Dynamic Structure and Stability of DNA Duplexes Bearing a Dinuclear Hg(II)-Mediated Base Pair
Abstract
1. Introduction
2. Computational Details
2.1. Static DFT Calculations
2.2. Topology Generation for Classical Molecular Dynamics
2.3. Classical Molecular Dynamics
2.4. Ab Initio Molecular Dynamics and Optimization
2.5. QM/MM Simulations
2.6. Free Energy Calculations
2.6.1. Dynamic Distance Constraint
2.6.2. Thermodynamic Integration
3. Model Structures
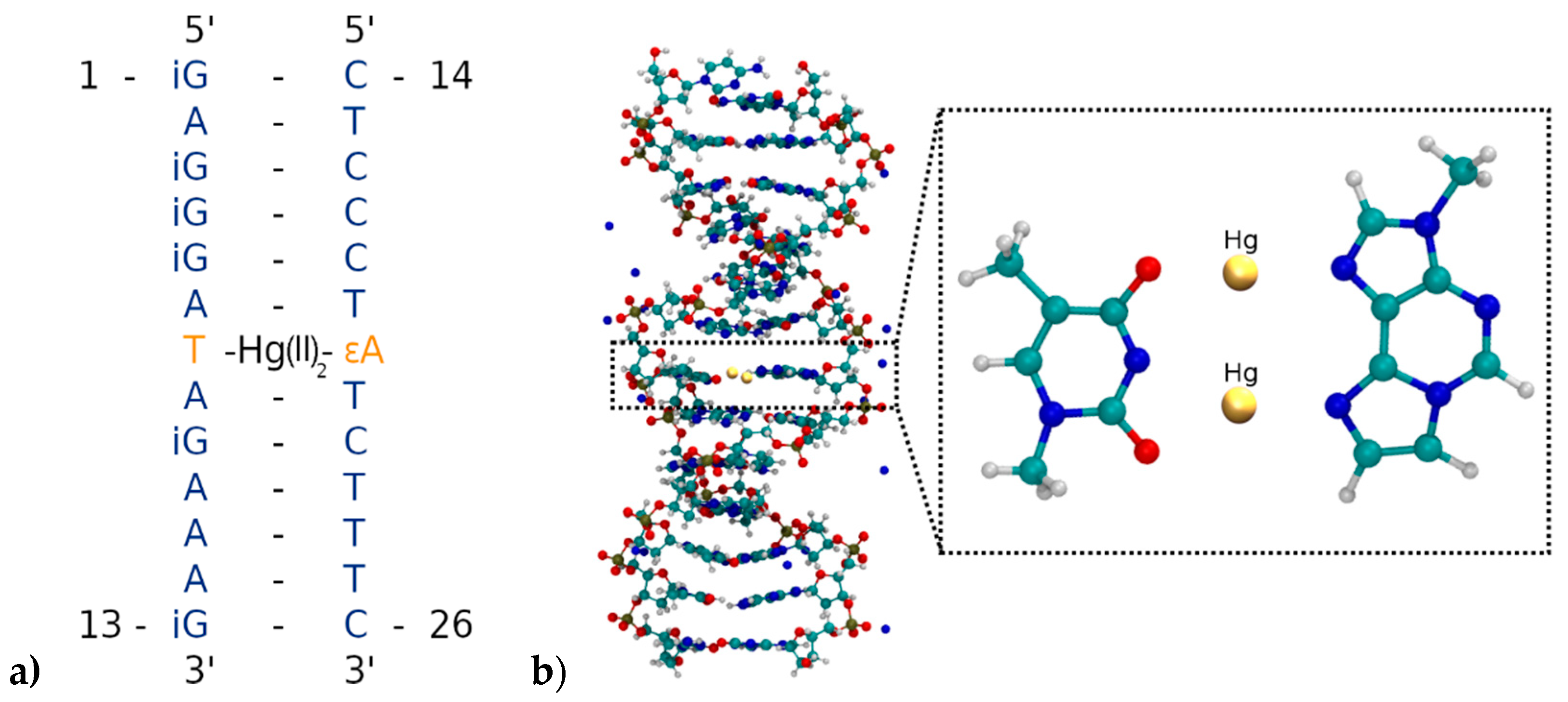
3.1. Generation of the Initial DNA Geometry
3.2. Zipping up the DNA by a Dynamic Distance Constraint
3.3. Relaxing the DNA Backbone by Geometry Optimization
3.4. Relaxing the DNA Structure at Finite Temperature
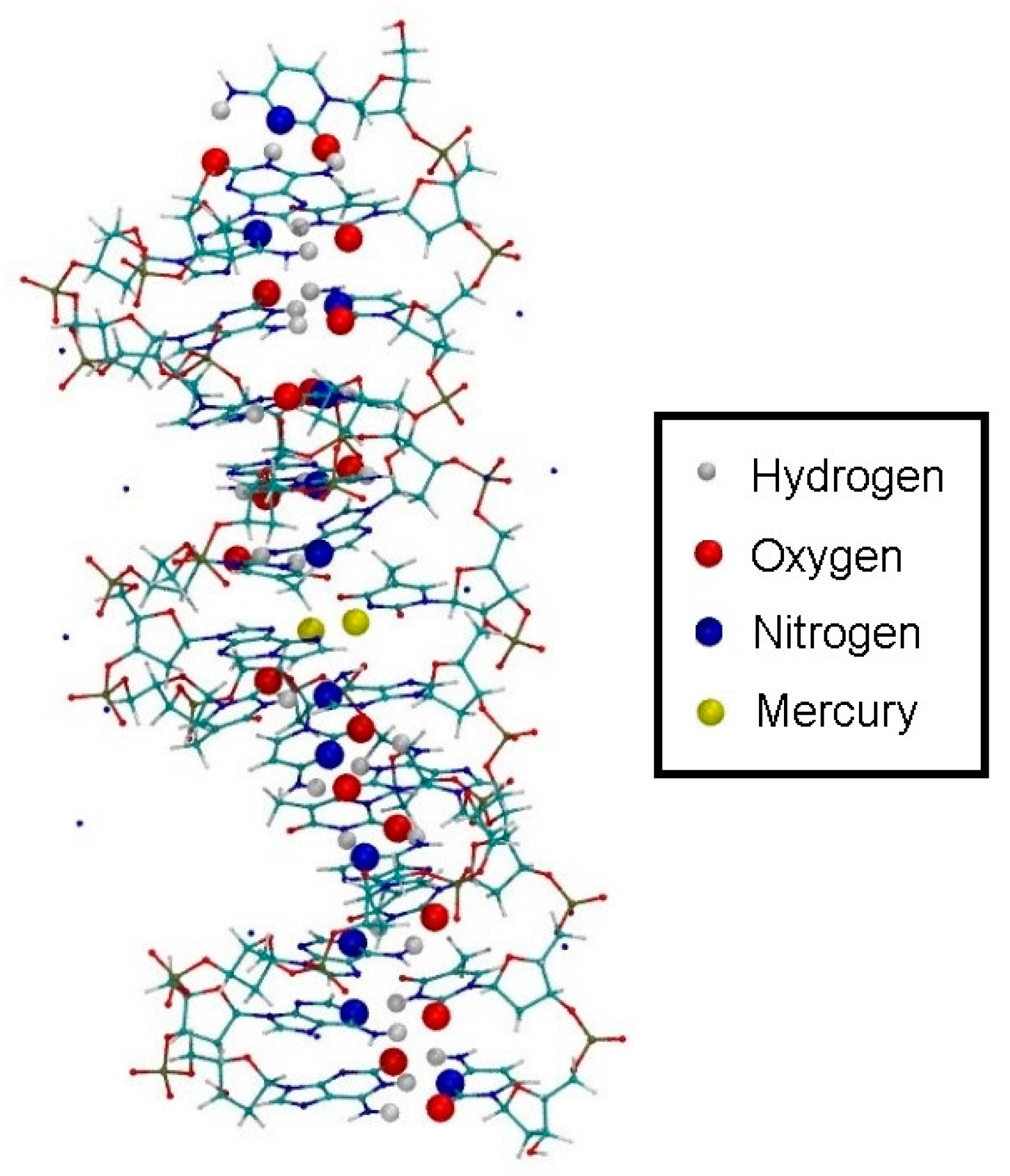
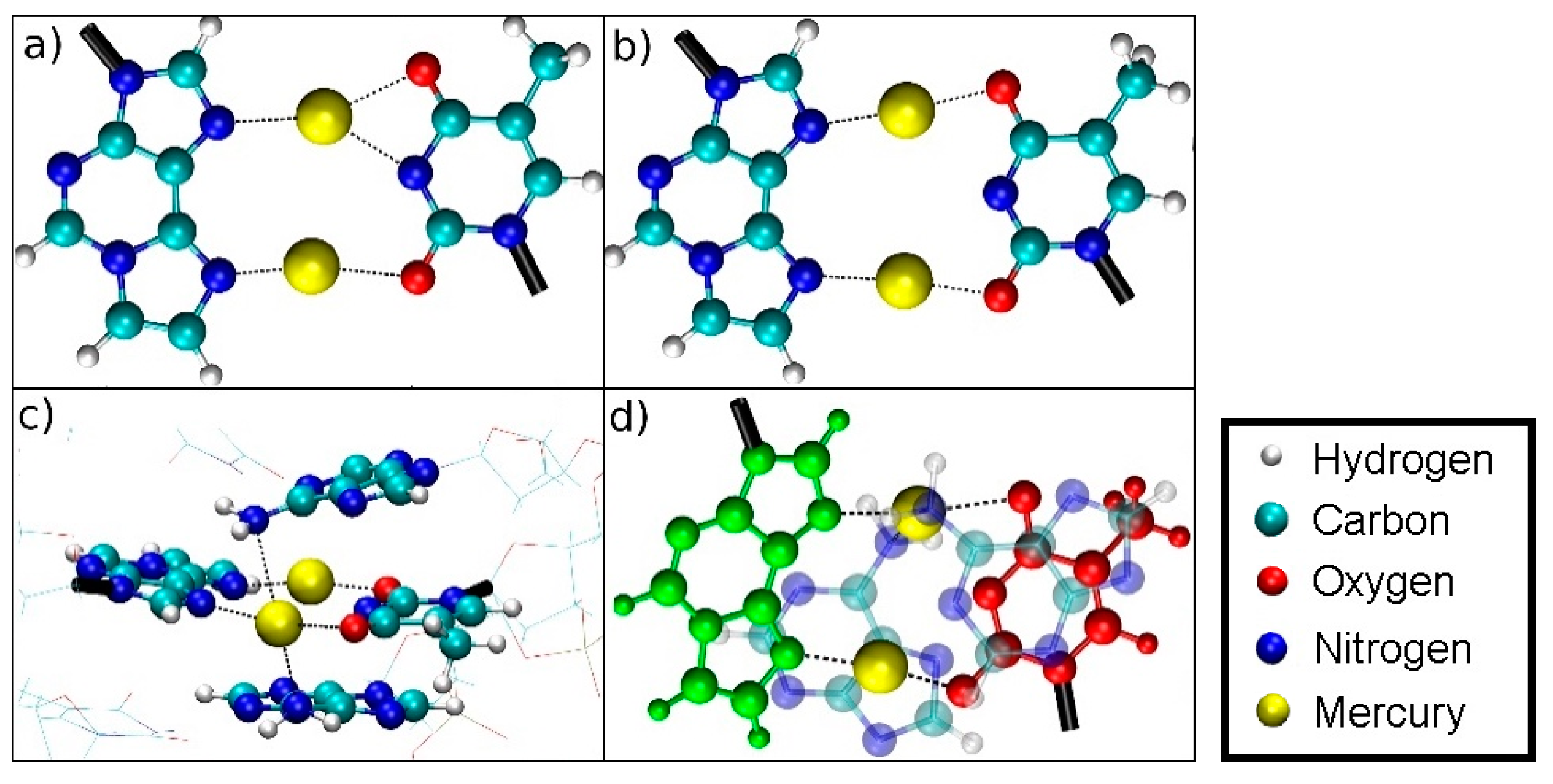
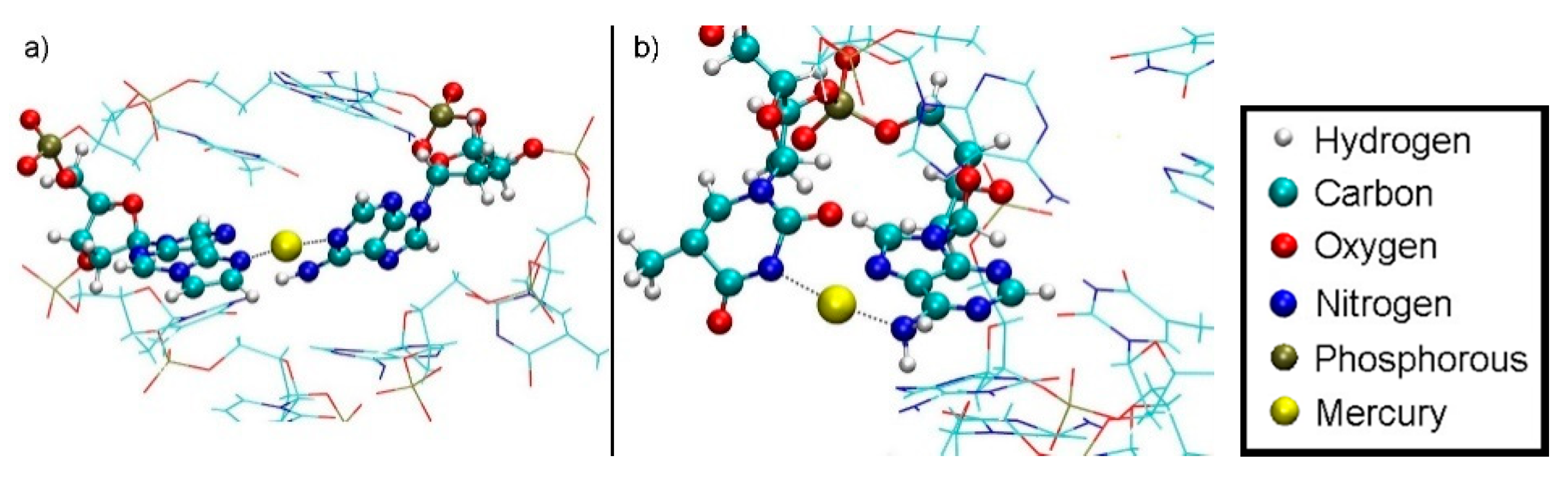
3.4.1. Structure 1
3.4.2. Structure 2
3.4.3. Structure 3
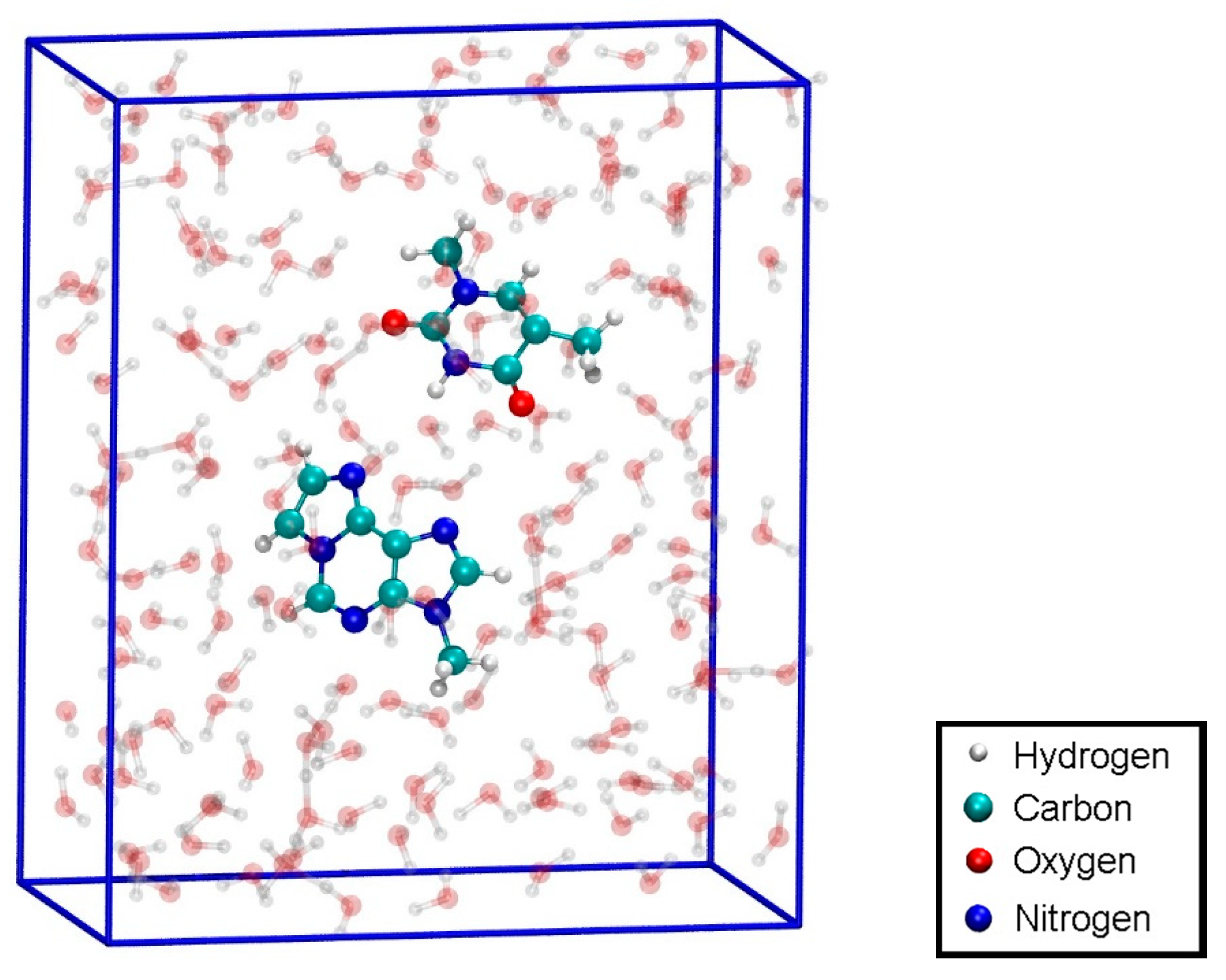
3.5. Ab Initio Simulations of Isolated Base Pairs in Solution
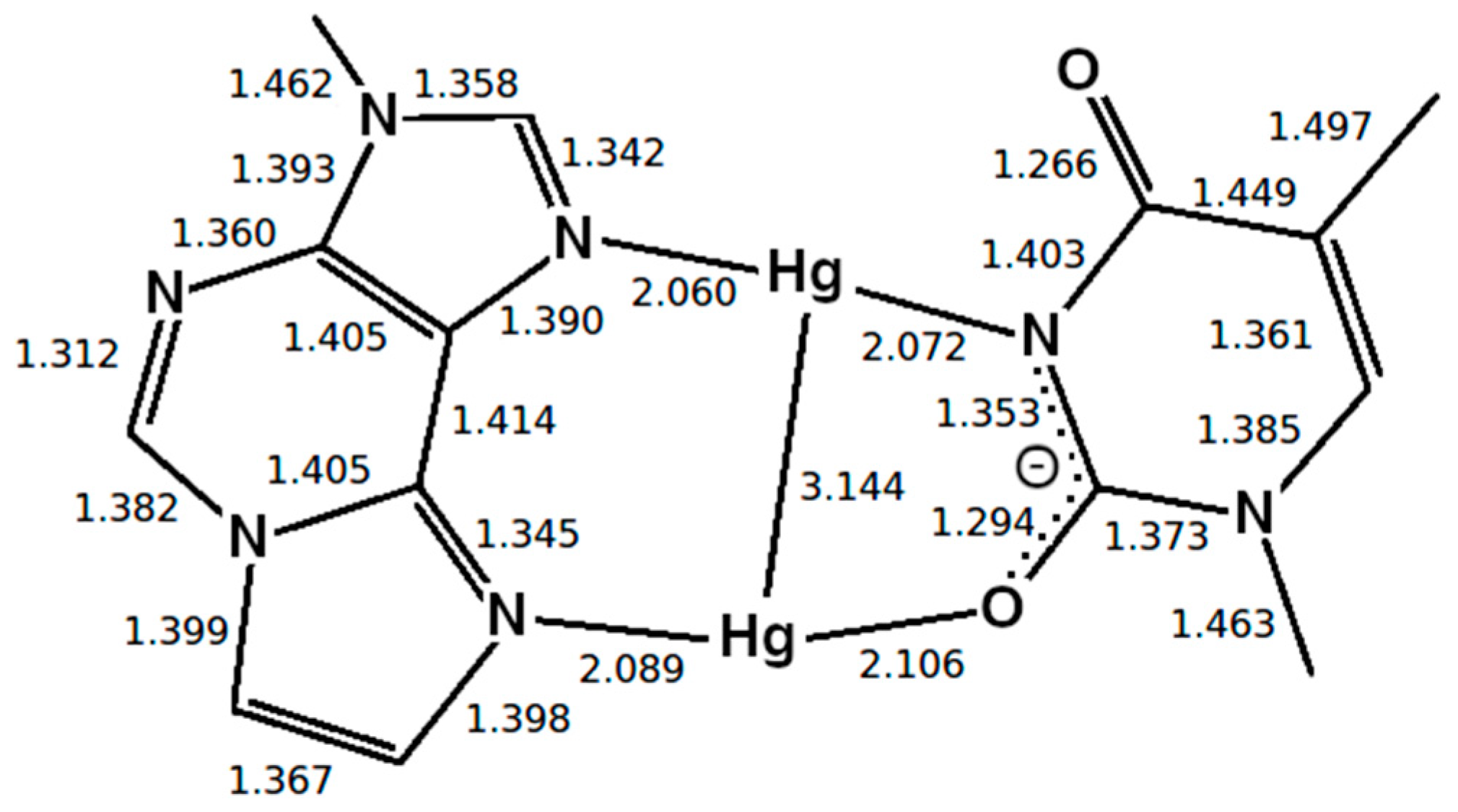
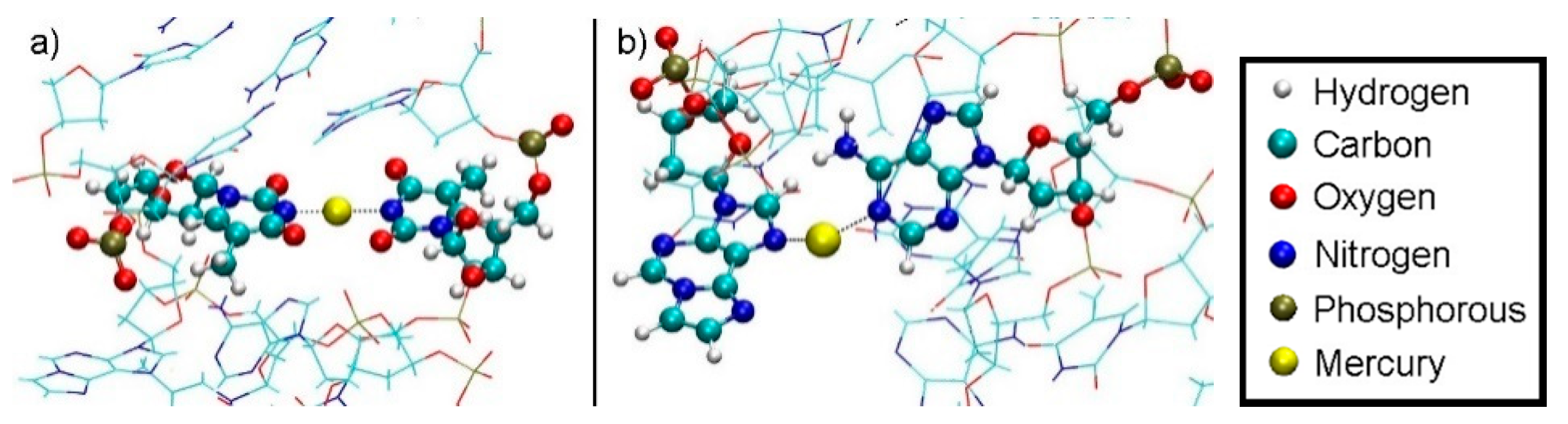
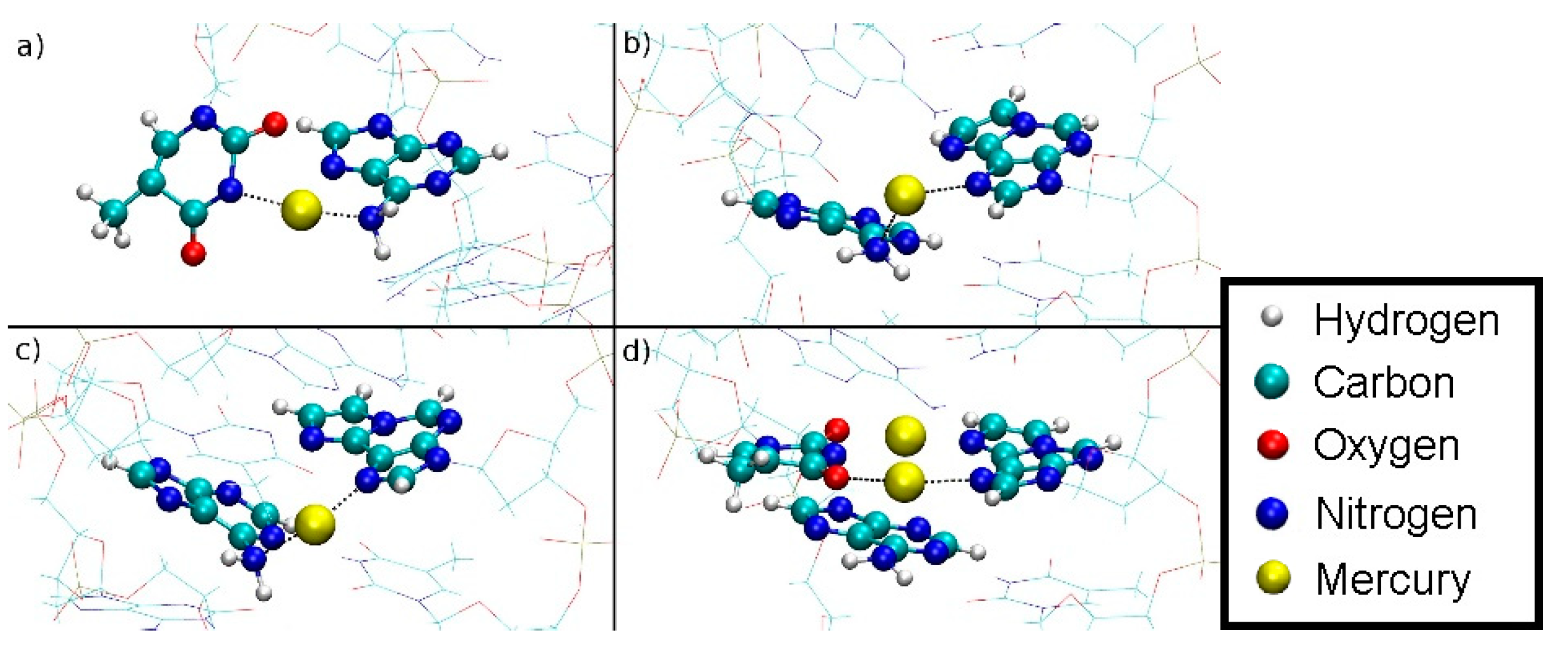
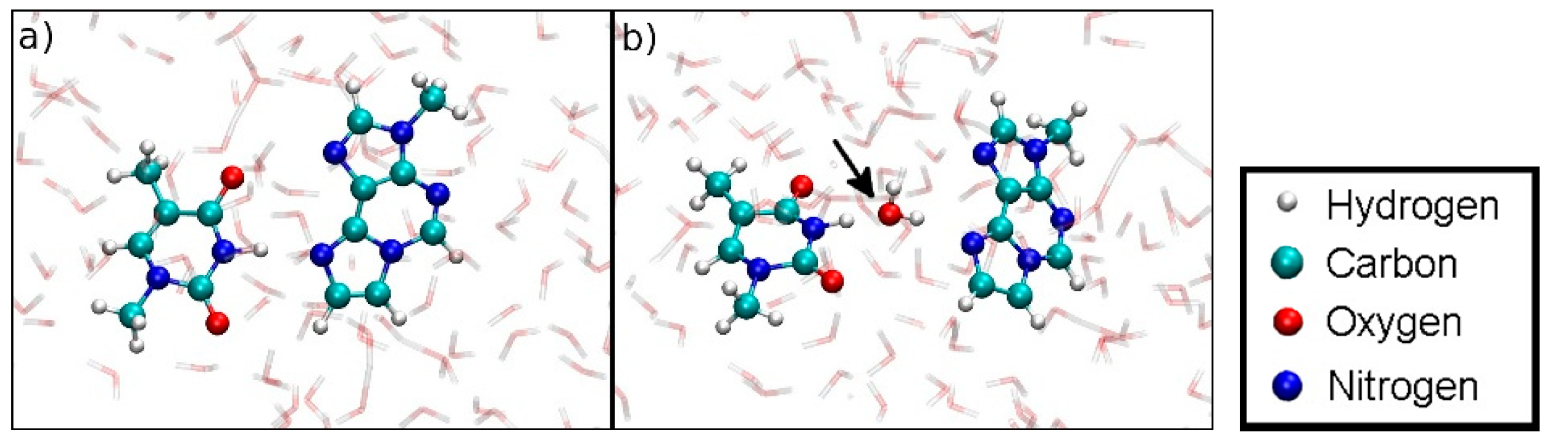
3.5.1. Structure 4
3.5.2. Structure 5
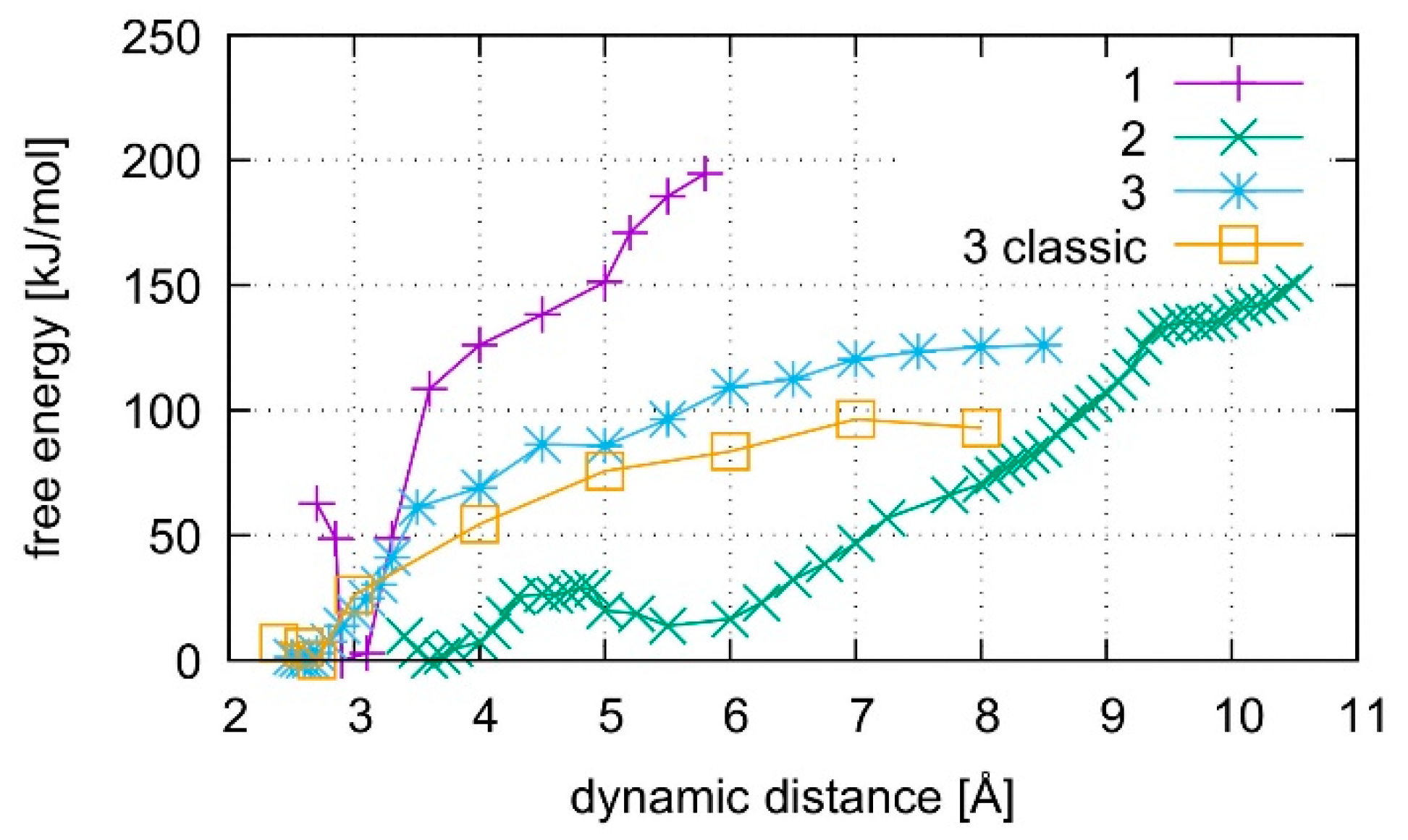
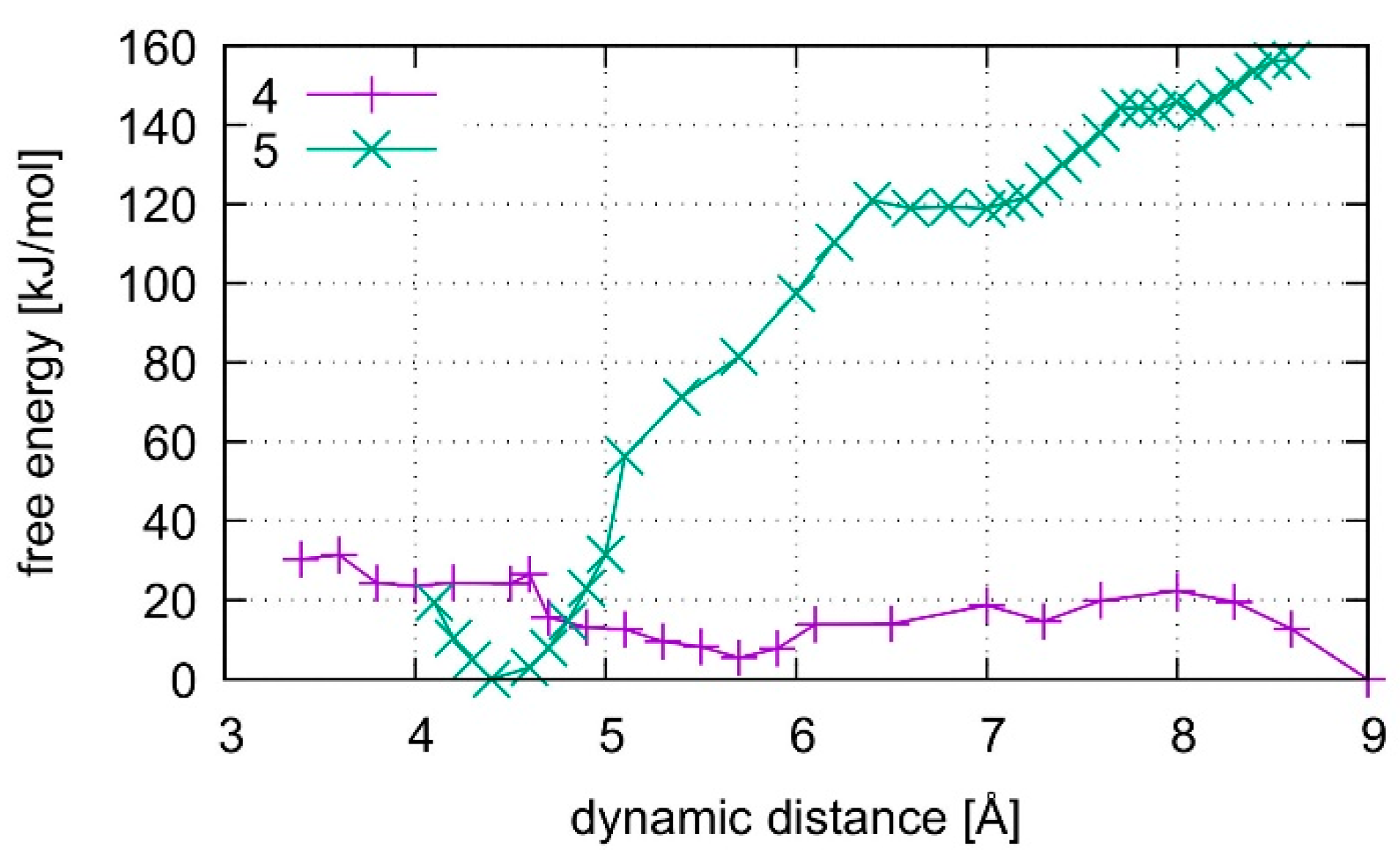
3.6. Free Energy Simulations
4. Discussion
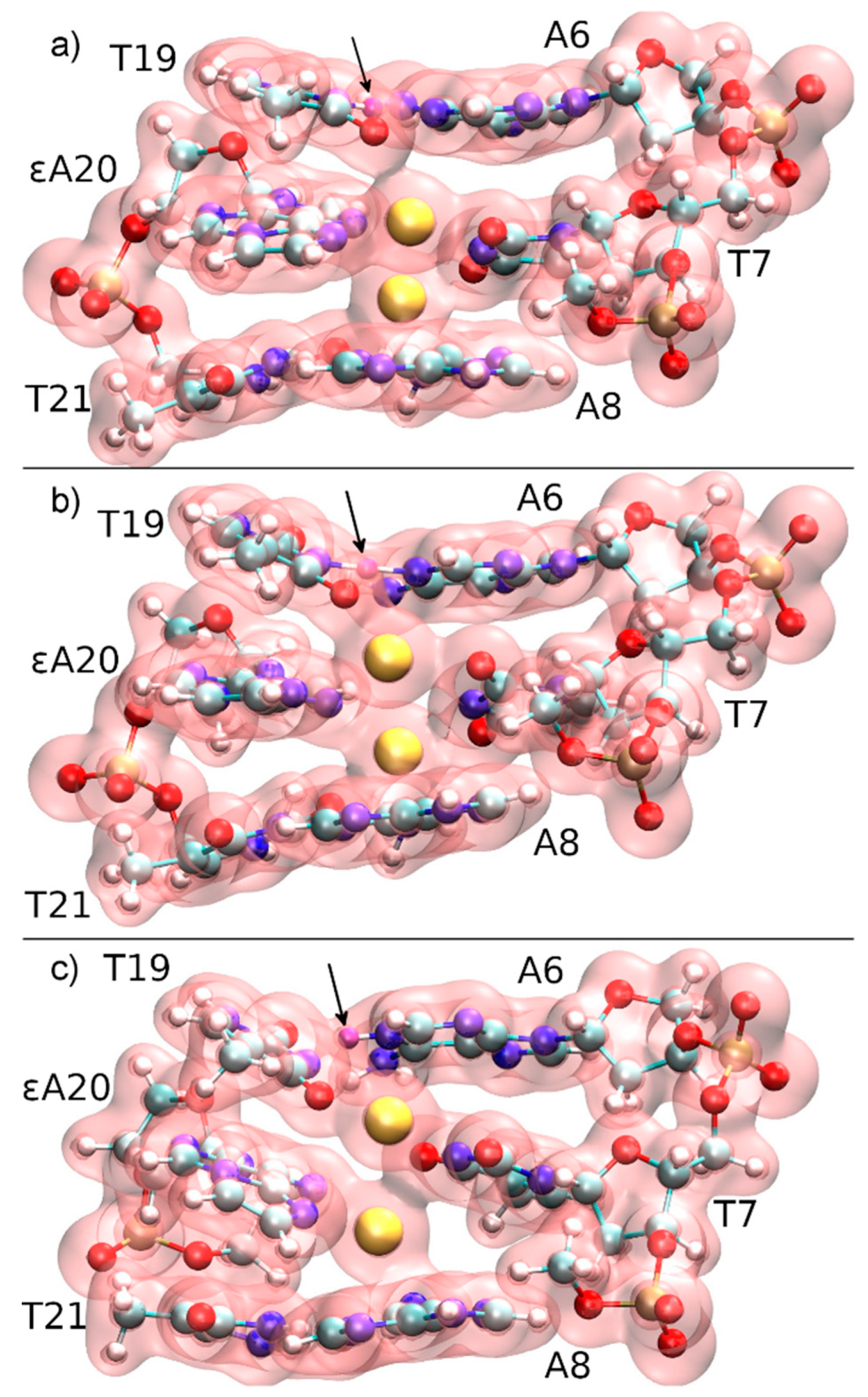
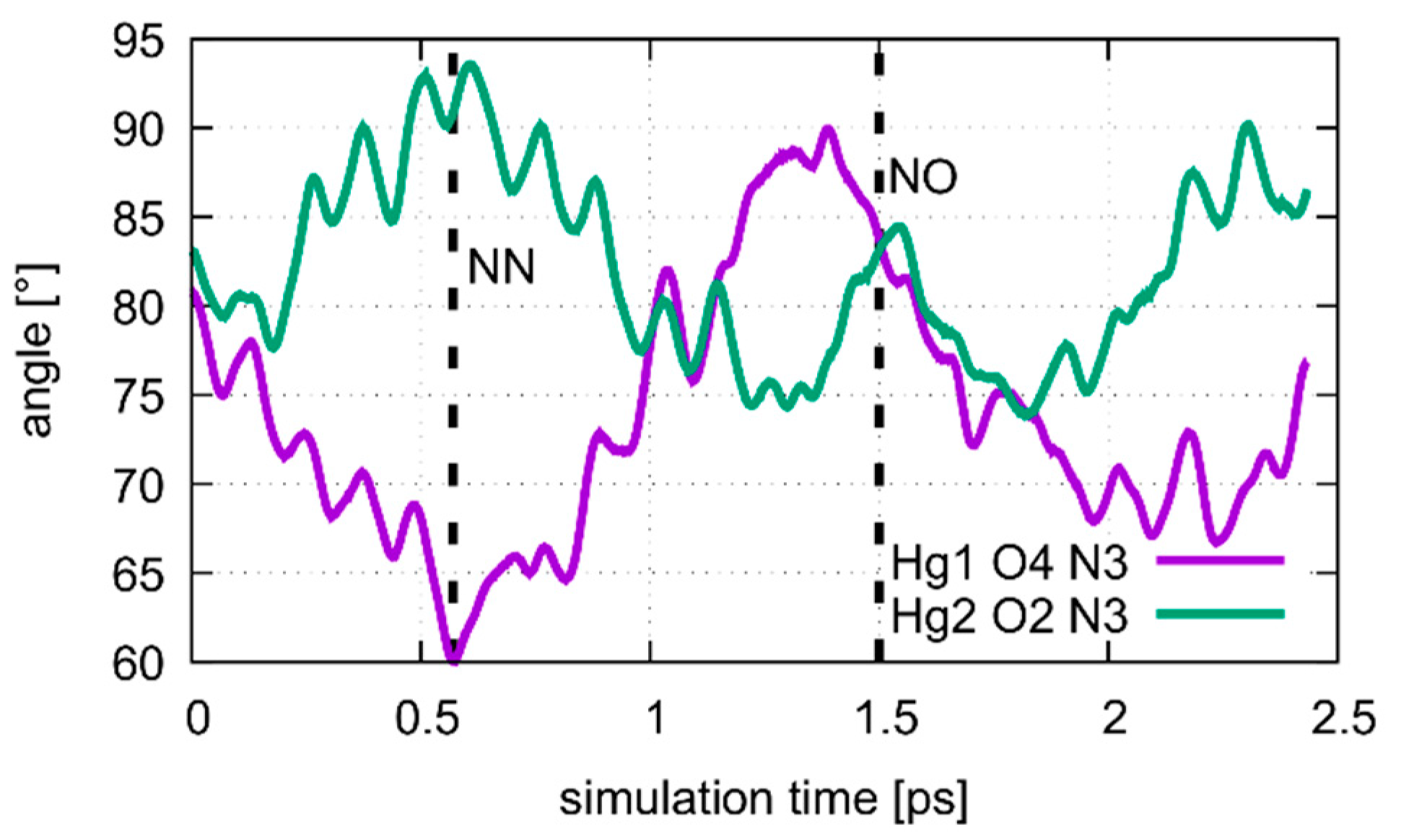
4.1. DNA Dissociation
4.2. Dissociation of the Isolated Base Pair
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stulz, E.; Clever, G.H. DNA in Supramolecular Chemistry and Nanotechnology; John Wiley & Sons: Chichester, UK, 2015. [Google Scholar]
- Jash, B.; Müller, J. Metal-Mediated Base Pairs: From Characterization to Application. Chem. Eur. J. 2017, 23, 17166–17178. [Google Scholar] [CrossRef]
- Takezawa, Y.; Müller, J.; Shionoya, M. Artificial DNA Base Pairing Mediated by Diverse Metal Ions. Chem. Lett. 2017, 46, 622–633. [Google Scholar] [CrossRef]
- Kondo, J.; Yamada, T.; Hirose, C.; Okamoto, I.; Tanaka, Y.; Ono, A. Crystal Structure of Metallo DNA Duplex Containing Consecutive Watson–Cric-klike T-HgII-T Base Pairs. Angew. Chem. Int. Ed. 2014, 53, 2385–2388. [Google Scholar] [CrossRef] [PubMed]
- Dairaku, T.; Furuita, K.; Sato, H.; Šebera, J.; Nakashima, K.; Kondo, J.; Yamanaka, D.; Kondo, Y.; Okamoto, I.; Ono, A.; et al. Structure Determination of an AgI-Mediated Cytosine–Cytosine Base Pair within DNA Duplex in Solution with 1H/15N/109Ag NMR Spectroscopy. Chem. Eur. J. 2016, 22, 13028–13031. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.P.; Jurt, S.; Johannsen, S.; Karimi, A.; Sigel, R.K.O.; Luedtke, N.W. Concerted dynamics of metallo-base pairs in an A/B-form helical transition. Nat. Commun. 2019, 10, 4818. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kondo, J.; Sychrovský, V.; Šebera, J.; Dairaku, T.; Saneyoshi, H.; Urata, H.; Torigoe, H.; Ono, A. Structures, physicochemical properties, and applications of T–HgII–T, C–AgI–C, and other metallo-base-pairs. Chem. Commun. 2015, 51, 17343–17360. [Google Scholar] [CrossRef] [PubMed]
- Naskar, S.; Müller, J. Light-induced formation of thymine-containing mercury(II)-mediated base pairs. Chem. Eur. J. 2019, 25, 16214–16218. [Google Scholar] [CrossRef] [PubMed]
- Naskar, S.; Hebenbrock, M.; Müller, J. Light-induced formation of silver(I)-mediated base pairs in DNA: Possibilities and limitations. Inorg. Chim. Acta 2020, 512, 119856. [Google Scholar] [CrossRef]
- Tanaka, K.; Tengeiji, A.; Kato, T.; Toyama, N.; Shionoya, M. A Discrete Self-Assembled Metal Array in Artificial DNA. Science 2003, 299, 1212–1213. [Google Scholar] [CrossRef]
- Clever, G.H.; Polborn, K.; Carell, T. A Highly DNA-Duplex-Stabilizing Metal-Salen Base Pair. Angew. Chem. Int. Ed. 2005, 44, 7204–7208. [Google Scholar] [CrossRef]
- Sandmann, N.; Defayay, D.; Hepp, A.; Müller, J. Metal-mediated base pairing in DNA involving the artificial nucleobase imidazole-4-carboxylate. J. Inorg. Biochem. 2019, 191, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, N.; Bachmann, J.; Hepp, A.; Doltsinis, N.L.; Müller, J. Copper(II)-mediated base pairing involving the artificial nucleobase 3H-imidazo[4,5-f]quinolin-5-ol. Dalton Trans. 2019, 48, 10505–10515. [Google Scholar] [CrossRef]
- Jash, B.; Müller, J. Stable Copper(I)-Mediated Base Pairing in DNA. Angew. Chem. Int. Ed. 2018, 57, 9524–9527. [Google Scholar] [CrossRef] [PubMed]
- Clever, G.H.; Carell, T. Controlled Stacking of 10 Transition-Metal Ions inside a DNA Duplex. Angew. Chem. Int. Ed. 2007, 46, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Switzer, C.; Sinha, S.; Kim, P.H.; Heuberger, B.D. A Purine-like Nickel(II) Base Pair for DNA. Angew. Chem. Int. Ed. 2005, 44, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Jash, B.; Müller, J. A stable zinc(II)-mediated base pair in a parallel-stranded DNA duplex. J. Inorg. Biochem. 2018, 186, 301–306. [Google Scholar] [CrossRef]
- Takezawa, Y.; Nishiyama, K.; Mashima, T.; Katahira, M.; Shionoya, M. Bifacial Base-Pairing Behaviors of 5-Hydroxyuracil DNA Bases through Hydrogen Bonding and Metal Coordination. Chem. Eur. J. 2015, 21, 14713–14716. [Google Scholar] [CrossRef]
- Räisälä, H.; Lönnberg, T. Covalently Palladated Oligonucleotides Through Oxidative Addition of Pd0. Chem. Eur. J. 2019, 25, 4751–4756. [Google Scholar] [CrossRef]
- Escher, D.; Müller, J. Silver(I) coordination in silver(I)-mediated homo base pairs of 6-pyrazolylpurine in DNA duplexes involves the Watson-Crick edge. Chem. Eur. J. 2020, 26. [Google Scholar] [CrossRef]
- Schönrath, I.; Tsvetkov, V.B.; Zatsepin, T.S.; Aralov, A.V.; Müller, J. Silver(I)-mediated base pairing in parallel-stranded DNA involving the luminescent cytosine analog 1,3-diaza-2-oxophenoxazine. J. Biol. Inorg. Chem. 2019, 24, 693–702. [Google Scholar] [CrossRef]
- Ukale, D.U.; Tähtinen, P.; Lönnberg, T. 1,8-Dimercuri-6-Phenyl-1H-Carbazole as a Monofacial Dinuclear Organometallic Nucleobase. Chem. Eur. J. 2020, 26, 2164–2168. [Google Scholar] [CrossRef]
- Méndez-Arriaga, J.M.; Maldonado, C.R.; Dobado, J.A.; Galindo, M.A. Silver(I)-Mediated Base Pairs in DNA Sequences Containing 7-Deazaguanine/Cytosine: Towards DNA with Entirely Metallated Watson–Crick Base Pairs. Chem. Eur. J. 2018, 24, 4583–4589. [Google Scholar] [CrossRef] [PubMed]
- Santamaría-Díaz, N.; Méndez-Arriaga, J.M.; Salas, J.M.; Galindo, M.A. Highly Stable Double-Stranded DNA Containing Sequential Silver(I)-Mediated 7-Deazaadenine/Thymine Watson–Crick Base Pairs. Angew. Chem. Int. Ed. 2016, 55, 6170–6174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Kondhare, D.; Leonard, P.; Seela, F. Anomeric 5-Aza-7-deaza-2’-deoxyguanosines in Silver-Ion-Mediated Homo and Hybrid Base Pairs: Impact of Mismatch Structure, Helical Environment, and Nucleobase Substituents on DNA Stability. Chem. Eur. J. 2019, 25, 10408–10419. [Google Scholar] [CrossRef]
- Guo, X.; Leonard, P.; Ingale, S.A.; Liu, J.; Mei, H.; Sieg, M.; Seela, F. 5-Aza-7-deaza-2’-deoxyguanosine and 2’-Deoxycytidine Form Programmable Silver-Mediated Base Pairs with Metal Ions in the Core of the DNA Double Helix. Chem. Eur. J. 2018, 24, 8883–8892. [Google Scholar] [CrossRef]
- Zhao, H.; Leonard, P.; Guo, X.; Yang, H.; Seela, F. Silver-Mediated Base Pairs in DNA Incorporating Purines,7-Deazapurines, and 8-Aza-7-deazapurines: Impact of Reduced Nucleobase Binding Sites and an Altered Glycosylation Position. Chem. Eur. J. 2017, 23, 5529–5540. [Google Scholar] [CrossRef]
- Liu, S.; Clever, G.H.; Takezawa, Y.; Kaneko, M.; Tanaka, K.; Guo, X.; Shionoya, M. Direct Conductance Measurement of Individual Metallo-DNA Duplexes within Single-Molecule Break Junctions. Angew. Chem. Int. Ed. 2011, 50, 8886–8890. [Google Scholar] [CrossRef]
- Ehrenschwender, T.; Schmucker, W.; Wellner, C.; Augenstein, T.; Carl, P.; Harmer, J.; Breher, F.; Wagenknecht, H.-A. Development of a Metal-Ion-Mediated Base Pair for Electron Transfer in DNA. Chem. Eur. J. 2013, 19, 12547–12552. [Google Scholar] [CrossRef]
- Al-Mahamad, L.L.G.; El-Zubir, O.; Smith, D.G.; Horrocks, B.R.; Houlton, A. A coordination polymer for the site-specific integration of semiconducting sequences into DNA-based materials. Nat. Commun. 2017, 8, 720. [Google Scholar] [CrossRef]
- Léon, J.C.; She, Z.; Kamal, A.; Shamsi, M.H.; Müller, J.; Kraatz, H.-B. DNA Films Containing the Artificial Nucleobase Imidazole Mediate Charge Transfer in a Silver(I)-Responsive Way. Angew. Chem. Int. Ed. 2017, 56, 6098–6102. [Google Scholar] [CrossRef]
- Hensel, S.; Eckey, K.; Scharf, P.; Megger, N.; Karst, U.; Müller, J. Excess Electron Transfer through DNA Duplexes Comprising a Metal-Mediated Base Pair. Chem. Eur. J. 2017, 23, 10244–10248. [Google Scholar] [CrossRef] [PubMed]
- Nakama, T.; Takezawa, Y.; Sasaki, D.; Shionoya, M. Allosteric Regulation of DNAzyme Activities through Intrastrand Transformation Induced by Cu(II)-Mediated Artificial Base Pairing. J. Am. Chem. Soc. 2020, 142, 10153–10162. [Google Scholar] [CrossRef] [PubMed]
- Takezawa, Y.; Nakama, T.; Shionoya, M. Enzymatic Synthesis of Cu(II)-Responsive Deoxyribozymes through Polymerase Incorporation of Artificial Ligand-Type Nucleotides. J. Am. Chem. Soc. 2019, 141, 19342–19350. [Google Scholar] [CrossRef] [PubMed]
- Takezawa, Y.; Yoneda, S.; Duprey, J.-L.H.A.; Nakama, T.; Shionoya, M. Metal-responsive structural transformation between artificial DNA duplexes and three-way junctions. Chem. Sci. 2016, 7, 3006–3010. [Google Scholar] [CrossRef]
- Léon, J.C.; González-Abradelo, D.; Strassert, C.A.; Müller, J. Fluorescent DNA-Templated Silver Nanoclusters from Silver(I)-Mediated Base Pairs. Chem. Eur. J. 2018, 24, 8320–8324. [Google Scholar] [CrossRef]
- Taherpour, S.; Golubev, O.; Lönnberg, T. On the feasibility of recognition of nucleic acid sequences by metal-ion-carrying oligonucleotides. Inorg. Chim. Acta 2016, 452, 43–49. [Google Scholar] [CrossRef]
- Jash, B.; Müller, J. Application of a Metal-Mediated Base Pair to the Detection of Medicinally Relevant Single Nucleotide Polymorphisms. Eur. J. Inorg. Chem. 2017, 3857–3861. [Google Scholar] [CrossRef]
- Jash, B.; Scharf, P.; Sandmann, N.; Fonseca Guerra, C.; Megger, D.A.; Müller, J. A metal-mediated base pair that discriminates between the canonical pyrimidine nucleobases. Chem. Sci. 2017, 8, 1337–1343. [Google Scholar] [CrossRef]
- Müller, J. Nucleic acid duplexes with metal-mediated base pairs and their structures. Coord. Chem. Rev. 2019, 393, 37–47. [Google Scholar] [CrossRef]
- Funai, T.; Miyazaki, Y.; Aotani, M.; Yamaguchi, E.; Nakagawa, O.; Wada, S.-i.; Torigoe, H.; Ono, A.; Urata, H. AgI Ion Mediated Formation of a C–A Mispair by DNA Polymerases. Angew. Chem. Int. Ed. 2012, 51, 6464–6466. [Google Scholar] [CrossRef]
- Funai, T.; Nakamura, J.; Miyazaki, Y.; Kiriu, R.; Nakagawa, O.; Wada, S.-I.; Ono, A.; Urata, H. Regulated Incorporation of Two Different Metal Ions into Programmed Sites in a Duplex by DNA Polymerase Catalyzed Primer Extension. Angew. Chem. Int. Ed. 2014, 53, 6624–6627. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Takezawa, Y.; Sakamoto, A.; Shionoya, M. Enzymatic synthesis of ligand-bearing DNAs for metal-mediated base pairing utilising a template-independent polymerase. Chem. Commun. 2016, 52, 3762–3765. [Google Scholar] [CrossRef] [PubMed]
- Kaul, C.; Müller, M.; Wagner, M.; Schneider, S.; Carell, T. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nat. Chem. 2011, 3, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Flamme, M.; Levi-Acobas, F.; Hensel, S.; Naskar, S.; Röthlisberger, P.; Sarac, I.; Gasser, G.; Müller, J.; Hollenstein, M. Effect of metal shielding on the enzymatic construction of artificial base pairs. ChemBioChem 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Levi-Acobas, F.; Sarac, I.; Marlière, P.; Herdewijn, P.; Hollenstein, M. Towards the enzymatic formation of artificial metal base pairs with a carboxy-imidazole-modified nucleotide. J. Inorg. Biochem. 2019, 191, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Levi-Acobas, F.; Sarac, I.; Marlière, P.; Herdewijn, P.; Hollenstein, M. On the enzymatic incorporation of an imidazole nucleotide into DNA. Org. Biomol. Chem. 2017, 15, 4449–4455. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, I.; Ono, T.; Sameshima, R.; Ono, A. Metal ion-binding properties of DNA duplexes containing thiopyrimidine base pairs. Chem. Commun. 2012, 48, 4347–4349. [Google Scholar] [CrossRef]
- Mandal, S.; Hepp, A.; Müller, J. Unprecedented dinuclear silver(I)-mediated base pair involving the DNA lesion 1,N6-ethenoadenine. Dalton Trans. 2015, 44, 3540–3543. [Google Scholar] [CrossRef]
- Mandal, S.; Hebenbrock, M.; Müller, J. Relative Strand Orientation in a DNA Duplex Controls the Nuclearity of a Metal-Mediated Base Pair. Chem. Eur. J. 2017, 23, 5962–5965. [Google Scholar] [CrossRef]
- Fujii, A.; Nakagawa, O.; Kishimoto, Y.; Okuda, T.; Nakatsuji, Y.; Nozaki, N.; Kasahara, Y.; Obika, S. 1,3,9-Triaza-2-oxophenoxazine artificial nucleobase forms highly stable self-base pairs with three AgI ions in a duplex. Chem. Eur. J. 2019, 25, 7443–7448. [Google Scholar] [CrossRef]
- Mandal, S.; Hebenbrock, M.; Müller, J. A Dinuclear Mercury(II)-Mediated Base Pair in DNA. Angew. Chem. Int. Ed. 2016, 55, 15520–15523. [Google Scholar] [CrossRef] [PubMed]
- Müller, J. Metal-mediated base pairs in parallel-stranded DNA. Beilstein J. Org. Chem. 2017, 13, 2671–2681. [Google Scholar] [CrossRef]
- Onyido, I.; Norris, A.R.; Buncel, E. Biomolecule-Mercury Interactions: Modalities of DNA Base-Mercury Binding Mechanisms. Remediation Strategies. Chem. Rev. 2004, 104, 5911–5929. [Google Scholar] [CrossRef]
- Burisch, C.; Markwick, P.R.L.; Doltsinis, N.L.; Schlitter, J. ‘Dynamic Distance’ Reaction Coordinate for Competing Bonds: Applications in Classical and Ab Initio Simulations. J. Chem. Theory Comput. 2008, 4, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CK, USA, 2013. [Google Scholar]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Perdew, J.P.; Ernzerhof, M.; Burke, K. Rationale for mixing exact exchange with density functional approximations. J. Chem. Phys. 1996, 105, 9982–9985. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Ernzerhof, M.; Scuseria, G.E. Assessment of the Perdew–Burke–Ernzerhof exchange-correlation functional. J. Chem. Phys. 1999, 110, 5029–5036. [Google Scholar] [CrossRef]
- Dunning, T.H., Jr.; Hay, P.J. Modern Theoretical Chemistry; Schaefer, H.F., III, Ed.; Plenum: New York, NY, USA, 1977; Volume 3, pp. 1–28. [Google Scholar]
- Andrae, D.; Häußermann, U.; Dolg, M.; Stoll, H.; Preuß, H. Energy-adjusted ab initio pseudopotentials for the second and third row transition elements. Theor. Chim. Acta 1990, 77, 123–141. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the Damping Function in Dispersion Corrected Density Functional Theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [PubMed]
- Cossi, M.; Barone, V.; Cammi, R.; Tomasi, J. Ab initio study of solvated molecules: A new implementation of the polarizable continuum model. Chem. Phys. Lett. 1996, 255, 327–335. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Van Lenthe, E.; Snijders, J.G.; Baerends, E.J. The zero-order regular approximation for relativistic effects: The effect of spin–orbit coupling in closed shell molecules. J. Chem. Phys. 1996, 105, 6505–6516. [Google Scholar] [CrossRef]
- Pantazis, D.A.; Chen, X.-Y.; Landis, C.R.; Neese, F. All-Electron Scalar Relativistic Basis Sets for Third-Row Transition Metal Atoms. J. Chem. Theory Comput. 2008, 4, 908–919. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Ivani, I.; Dans, P.D.; Noy, A.; Pérez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goñi, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef]
- Fuchs, J.-F.; Nedev, H.; Poger, D.; Ferrand, M.; Brenner, V.; Dognon, J.-P.; Crouzy, S. New model potentials for sulfur–copper(I) and sulfur–mercury(II) interactions in proteins: From ab initio to molecular dynamics. J. Comput. Chem. 2006, 27, 837–856. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.D.; Kollman, P.A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Kühne, T.D.; Iannuzzi, M.; Del Ben, M.; Rybkin, V.V.; Seewald, P.; Stein, F.; Laino, T.; Khaliullin, R.Z.; Schütt, O.; Schiffmann, F.; et al. CP2K: An electronic structure and molecular dynamics software package—Quickstep: Efficient and accurate electronic structure calculations. J. Chem. Phys. 2020, 152, 194103. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, I.C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Lippert, G.; Hutter, J.; Parrinello, M. A hybrid Gaussian and plane wave density functional scheme. Mol. Phys. 1997, 92, 477–487. [Google Scholar] [CrossRef]
- Hutter, J.; Iannuzzi, M.; Schiffmann, F.; VandeVondele, J. CP2K: Atomistic simulations of condensed matter systems. WIREs Comput. Mol. Sci. 2014, 4, 15–25. [Google Scholar] [CrossRef]
- VandeVondele, J.; Hutter, J. Gaussian basis sets for accurate calculations on molecular systems in gas and condensed phases. J. Chem. Phys. 2007, 127, 114105. [Google Scholar] [CrossRef]
- Krack, M. Pseudopotentials for H to Kr optimized for gradient-corrected exchange-correlation functionals. Theor. Chem. Acc. 2005, 114, 145–152. [Google Scholar] [CrossRef]
- Hartwigsen, C.; Goedecker, S.; Hutter, J. Relativistic separable dual-space Gaussian pseudopotentials from H to Rn. Phys. Rev. B 1998, 58, 3641–3662. [Google Scholar] [CrossRef]
- Goedecker, S.; Teter, M.; Hutter, J. Separable dual-space Gaussian pseudopotentials. Phys. Rev. B 1996, 54, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Maseras, F.; Morokuma, K. IMOMM: A new integrated ab initio+ molecular mechanics geometry optimization scheme of equilibrium structures and transition states. J. Comput. Chem. 1995, 16, 1170–1179. [Google Scholar] [CrossRef]
- Lu, X.-J.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef]
- Laino, T.; Mohamed, F.; Laio, A.; Parrinello, M. An Efficient Real Space Multigrid QM/MM Electrostatic Coupling. J. Chem. Theory Comput. 2005, 1, 1176–1184. [Google Scholar] [CrossRef]
- Laino, T.; Mohamed, F.; Laio, A.; Parrinello, M. An Efficient Linear-Scaling Electrostatic Coupling for Treating Periodic Boundary Conditions in QM/MM Simulations. J. Chem. Theory Comput. 2006, 2, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, V.R.; Bhaumik, S.R.; Chary, K.V.R.; Govil, G.; Liu, K.; Howard, F.B.; Miles, H.T. NMR structure of a parallel-stranded DNA duplex at atomic resolution. Nucleic Acids Res. 2002, 30, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Kanazawa, H.; Ito, H.; Goto, M.; Nakamura, K.; Saneyoshi, H.; Kondo, J. A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs. Angew. Chem. Int. Ed. 2019, 58, 16835–16838. [Google Scholar] [CrossRef]
- Szatyłowicz, H.; Sadlej-Sosnowska, N. Characterizing the Strength of Individual Hydrogen Bonds in DNA Base Pairs. J. Chem. Inf. Model. 2010, 50, 2151–2161. [Google Scholar] [CrossRef]
- Silaghi-Dumitrescu, R.; Mihály, B.; Mihály, T.; Attia, A.A.A.; Sanz Miguel, P.J.; Lippert, B. The exocyclic amino group of adenine in PtII and PdII complexes: A critical comparison of the X-ray crystallographic structural data and gas phase calculations. J. Biol. Inorg. Chem. 2017, 22, 567–579. [Google Scholar] [CrossRef] [PubMed]
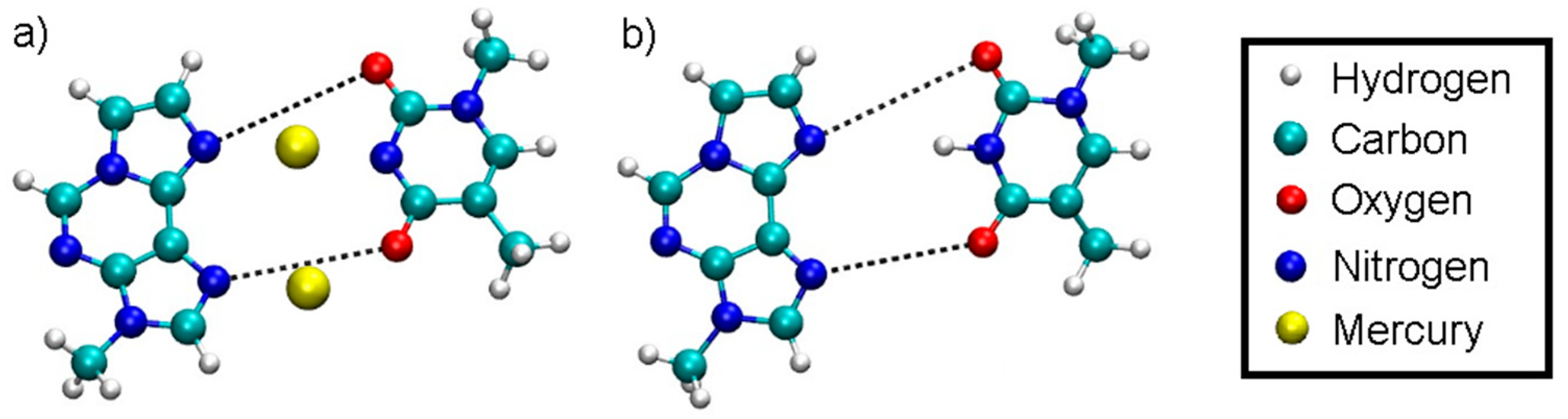
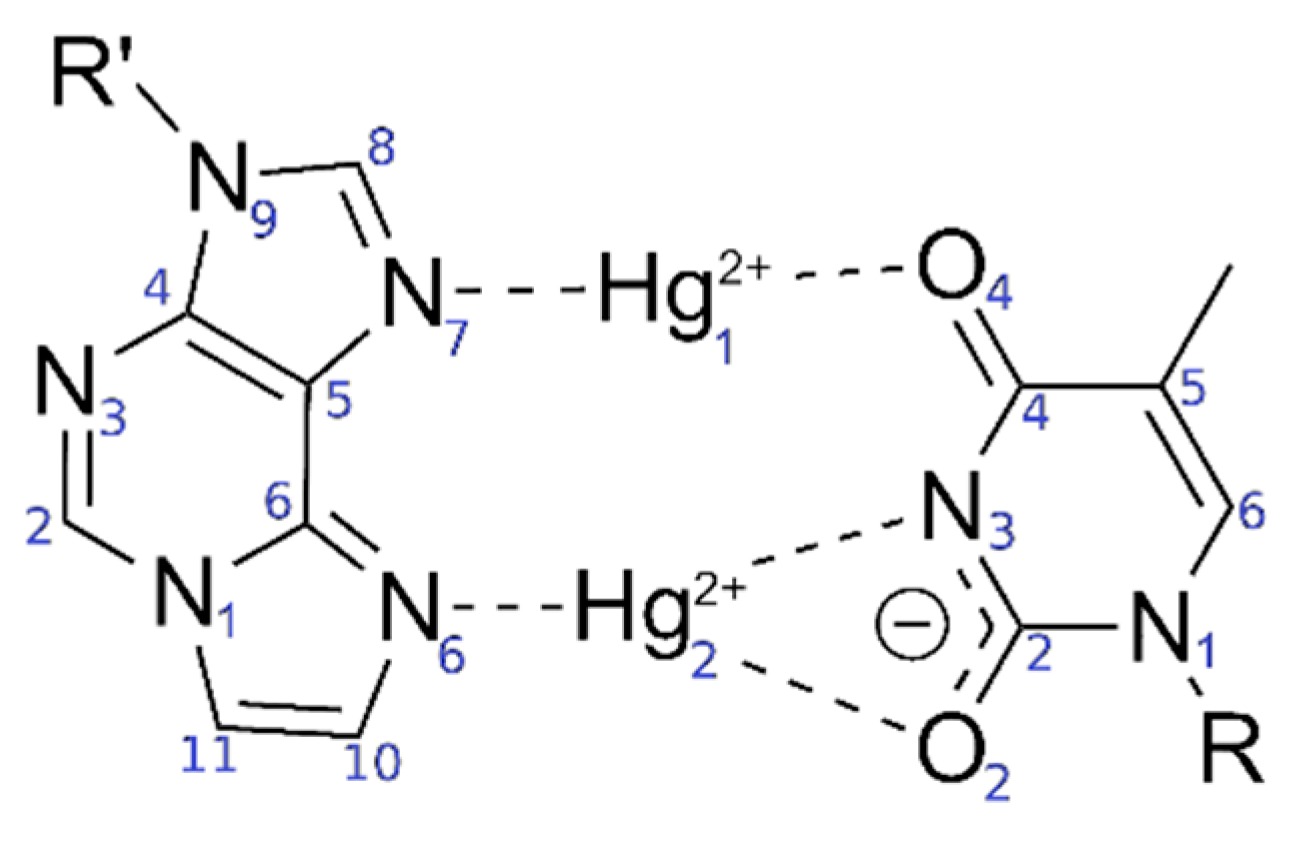
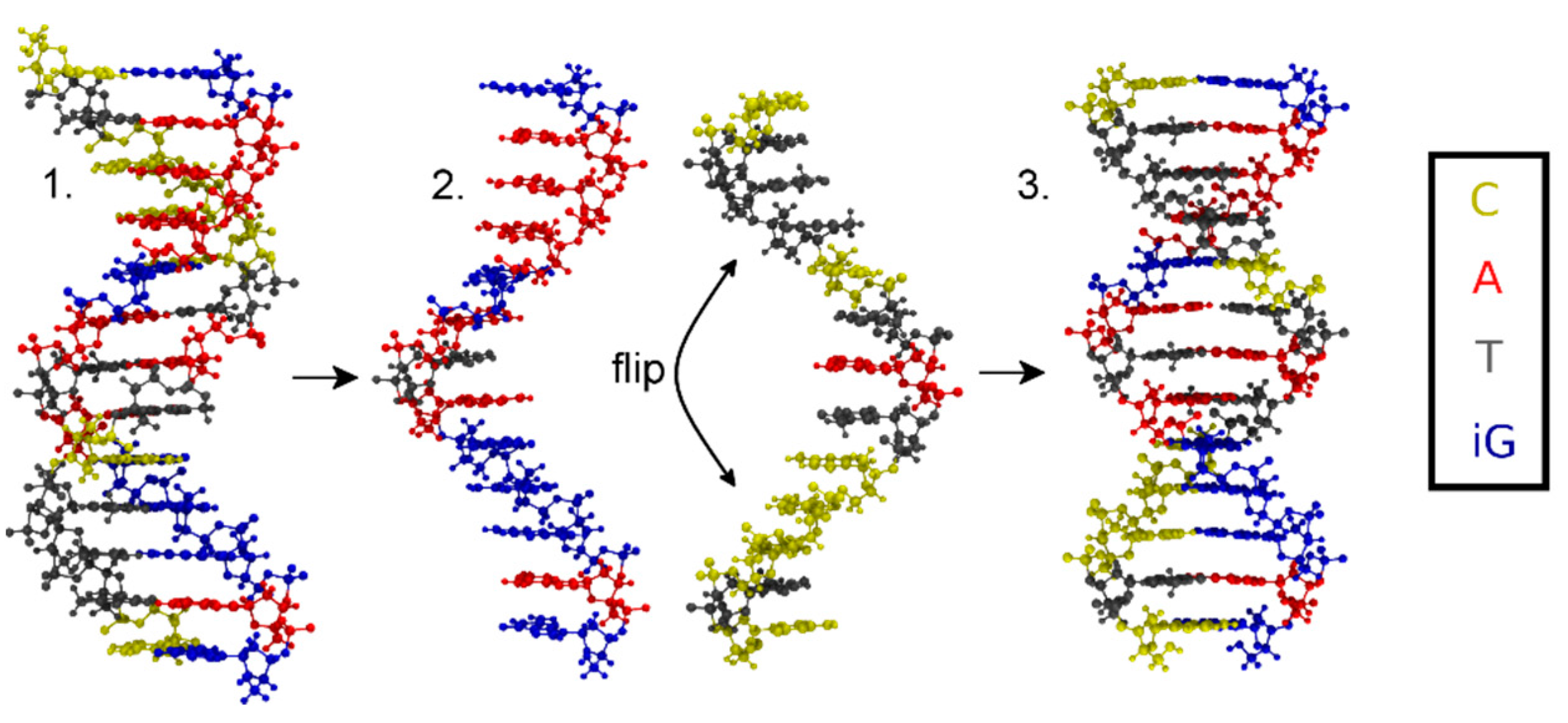
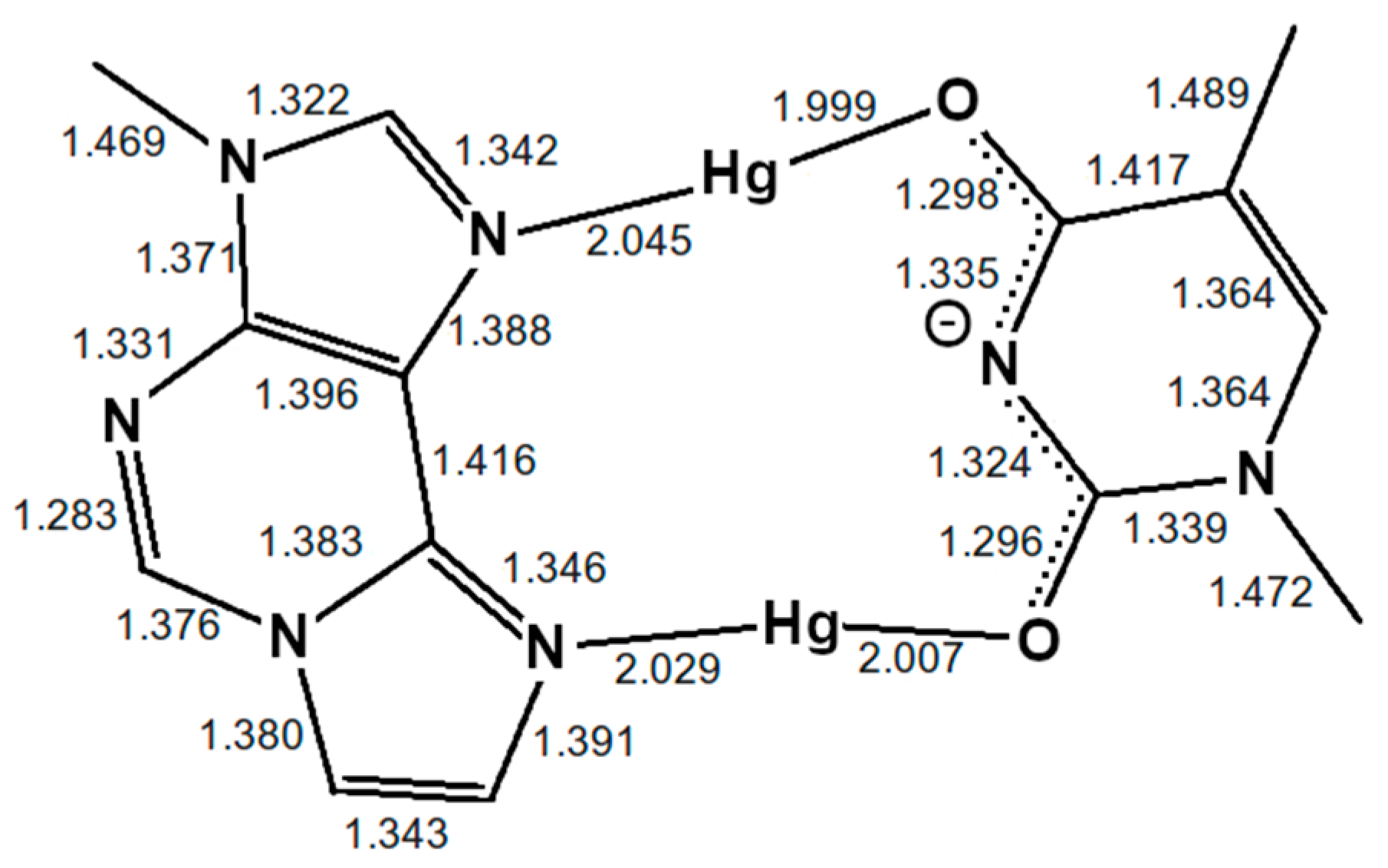



Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bachmann, J.; Schönrath, I.; Müller, J.; Doltsinis, N.L. Dynamic Structure and Stability of DNA Duplexes Bearing a Dinuclear Hg(II)-Mediated Base Pair. Molecules 2020, 25, 4942. https://doi.org/10.3390/molecules25214942
Bachmann J, Schönrath I, Müller J, Doltsinis NL. Dynamic Structure and Stability of DNA Duplexes Bearing a Dinuclear Hg(II)-Mediated Base Pair. Molecules. 2020; 25(21):4942. https://doi.org/10.3390/molecules25214942
Chicago/Turabian StyleBachmann, Jim, Isabell Schönrath, Jens Müller, and Nikos L. Doltsinis. 2020. "Dynamic Structure and Stability of DNA Duplexes Bearing a Dinuclear Hg(II)-Mediated Base Pair" Molecules 25, no. 21: 4942. https://doi.org/10.3390/molecules25214942
APA StyleBachmann, J., Schönrath, I., Müller, J., & Doltsinis, N. L. (2020). Dynamic Structure and Stability of DNA Duplexes Bearing a Dinuclear Hg(II)-Mediated Base Pair. Molecules, 25(21), 4942. https://doi.org/10.3390/molecules25214942





