J., Ed.; Elsevier: Amsterdam, The Netherlands, 1976. [Google Scholar]
- Zhang, Z.; Fleissner, M.R.; Tipikin, D.S.; Liang, Z.; Moscicki, J.K.; Earle, K.A.; Hubbell, W.L.; Freed, J.H. Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J. Phys. Chem. B 2010, 114, 5503–5521. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Lorigan, G.A. Site-Directed Spin Labeling EPR for Studying Membrane Proteins. BioMed Res. Int. 2018, 2018, 3248289. [Google Scholar] [CrossRef] [PubMed]
- Budil, D.E.; Lee, S.; Saxena, S.; Freed, J.H. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimentions using a modified Levenberg-Marquard algorithm. J. Magn. Reson. 1996, 120, 155–189. [Google Scholar] [CrossRef]
- Borbat, P.P.; Costa-Filho, A.J.; Earle, K.A.; Moscicki, J.K.; Freed, J.H. Electron spin resonance in studies of membranes and proteins. Science 2001, 291, 266–269. [Google Scholar] [CrossRef]
- Freed, J.H. New technologies in electron spin resonance. Annu. Rev. Phys. Chem. 2000, 51, 655–689. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Lee, J. Active site structure and stability of the thiol protease papain studied by electron paramagnetic resonance employing a methanethiosulfonate spin label. Arch. Biochem. Biophys. 1994, 310, 167–171. [Google Scholar] [CrossRef]
- Altenbach, C.; Froncisz, W.; Hemker, R.; McHaourab, H.; Hubbell, W.L. Accessibility of nitroxide side chains: Absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 2005, 89, 2103–2112. [Google Scholar] [CrossRef]
- Borbat, P.P.; Freed, J.H. Measuring distances by pulsed dipolar ESR spectroscopy: Spin-labeled histidine kinases. Methods Enzymol. 2007, 423, 52–116. [Google Scholar]
- Borbat, P.P.; Freed, J.H. Pros and Cons of Pulse Dipolar ESR: DQC and DEER. EPR Newsl. 2007, 17, 21–33. [Google Scholar]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. 2000. J. Magn. Reson. 2011, 213, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Walti, M.A.; Baber, J.L.; Hustedt, E.J.; Clore, G.M. Long Distance Measurements up to 160 A in the GroEL Tetradecamer Using Q-Band DEER EPR Spectroscopy. Angew. Chem. Int. Ed. Engl. 2016, 55, 15905–15909. [Google Scholar] [CrossRef] [PubMed]
- Borbat, P.P.; Georgieva, E.R.; Freed, J.H. Improved Sensitivity for Long-Distance Measurements in Biomolecules: Five-Pulse Double Electron-Electron Resonance. J. Phys. Chem. Lett. 2013, 4, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Borbat, P.P.; McHaourab, H.S.; Freed, J.H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: Study of T4 lysozyme. J. Am. Chem. Soc. 2002, 124, 5304–5314. [Google Scholar] [CrossRef] [PubMed]
- Ruthstein, S.; Ji, M.; Mehta, P.; Jen-Jacobson, L.; Saxena, S. Sensitive Cu2+-Cu2+ distance measurements in a protein-DNA complex by double-quantum coherence ESR. J. Phys. Chem. B 2013, 117, 6227–6230. [Google Scholar] [CrossRef] [PubMed]
- Reginsson, G.W.; Schiemann, O. Pulsed electron-electron double resonance: Beyond nanometre distance measurements on biomacromolecules. Biochem. J. 2011, 434, 353–363. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Roy, A.S.; Grigoryants, V.M.; Borbat, P.P.; Earle, K.A.; Scholes, C.P.; Freed, J.H. Effect of freezing conditions on distances and their distributions derived from Double Electron Electron Resonance (DEER): A study of doubly-spin-labeled T4 lysozyme. J. Magn. Reson. 2012, 216, 69–77. [Google Scholar] [CrossRef]
- Huang, X.; de Vera, I.M.; Veloro, A.M.; Blackburn, M.E.; Kear, J.L.; Carter, J.D.; Rocca, J.R.; Simmerling, C.; Dunn, B.M.; Fanucci, G.E. Inhibitor-induced conformational shifts and ligand-exchange dynamics for HIV-1 protease measured by pulsed EPR and NMR spectroscopy. J. Phys. Chem. B 2012, 116, 14235–14244. [Google Scholar] [CrossRef]
- Jeschke, G. The contribution of modern EPR to structural biology. Emerg. Top. Life Sci. 2018, 2, 9–18. [Google Scholar]
- Schultz, K.M.; Fischer, M.A.; Noey, E.L.; Klug, C.S. Disruption of the E. coli LptC dimerization interface and characterization of lipopolysaccharide and LptA binding to monomeric LptC. Protein Sci. 2018, 27, 1407–1417. [Google Scholar] [CrossRef]
- Selmke, B.; Borbat, P.P.; Nickolaus, C.; Varadarajan, R.; Freed, J.H.; Trommer, W.E. Open and Closed Form of Maltose Binding Protein in Its Native and Molten Globule State as Studied by Electron Paramagnetic Resonance Spectroscopy. Biochemistry 2018, 57, 5507–5512. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.M.; Hammler, D.; Peter, M.F.; Marx, A.; Schmitz, A.; Hagelueken, G. Inhibitor-Directed Spin Labelling—A High Precision and Minimally Invasive Technique to Study the Conformation of Proteins in Solution. Chemistry 2018, 24, 6665–6671. [Google Scholar] [CrossRef] [PubMed]
- Borbat, P.; Ramlall, T.F.; Freed, J.H.; Eliezer, D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J. Am. Chem. Soc. 2006, 128, 10004–10005. [Google Scholar] [CrossRef] [PubMed]
- Bortolus, M.; Tombolato, F.; Tessari, I.; Bisaglia, M.; Mammi, S.; Bubacco, L.; Ferrarini, A.; Maniero, A.L. Broken helix in vesicle and micelle-bound alpha-synuclein: Insights from site-directed spin labeling-EPR experiments and MD simulations. J. Am. Chem. Soc. 2008, 130, 6690–6691. [Google Scholar] [CrossRef]
- Bordignon, E. Site-directed spin labeling of membrane proteins. Top. Curr. Chem. 2012, 321, 121–157. [Google Scholar]
- Claxton, D.P.; Kazmier, K.; Mishra, S.; McHaourab, H.S. Navigating Membrane Protein Structure, Dynamics, and Energy Landscapes Using Spin Labeling and EPR Spectroscopy. Methods Enzymol. 2015, 564, 349–387. [Google Scholar]
- Joseph, B.; Sikora, A.; Cafiso, D.S. Ligand Induced Conformational Changes of a Membrane Transporter in E. coli Cells Observed with DEER/PELDOR. J. Am. Chem. Soc. 2016, 138, 1844–1847. [Google Scholar] [CrossRef]
- Kumar, P.; van Son, M.; Zheng, T.; Valdink, D.; Raap, J.; Kros, A.; Huber, M. Coiled-coil formation of the membrane-fusion K/E peptides viewed by electron paramagnetic resonance. PLoS ONE 2018, 13, e0191197. [Google Scholar] [CrossRef]
- Meyer, V.; Swanson, M.A.; Clouston, L.J.; Boratynski, P.J.; Stein, R.A.; McHaourab, H.S.; Rajca, A.; Eaton, S.S.; Eaton, G.R. Room-temperature distance measurements of immobilized spin-labeled protein by DEER/PELDOR. Biophys. J. 2015, 108, 1213–1219. [Google Scholar] [CrossRef][Green Version]
- Snead, D.; Lai, A.L.; Wragg, R.T.; Parisotto, D.A.; Ramlall, T.F.; Dittman, J.S.; Freed, J.H.; Eliezer, D. Unique Structural Features of Membrane-Bound C-Terminal Domain Motifs Modulate Complexin Inhibitory Function. Front. Mol. Neurosci. 2017, 10, 154. [Google Scholar] [CrossRef]
- Tao, M.; Pandey, N.K.; Barnes, R.; Han, S.; Langen, R. Structure of Membrane-Bound Huntingtin Exon 1 Reveals Membrane Interaction and Aggregation Mechanisms. Structure 2019, 27, 1570–1580.e4. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Isas, J.M.; Langen, R. Annexin B12 Trimer Formation is Governed by a Network of Protein-Protein and Protein-Lipid Interactions. Sci. Rep. 2020, 10, 5301. [Google Scholar] [CrossRef]
- Bates, I.R.; Feix, J.B.; Boggs, J.M.; Harauz, G. An immunodominant epitope of myelin basic protein is an amphipathic alpha-helix. J. Biol. Chem. 2004, 279, 5757–5764. [Google Scholar] [CrossRef] [PubMed]
- Rawat, A.; Langen, R.; Varkey, J. Membranes as modulators of amyloid protein misfolding and target of toxicity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1863–1875. [Google Scholar] [CrossRef]
- Rawat, A.; Maity, B.K.; Chandra, B.; Maiti, S. Aggregation-induced conformation changes dictate islet amyloid polypeptide (IAPP) membrane affinity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1734–1740. [Google Scholar] [CrossRef]
- Pan, Y.; Neupane, S.; Farmakes, J.; Bridges, M.; Froberg, J.; Rao, J.; Qian, S.Y.; Liu, G.; Choi, Y.; Yang, Z. Probing the structural basis and adsorption mechanism of an enzyme on nano-sized protein carriers. Nanoscale 2017, 9, 3512–3523. [Google Scholar] [CrossRef]
- Jahnke, J.P.; Idso, M.N.; Hussain, S.; Junk, M.J.N.; Fisher, J.M.; Phan, D.D.; Han, S.; Chmelka, B.F. Functionally Active Membrane Proteins Incorporated in Mesostructured Silica Films. J. Am. Chem. Soc. 2018, 140, 3892–3906. [Google Scholar] [CrossRef]
- Berliner, L.J.; Miller, S.T.; Uy, R.; Royer, G.P. An ESR study of the active-site conformations of free and immobilized trypsin. Biochim. Biophys. Acta 1973, 315, 195–199. [Google Scholar] [CrossRef]
- Clark, D.S.; Bailey, J.E. Characterization of heterogeneous immobilized enzyme subpopulations using EPR spectroscopy. Biotechnol. Bioeng. 1984, 26, 231–238. [Google Scholar] [CrossRef]
- Skerker, P.S.; Miller, R.R.; Millhauser, G.L.; Clark, D.S. Structural and functional responses of enzymes to immobilization: New insights from EPR spectroscopy. Ann. N. Y. Acad. Sci. 1987, 501, 80–84. [Google Scholar] [CrossRef]
- Marg, G.A.; Millhauser, G.L.; Skerker, P.S.; Clark, D.S. Aplication of EPR method in studies of immobilized enzyme systems. Ann. N. Y. Acad. Sci. 1987, 469, 253–258. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, K.C.; Bailey, J.E. ESR investigations of free and immobilized glutamate dehydrogenase. Biotechnol. Bioeng. 1989, 34, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Snead, D.; Eliezer, D. Intrinsically disordered proteins in synaptic vesicle trafficking and release. J. Biol. Chem. 2019, 294, 3325–3342. [Google Scholar] [CrossRef] [PubMed]
- Sulzer, D.; Edwards, R.H. The physiological role of alpha-synuclein and its relationship to Parkinson’s Disease. J. Neurochem. 2019, 150, 475–486. [Google Scholar] [CrossRef]
- Delenclos, M.; Burgess, J.D.; Lamprokostopoulou, A.; Outeiro, T.F.; Vekrellis, K.; McLean, P.J. Cellular models of alpha-synuclein toxicity and aggregation. J. Neurochem. 2019, 150, 566–576. [Google Scholar] [CrossRef]
- Fauvet, B.; Mbefo, M.K.; Fares, M.B.; Desobry, C.; Michael, S.; Ardah, M.T.; Tsika, E.; Coune, P.; Prudent, M.; Lion, N.; et al. Alpha-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 2012, 287, 15345–15364. [Google Scholar] [CrossRef]
- Eliezer, D.; Kutluay, E.; Bussell, R., Jr.; Browne, G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001, 307, 1061–1073. [Google Scholar] [CrossRef]
- Maiorano, J.N.; Davidson, W.S. The orientation of helix 4 in apolipoprotein A-I-containing reconstituted high density lipoproteins. J. Biol. Chem. 2000, 275, 17374–17380. [Google Scholar] [CrossRef]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Weickert, S.; Cattani, J.; Drescher, M. Intrinsically disordered proteins (IDPs) studied by EPR and in-cell EPR. In Electron Paramagnetic Resonance; Chechik, V., Murphy, D.M., Eds.; Royal Society of Chemistry: London, UK, 2018; Volume 26, pp. 1–37. [Google Scholar]
- Jao, C.C.; Hegde, B.G.; Chen, J.; Haworth, I.S.; Langen, R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA 2008, 105, 19666–19671. [Google Scholar] [CrossRef]
- Drescher, M.; van Rooijen, B.D.; Veldhuis, G.; Subramaniam, V.; Huber, M. A stable lipid-induced aggregate of alpha-synuclein. J. Am. Chem. Soc. 2010, 132, 4080–4082. [Google Scholar] [CrossRef] [PubMed]
- Alza, N.P.; Iglesias Gonzalez, P.A.; Conde, M.A.; Uranga, R.M.; Salvador, G.A. Lipids at the Crossroad of alpha-Synuclein Function and Dysfunction: Biological and Pathological Implications. Front. Cell Neurosci. 2019, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, C.G.; Walter, E.D.; Abrams, B.S.; Jurica, M.S.; Millhauser, G.L. Coordination of copper to the membrane-bound form of alpha-synuclein. Biochemistry 2013, 52, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Deligiannakis, Y.; Louloudi, M.; Hadjiliadis, N. Electron spin echo envelope modulation (ESEEM) spectroscopy as a tool to investigate the coordination environment of metal centers. Coord. Chem. Rev. 2000, 204, 1–112. [Google Scholar] [CrossRef]
- Cleveland, D.W.; Hwo, S.Y.; Kirschner, M.W. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J. Mol. Biol. 1977, 116, 207–225. [Google Scholar] [CrossRef]
- Cleveland, D.W.; Hwo, S.Y.; Kirschner, M.W. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J. Mol. Biol. 1977, 116, 227–247. [Google Scholar] [CrossRef]
- Honson, N.S.; Kuret, J. Tau aggregation and toxicity in tauopathic neurodegenerative diseases. J. Alzheimers Dis. 2008, 14, 417–422. [Google Scholar] [CrossRef]
- Brandt, R.; Leger, J.; Lee, G. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J. Cell Biol. 1995, 131, 1327–1340. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Ramlall, T.; Rhoades, E. The role of the lipid bilayer in tau aggregation. Biophys. J. 2010, 98, 2722–2730. [Google Scholar] [CrossRef]
- Mena, R.; Edwards, P.C.; Harrington, C.R.; Mukaetova-Ladinska, E.B.; Wischik, C.M. Staging the pathological assembly of truncated tau protein into paired helical filaments in Alzheimer’s disease. Acta Neuropathol. 1996, 91, 633–641. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Eliezer, D.; Barre, P.; Kobaslija, M.; Chan, D.; Li, X.; Heend, L. Residual structure in the repeat domain of tau: Echoes of microtubule binding and paired helical filament formation. Biochemistry 2005, 44, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Barre, P.; Eliezer, D. Folding of the repeat domain of tau upon binding to lipid surfaces. J. Mol. Biol. 2006, 362, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Barre, P.; Eliezer, D. Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci. 2013, 22, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef] [PubMed]
- Bleicken, S.; Classen, M.; Padmavathi, P.V.; Ishikawa, T.; Zeth, K.; Steinhoff, H.J.; Bordignon, E. Molecular details of Bax activation, oligomerization, and membrane insertion. J. Biol. Chem. 2010, 285, 6636–6647. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Zhao, Y.; Emr, S.D. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 2012, 151, 356–371. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Hurley, J.H.; Hanson, P.I. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 2010, 11, 556–566. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Lee, J.B.; Glauner, K.; Ganapathi, S.; Bhattacharyya, D.; Butterfield, D.A. Biofunctional membranes: An EPR study of active site structure and stability of papain non-covalently immobilized on the surface of modified poly(ether) sulfone membranes through the avidin-biotin linkage. J. Membr. Sci. 1996, 119, 241–252. [Google Scholar] [CrossRef]
- Ballet, T.; Boulange, L.; Brechet, Y.; Bruckert, F.; Weidenhaupt, M. Protein conformational changes induced by adsorption onto material surfaces: An important issue for biomedical applications of material science. Bull. Pol. Acad. Sci. 2010, 58, 303–315. [Google Scholar] [CrossRef]
- Dee, K.C.; Puleo, D.A.; Bizios, R. Protein-Surface Interactions. In An Introduction to Tissue-Biomaterial Interactions: Tissue-Biomaterial; Dee, K.C., Puleo, D.A., Bizios, R., Eds.; Wiley-Liss, Inc.: New York, NY, USA, 2002; pp. 37–52. [Google Scholar]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszynska, D. Immobilization as a strategy for improving enzyme properties-application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.S.; Skerker, P.S.; Fernandez, E.J.; Jagoda, R.B. Spectroscopic studies of structure-function relationships in free and immobilized alcohol dehydrogenase. Ann. N. Y. Acad. Sci. 1987, 506, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Lee, J.; Ganapathi, S.; Bhattacharyya, D. Biofunctional membranes 4. Active-site structure and stability of an immobilized enzyme, papain, on modified polysulfone membranes studied by electron-paramagnetic-resonance and kinetics. J. Membr. Sci. 1994, 91, 47–64. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva, E.R. Protein Conformational Dynamics upon Association with the Surfaces of Lipid Membranes and Engineered Nanoparticles: Insights from Electron Paramagnetic Resonance Spectroscopy. Molecules 2020, 25, 5393. https://doi.org/10.3390/molecules25225393
Georgieva ER. Protein Conformational Dynamics upon Association with the Surfaces of Lipid Membranes and Engineered Nanoparticles: Insights from Electron Paramagnetic Resonance Spectroscopy. Molecules. 2020; 25(22):5393. https://doi.org/10.3390/molecules25225393
Chicago/Turabian StyleGeorgieva, Elka R. 2020. "Protein Conformational Dynamics upon Association with the Surfaces of Lipid Membranes and Engineered Nanoparticles: Insights from Electron Paramagnetic Resonance Spectroscopy" Molecules 25, no. 22: 5393. https://doi.org/10.3390/molecules25225393
APA StyleGeorgieva, E. R. (2020). Protein Conformational Dynamics upon Association with the Surfaces of Lipid Membranes and Engineered Nanoparticles: Insights from Electron Paramagnetic Resonance Spectroscopy. Molecules, 25(22), 5393. https://doi.org/10.3390/molecules25225393




