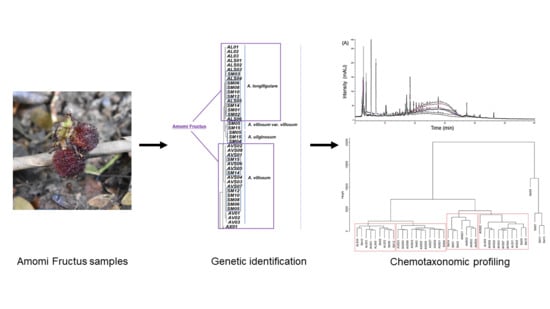
Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis
Abstract
1. Introduction
2. Results
2.1. ITS Regions of the Nuclear Ribosomal Cistron
2.2. Chloroplast Genome-Based DNA Barcode Sequence Analysis
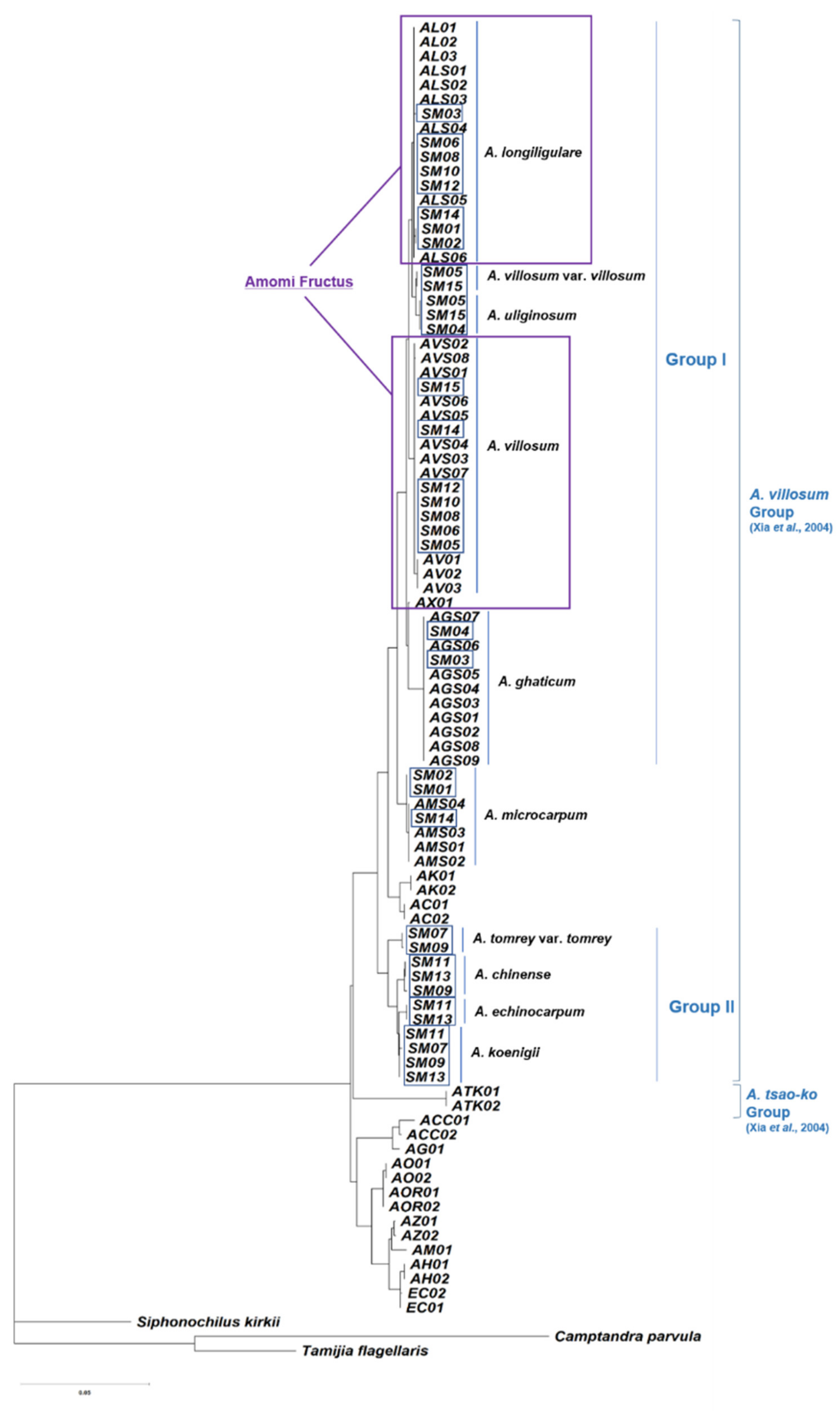
2.3. Phylogenetic Analysis
2.4. Optimization of Sample Extraction and Analytical Conditions
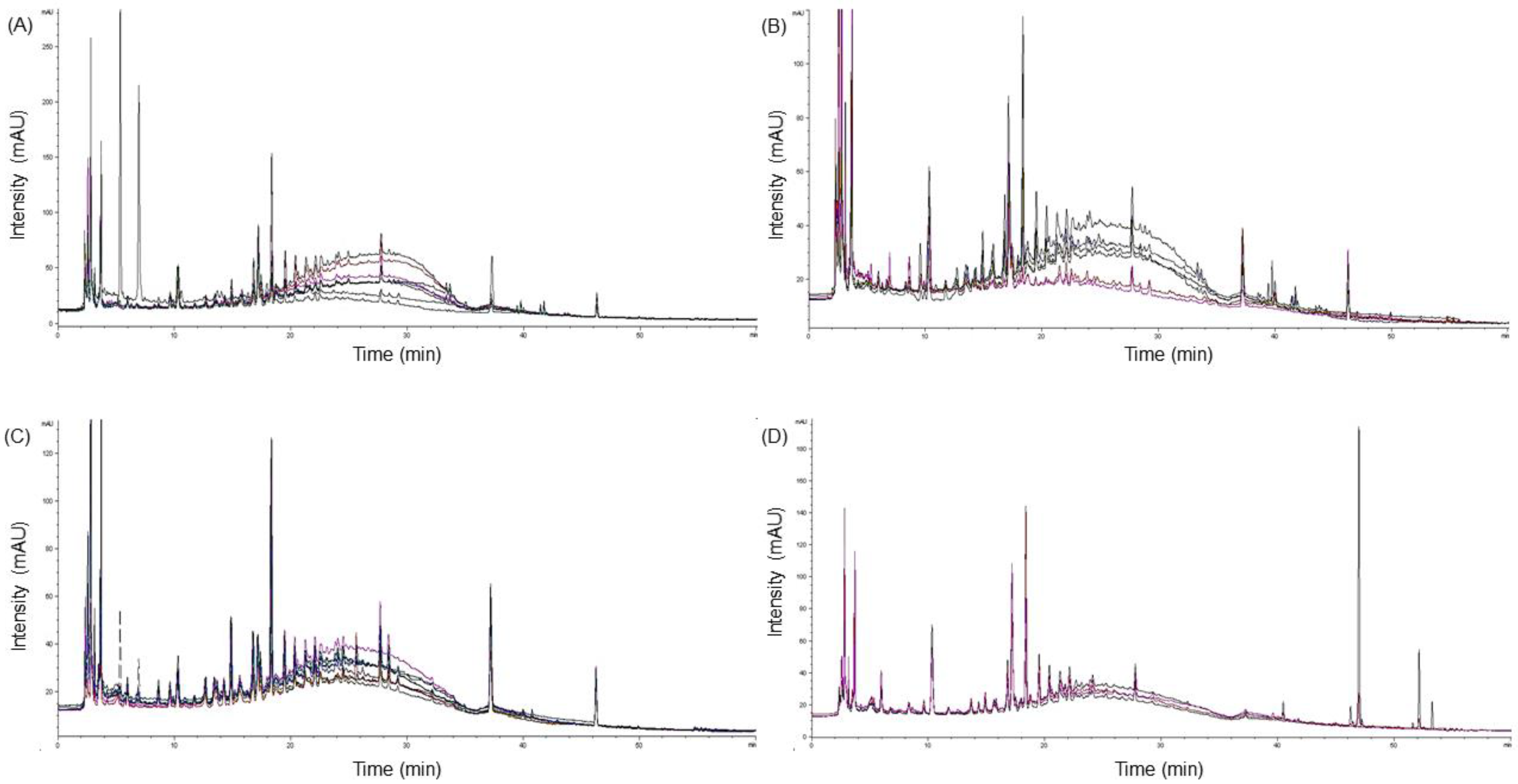
2.5. Chromatographic Profiling of Amomi Fructus Samples
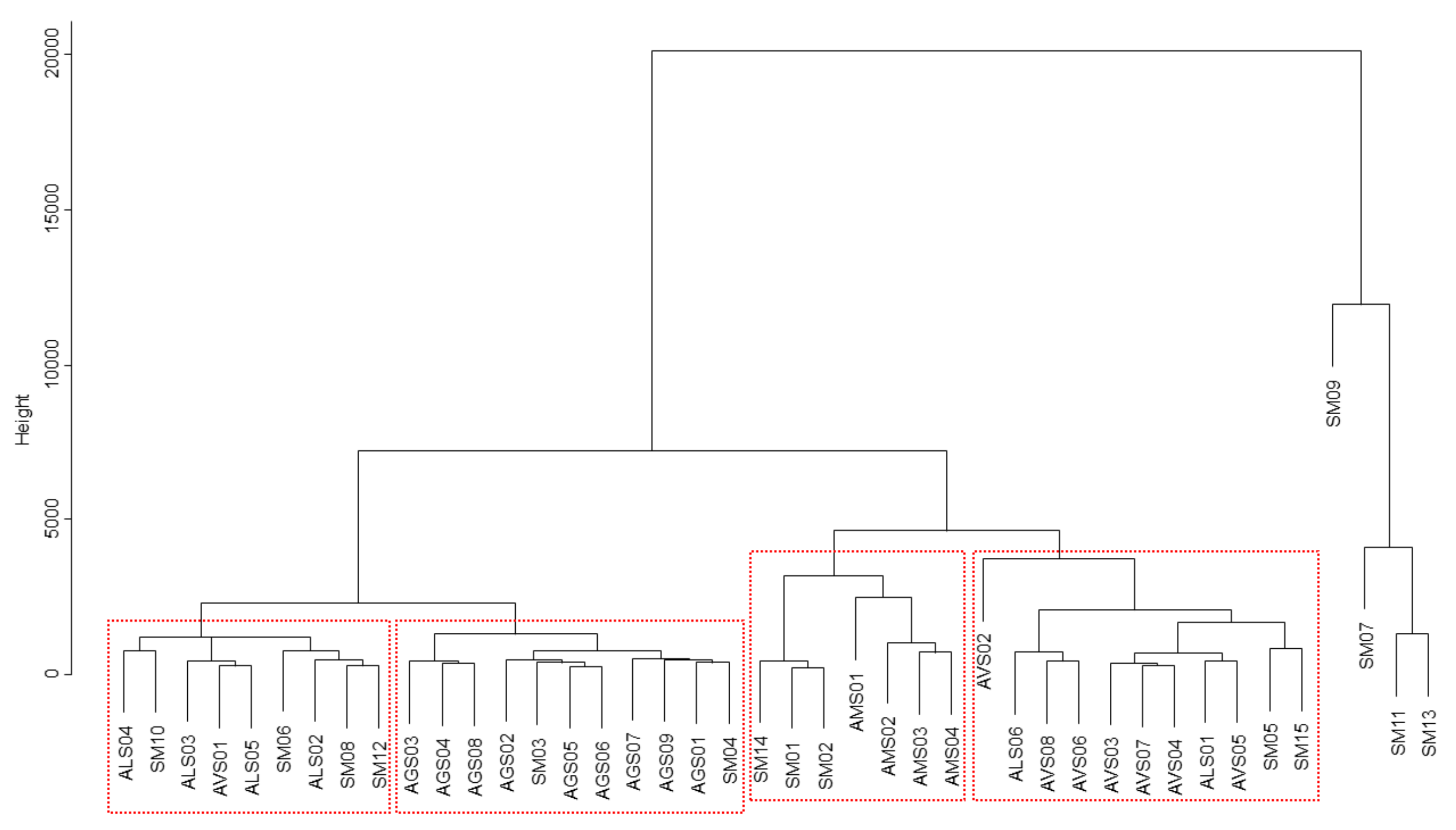
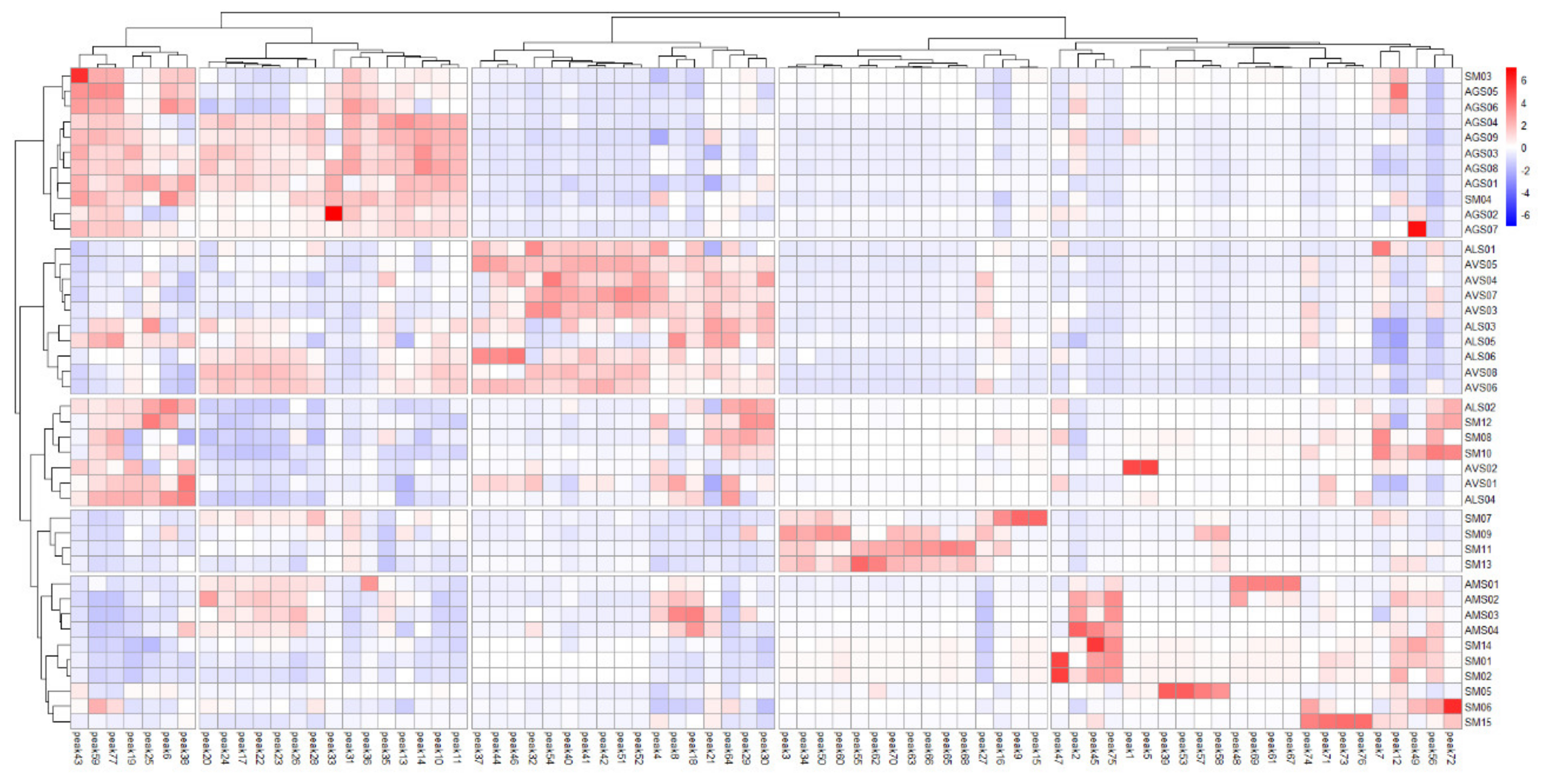
2.6. Clustering Analysis of Amomi Fructus Samples Using Chemometric Statistical Methods
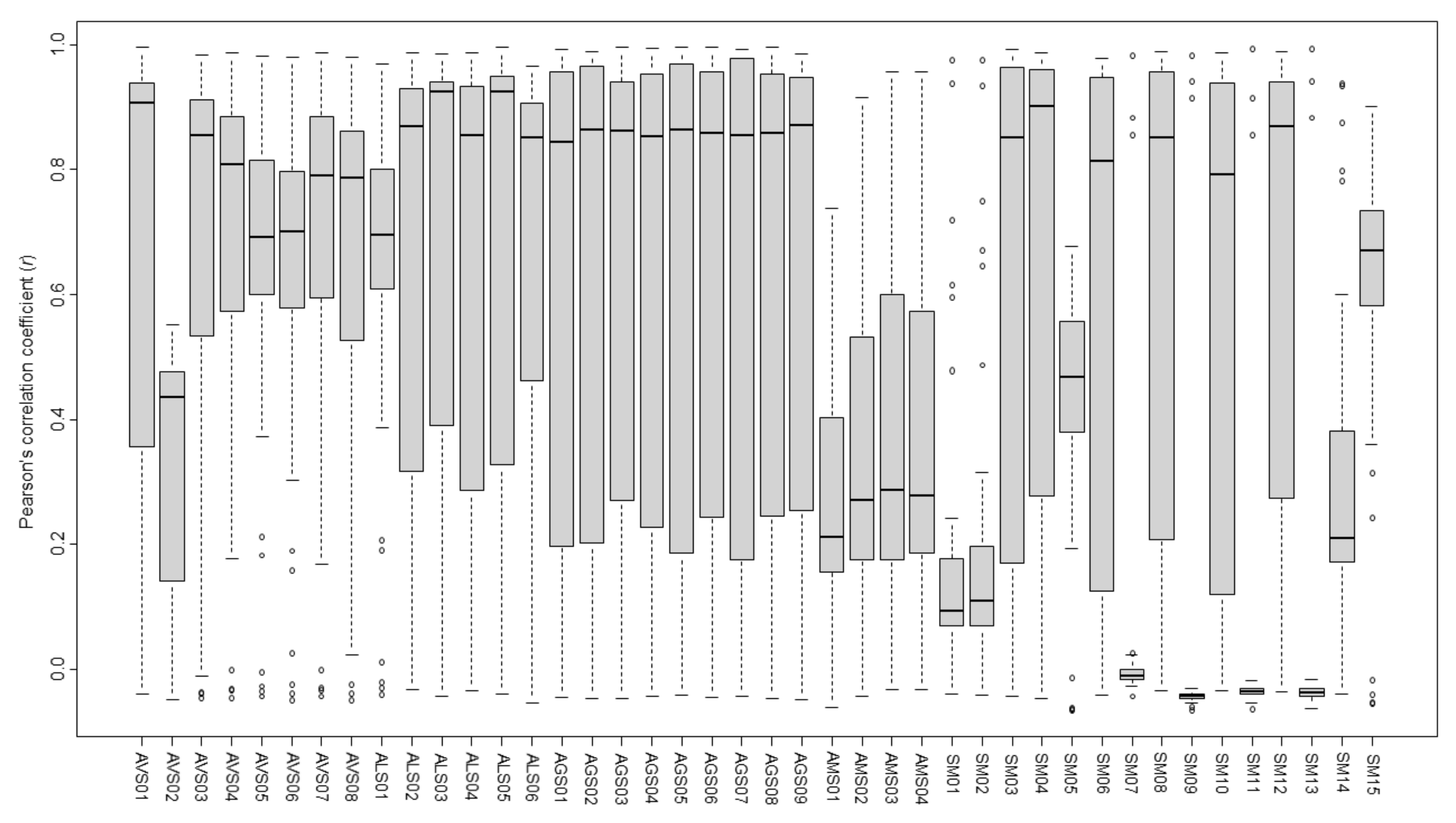
2.7. Similarity Evaluation of Amomi Fructus Samples Using the Pearson’s Correlation Coefficient
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Reagents
4.2. Preparation of Genomic DNA
4.3. Polymerase Chain Reaction (PCR) Amplification for DNA Barcode Analysis
4.4. Determination of DNA Sequences of PCR Product
4.5. Analysis of DNA Sequences and Preparation of the Dendrogram
4.6. Preparation of Samples for HPLC Analysis
4.7. HPLC Conditions for Chromatographic Profiling
4.8. Chemometric Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- The Ministry of Korean Food and Drug Safety. The Korean Pharmacopoeia, 12th ed.; The Ministry of Korean Food and Drug Safety, Ed.; KFDA Notification No. 2019-11, Monographs, Part II, 52; Ministry of Food and Drug Safety: Cheongju-si, Korea, 2019.
- Commission of Chinese Pharmacopeia. Pharmacopoeia of the Peoples Republic of China; China Medical Science Press: Beijing, China, 2015.
- Doh, E.J.; Kim, J.H.; Lee, G. Identification and monitoring of Amomi Fructus and its adulterants based on DNA barcoding analysis and designed DNA markers. Molecules 2019, 24, 4193. [Google Scholar] [CrossRef] [PubMed]
- Ao, H.; Wang, J.; Chen, L.; Li, S.M.; Dai, C.M. Comparison of volatile oil between the fruits of Amomum villosum Lour. and Amomum villosum Lour. var. xanthioides T. L. Wu et Senjen based on GC-MS and chemometric techniques. Molecules 2019, 24, 1663. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.M.; Kress, W.J.; Prince, L.M. Phylogenetic analyses of amomum (Alpinioideae: Zingiberaceae) using ITS and matK DNA sequence data. Syst. Bot. 2004, 29, 334–344. [Google Scholar] [CrossRef]
- Qiao, C.F.; Han, Q.B.; Zhao, Z.L.; Wang, Z.T.; Xu, L.S.; Xu, H.X. Sequence analysis based on ITS1 region of nuclear ribosomal DNA of Amomum villosum and ten species of Alpinia. J. Food Drug Anal. 2009, 17, 142–145. [Google Scholar] [CrossRef]
- Huang, Q.L.; Duan, Z.G.; Yang, J.F.; Ma, X.Y.; Zhan, R.T.; Xu, H.; Chen, W.W. SNP typing for germplasm identification of Amomum villosum Lour. based on DNA barcoding markers. PLoS ONE 2014, 9, e114940. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.X.; Chen, X.L.; Nie, L.P.; Sun, W.; Hu, H.Y.; Lin, Y.L.; Li, H.T.; Zheng, X.L.; Song, J.Y.; Yao, H. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 2019, 20, 4040. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W.; Zhang, W.X.; Zeng, P.; Hu, H.; Wang, Y.T.; Cai, J. The complete chloroplast genome of the essential medicinal herb, Amomum villosum (Zingiberaceae). Mitochondrial DNA B 2019, 4, 1798–1799. [Google Scholar] [CrossRef]
- Li, D.M.; Zhao, C.Y.; Zhu, G.F.; Xu, Y.C. Complete chloroplast genome sequence of Amomum villosum. Mitochondrial DNA B 2019, 4, 2673–2674. [Google Scholar] [CrossRef]
- Deng, C.H.; Wang, A.Q.; Shen, S.; Fu, D.X.; Chen, J.K.; Zhang, X.M. Rapid analysis of essential oil from Fructus Amomi by pressurized hot water extraction followed by solid-phase microextraction and gas chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2005, 38, 326–331. [Google Scholar] [CrossRef]
- Kang, W.Y.; Zhang, F.; Su, Y.; Guo, Y.L. Application of gas chromatography-quadrupole-time-of-flight-mass spectrometry for post-target analysis of volatile compounds in Fructus Amomi. Eur. J. Mass Spectrom. 2013, 19, 103–110. [Google Scholar] [CrossRef]
- Dai, D.N.; Huong, L.T.; Thang, T.D.; Ogunwande, I.A. Chemical composition of essential oils of Amomum villosum Lour. Am. J. Essent. Oils Nat. Prod. 2016, 4, 8–11. [Google Scholar]
- Yu, G.W.; Cheng, Q.; Nie, J.; Wang, X.J.; Wang, P.; Li, Z.G.; Lee, M.R. Microwave hydrodistillation based on deep eutectic solvent for extraction and analysis of essential oil from three Amomum species using gas chromatography–mass spectrometry. Chromatographia 2018, 81, 657–667. [Google Scholar] [CrossRef]
- An, X.Q.; Li, Z.Z.; Shen, L.G.; Si, J.Y. Chemical constituents of Amomum villosum Lour. Nat. Prod. Res. Dev. 2011, 23, 1021–1024. [Google Scholar]
- Chen, C.; Fu, C.; Ye, W.C.; Zhou, G.X. Study on phenolic constituents of Amomum villosum. J. Chin. Med. Mat. 2012, 35, 571–573. [Google Scholar]
- Wang, Y.; Shen, L.; Jiang, K.; Yin, G.; Wang, J.; Lu, Y.; Xiang, R.W.; Wang, T.J. Quality assessment of the traditional Chinese medicine Amomum villosum by chemical pattern recognition techniques. Chin. J. Pharm. Anal. 2016, 36, 1863–1869. [Google Scholar]
- Shen, L.; Wang, Y.; Jiang, K.; Yin, G.; Wang, J.; Lu, Y.; Wang, T.J. Comparison of HPLC fingerprints of Amomum villosum Lour., Amomum villosum Lour. var. xanthioides T. L. Wu et Senjen and Amomum longiligulare T. L. Wu. Chin. Pharm. J. 2016, 51, 1039–1043. [Google Scholar]
- Palhares, R.M.; Drummond, M.G.; Brasil, B.D.A.F.; Cosenza, G.P.; Brandao, M.D.L.; Oliveira, G. Medicinal plants recommended by the World Health Organization: DNA barcode identification associated with chemical analyses guarantees their quality. PLoS ONE 2015, 10, 0127866. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, B.M.; Salleh, F.M.; Omar, M.S.S.; Wagiran, A. Review: DNA barcoding and chromatography fingerprints for the authentication of botanicals in herbal medicinal products. Evid.-Based Complementary Altern. Med. 2017, 2017, 1352948. [Google Scholar]
- Han, J.P.; Shi, L.C.; Li, M.H.; Yao, H.; Song, J.Y.; Xu, H.X.; Sun, C.; Chen, S.L. Relationship between DNA barcoding and chemical classification of Salvia L. medicinal herbs. Planta Med. 2009, 75, 416–417. [Google Scholar] [CrossRef]
- Zhang, Z.P.; Zhang, Y.; Zhang, Z.; Yao, H.; Liu, H.T.; Zhang, B.G.; Liao, Y.H. Comparative analysis of DNA barcoding and HPLC fingerprint to trace species of Phellodendri Cortex, an important traditional Chinese medicine from multiple sources. Biol. Pharm. Bull. 2016, 39, 1325–1330. [Google Scholar] [CrossRef]
- Li, Y.P.; Geng, L.; Liu, Y.Y.; Chen, M.Y.; Mu, Q.R.; Zhang, X.; Zhang, Z.Y.; Ren, G.X.; Liu, C.S. Identification of three Daphne species by DNA barcoding and HPLC fingerprint analysis. PLoS ONE 2018, 13, 0201711. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Doh, E.J.; Lee, G. Evaluation of medicinal categorization of Atractylodes japonica Koidz. by using internal transcribed spacer sequencing analysis and HPLC fingerprinting combined with statistical tools. Evid.-Based Complementary Altern. Med. 2016, 2016, 2926819. [Google Scholar]
- Kim, J.H.; Doh, E.J.; Lee, G. Chemical differentiation of genetically identified Atractylodes japonica, A. macrocephala, and A. chinensis rhizomes using high-performance liquid chromatography with chemometric analysis. Evid.-Based Complementary Altern. Med. 2018, 2018, 4860371. [Google Scholar] [CrossRef] [PubMed]
- Larsen, K.; Lock, J.M.; Maas, H.; Maas, P.J.M. Zingiberaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Huber, H., Rudall, P.J., Stevens, P.S., Stutzei, T., Eds.; Springer: Heidelberg, Germany, 1998. [Google Scholar]
- Tong, S.Q. Zingiberaceae. In Flora Yunnanica, Tomus 8; Wu, Z.Y., Ed.; Science Press: Beijing, China, 1999. [Google Scholar]
- Droop, A.J.; Newman, M.F. A revision of Amomum (Zingiberaceae) in Sumatra. Edinb. J. Bot. 2014, 71, 193–258. [Google Scholar] [CrossRef]
- Hauke, J.; Kossowski, T. Comparison of values of Pearson’s and Spearman’s correlation coefficients on the same sets of data. Quaest. Geogr. 2011, 30, 87–93. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990. [Google Scholar]
- Fay, M.F.; Swensen, S.M.; Chase, M.W. Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull. 1997, 52, 111–201. [Google Scholar] [CrossRef]
- Cuenoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; Stahl, J.V.; Kelly, L.J.; Wikstrom, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Tate, J.A. Systematics and Evolution of Tarasa (Malvaceae): An Enigmatic Andean Polyploid Genus. Ph.D. Thesis, The University of Texas, Austin, TX, USA, 2002. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny. fr: New generation phylogenetic services for non-specialists. Nucleic Acid Res. 2019, 47, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kress, W.J.; Prince, L.M.; Williams, K.J. The phylogeny and a new classification of the gingers (Zingiberaceae): Evidence from molecular data. Am. J. Bot. 2002, 89, 1682–1696. [Google Scholar] [CrossRef]
Sample Availability: Samples are not available from the authors. |

| Code | Geographic Origin | Sample Type | Species Identification | Re-Code |
|---|---|---|---|---|
| AF01 | China | F | Amomum longiligulare | ALS01 |
| AF02 | China | F | A. ghaticum | AGS01 |
| AF03 | China | F | A. longiligulare | ALS02 |
| AF11 | Myanmar | S.P. | A. ghaticum | AGS02 |
| AF12 | China | F | A. villosum | AVS01 |
| AF14 | Myanmar | F | A. ghaticum | AGS03 |
| AF15 | Vietnam | S | A. microcarpum/A. longiligulare | SM01 |
| AF16 | Vietnam | S | A. microcarpum/A. longiligulare | SM02 |
| AF17 | Myanmar | S | A. ghaticum | AGS04 |
| AF21 | China | F | A. longiligulare | ALS03 |
| AF23 | Myanmar | S | A. ghaticum | AGS05 |
| AF24 | Myanmar | S | A. longiligulare/A. ghaticum | SM03 |
| AF25 | Myanmar | S | A. microcarpum | AMS01 |
| AF26 | Myanmar | S | A. ghaticum | AGS06 |
| AF27 | Myanmar | S.P. | A. ghaticum/A. uliginosum | SM04 |
| AF28 | Myanmar | S | A. longiligulare | ALS04 |
| AF29 | China | S | A. villosum | AVS02 |
| AF30 | Myanmar | S | A. microcarpum | AMS02 |
| AF31 | Myanmar | S | A. ghaticum | AGS07 |
| AF32 | Myanmar | S | A. ghaticum | AGS08 |
| AF33 | Myanmar | S | A. microcarpum | AMS03 |
| AF34 | Myanmar | S | A. ghaticum | AGS09 |
| AF35 | Vietnam | P | A. villosum/A. uliginosum/A. villosum var. villosum | SM05 |
| AF41 | Vietnam | F | A. longiligulare/A. villosum | SM06 |
| AF42 | Vietnam | F | A. tomrey var. tomrey/A. koenigii | SM07 |
| AF43 | Vietnam | F | A. longiligulare/A. villosum | SM08 |
| AF44 | Vietnam | F | A. koenigii/A. tomrey var. tomrey/A. chinense | SM09 |
| AF45 | Vietnam | F | A. longiligulare/A. villosum | SM10 |
| AF46 | Vietnam | F | A. koenigii/A. echinocarpum/A. chinense | SM11 |
| AF47 | Vietnam | F | A. longiligulare/A. villosum | SM12 |
| AF48 | Vietnam | F | A. koenigii/A. echinocarpum/A. chinense | SM13 |
| AF50 | China | F | A. villosum | AVS07 |
| AF51 | China | F | A. villosum | AVS08 |
| AF52 | China | F | A. longiligulare | ALS05 |
| AF53 | China | F | A. villosum | AVS03 |
| AF54 | China | F | A. villosum | AVS04 |
| AF55 | Myanmar | S.P. | A. longiligulare/A. microcarpum/A. villosum | SM14 |
| AF56 | China | F | A. villosum | AVS05 |
| AF57 | China | F | A. villosum | AVS06 |
| AF58 | China | F | A. longiligulare | ALS06 |
| AF59 | Vietnam | S | A. villosum/A. uliginosum/A. villosum var. villosum | SM15 |
| AF60 | China | S | A. microcarpum | AMS04 |
| No. | Accession Code | Scientific Name | Medicinal Name |
|---|---|---|---|
| 1 | AV01 | Amomum villosum Lour. (= Wurfbainia villosa (Lour.) Skornick. and A.D. Poulsen) | Amomi Fructus a |
| 2 | AV02 | ||
| 3 | AV03 | ||
| 4 | AL01 | Amomum longiligulare T.L. Wu (= Wurfbainia longiligularis (T.L. Wu) Skornick. and A.D. Poulsen) | Amomi Fructus b |
| 5 | AL02 | ||
| 6 | AL03 | ||
| 7 | AK01 | Amomum verum Blackw. (= Amomum krervanh Pierre ex Gagnep.) | Amomi Fructus Rotundus |
| 8 | AK02 | ||
| 9 | AC01 | Amomum compactum Sol. ex Maton | |
| 10 | AC02 | ||
| 11 | ATK01 | Amomum tsao-ko Crevost and Lemarié (= Amomum tsaoko) | Amomi tsao-ko Fructus |
| 12 | ATK02 | ||
| 13 | AH01 | Alpinia hainanensis K. Schum. (= Alpinia katsumadae Hayata) | Alpiniae katsumadai Semen |
| 14 | AH02 | ||
| 15 | AO01 | Alpinia oxyphylla Miq. | Alpiniae oxyphyllae Fructus |
| 16 | AO02 | ||
| 17 | AOR01 | Alpinia officinarum Hanc | Alpiniae officinari Rhizoma |
| 18 | AOR02 | ||
| 19 | ACC01 | Alpinia conchigera | jie bian shan jiang c |
| 20 | ACC02 | ||
| 21 | AZ01 | Alpinia zerumbet | yan shan jiang c |
| 22 | AZ02 | ||
| 23 | AM01 | Alpinia malaccensis(N.L.Burman) Roscoe | mao ban shan jiang c |
| 24 | AG01 | Alpinia galanga (L.) Willd. | Galangae Fructus |
| 25 | EC01 | Elettaria cardamomum (L.) Maton (= Amomum cardamomum L., Alpinia cardamomum (L.) Roxb.) | Cardamomi Fructus |
| 26 | EC02 |
| Barcode Target | Amplicon Size (bp) | Aligned Length (bp) | Conserved Sites | Variable Sites | Parsimony Informative Sites | Singleton Site |
|---|---|---|---|---|---|---|
| ITS | 670 | 645–665 | 492 | 185 | 164 | 21 |
| matk | 940 | 933 | 885 | 48 | 48 | none |
| rbcL | 750 | 743 | 657 | 86 | 12 | 74 |
| psbA-trnH | 830 | 800–836 | 820 | 49 | 35 | 14 |
| trnL-F intergenic spacer | 420 | 395–415 | 368 | 55 | 19 | 36 |
| matk + rbcL | - | 1676 | 1542 | 134 | 60 | 74 |
| psbA-trnH + trnL-F intergenic spacer | - | 1195-1251 | 1188 | 104 | 54 | 50 |
| mark + rbcL + psbA-trnH | - | 2479–2512 | 2362 | 183 | 95 | 88 |
| mark + rbcL + trnL-F intergenic spacer | - | 2071–2091 | 1910 | 189 | 79 | 110 |
| Four plastid targets | - | 2874–2927 | 2730 | 238 | 114 | 124 |
| Sample | Value | AVS | ALS | AGS | AMS | SM |
|---|---|---|---|---|---|---|
| AVS | Mean | 0.785 | ||||
| Median | 0.888 | |||||
| Max | 0.987 | |||||
| Min | 0.303 | |||||
| ALS | Mean | 0.829 | 0.895 | |||
| Median | 0.870 | 0.934 | ||||
| Max | 0.995 | 0.988 | ||||
| Min | 0.385 | 0.769 | ||||
| AGS | Mean | 0.767 | 0.883 | 0.980 | ||
| Median | 0.802 | 0.911 | 0.982 | |||
| Max | 0.949 | 0.963 | 0.995 | |||
| Min | 0.436 | 0.680 | 0.950 | |||
| AMS | Mean | 0.449 | 0.352 | 0.199 | 0.802 | |
| Median | 0.514 | 0.323 | 0.197 | 0.827 | ||
| Max | 0.711 | 0.645 | 0.271 | 0.958 | ||
| Min | 0.059 | 0.187 | 0.154 | 0.631 | ||
| SM | Mean | 0.427 | 0.466 | 0.474 | 0.267 | 0.306 |
| Median | 0.474 | 0.464 | 0.507 | 0.180 | 0.091 | |
| Max | 0.959 | 0.985 | 0.992 | 0.874 | 0.994 | |
| Min | −0.049 | −0.053 | −0.048 | −0.060 | −0.065 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doh, E.-J.; Lee, G.; Jung, H.-J.; Kwon, K.-B.; Kim, J.-H. Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis. Molecules 2020, 25, 4581. https://doi.org/10.3390/molecules25194581
Doh E-J, Lee G, Jung H-J, Kwon K-B, Kim J-H. Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis. Molecules. 2020; 25(19):4581. https://doi.org/10.3390/molecules25194581
Chicago/Turabian StyleDoh, Eui-Jeong, Guemsan Lee, Hyun-Jong Jung, Kang-Beom Kwon, and Jung-Hoon Kim. 2020. "Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis" Molecules 25, no. 19: 4581. https://doi.org/10.3390/molecules25194581
APA StyleDoh, E.-J., Lee, G., Jung, H.-J., Kwon, K.-B., & Kim, J.-H. (2020). Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis. Molecules, 25(19), 4581. https://doi.org/10.3390/molecules25194581