Modeling the Antileukemia Activity of Ellipticine-Related Compounds: QSAR and Molecular Docking Study
Abstract
1. Introduction
2. Results
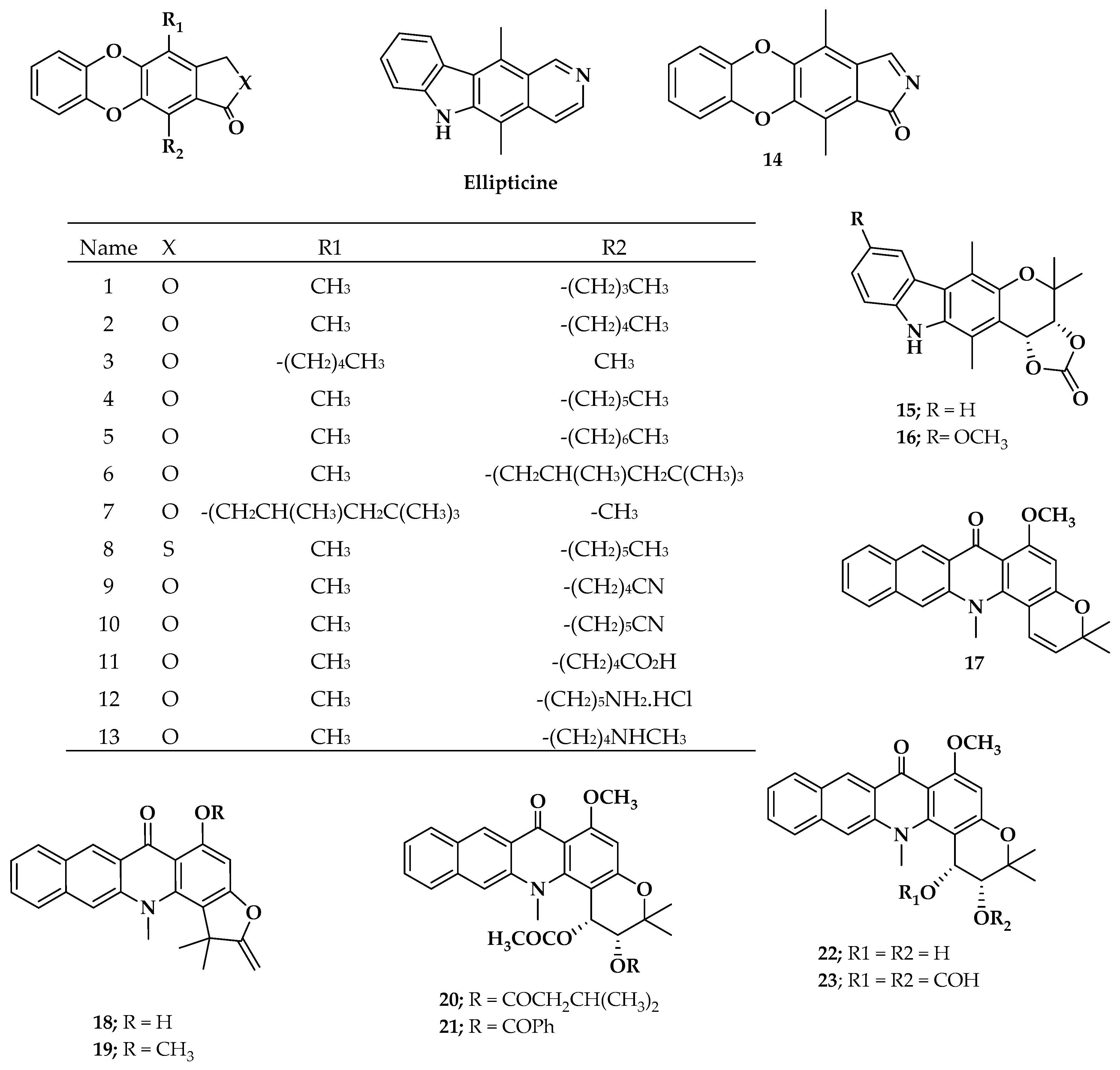
2.1. Molecular Modeling
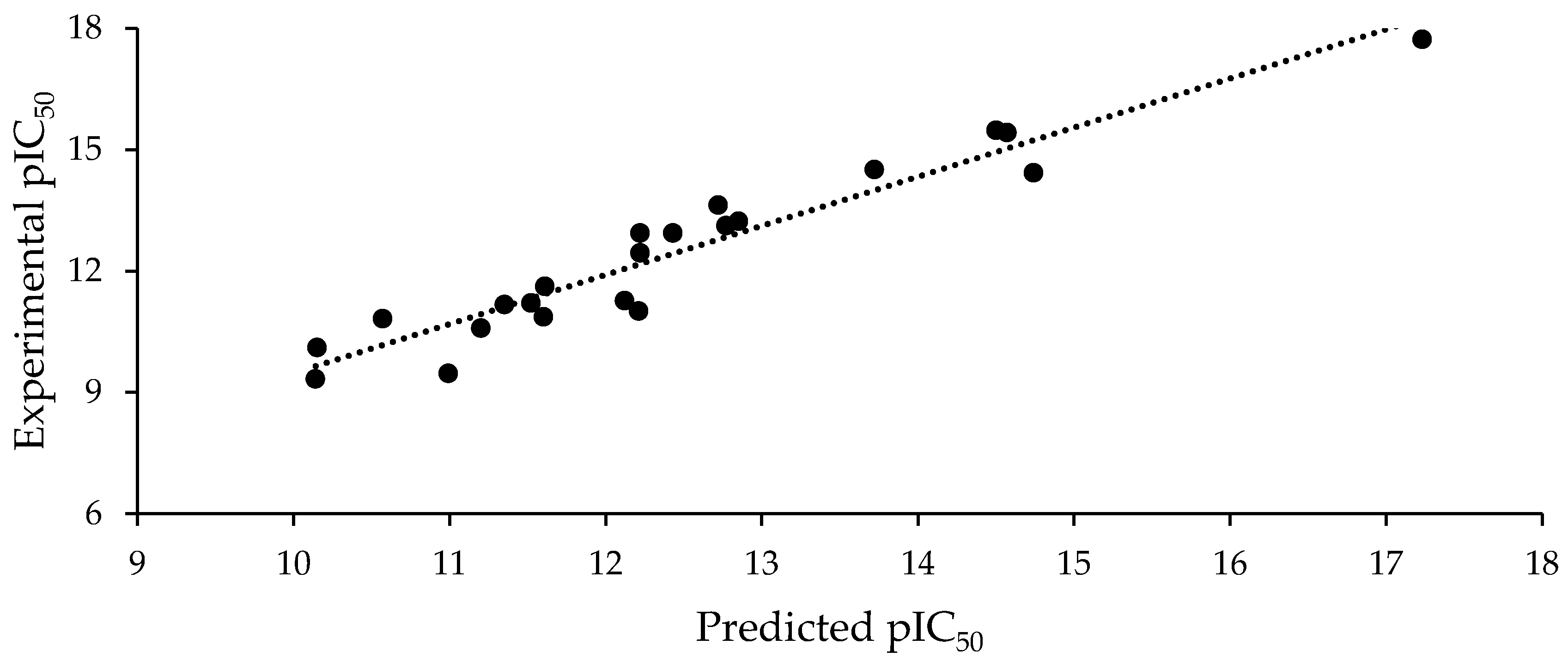
2.2. Statistical Analysis
QSAR Model
R2 = 0.798, F = 25.0, s = 1.023, Q2 = 0.650, a(R2) = 0.064, a(Q2) = −0.453
R2 = 0.652, F = 11.9, s = 1.342, Q2 = 0.506, a(R2) = 0.036, a(Q2) = −0.287
R2 = 0.836, F = 23.0, s = 0.946, Q2 = 0.729, a(R2) = 0.126, a(Q2) = −0.505
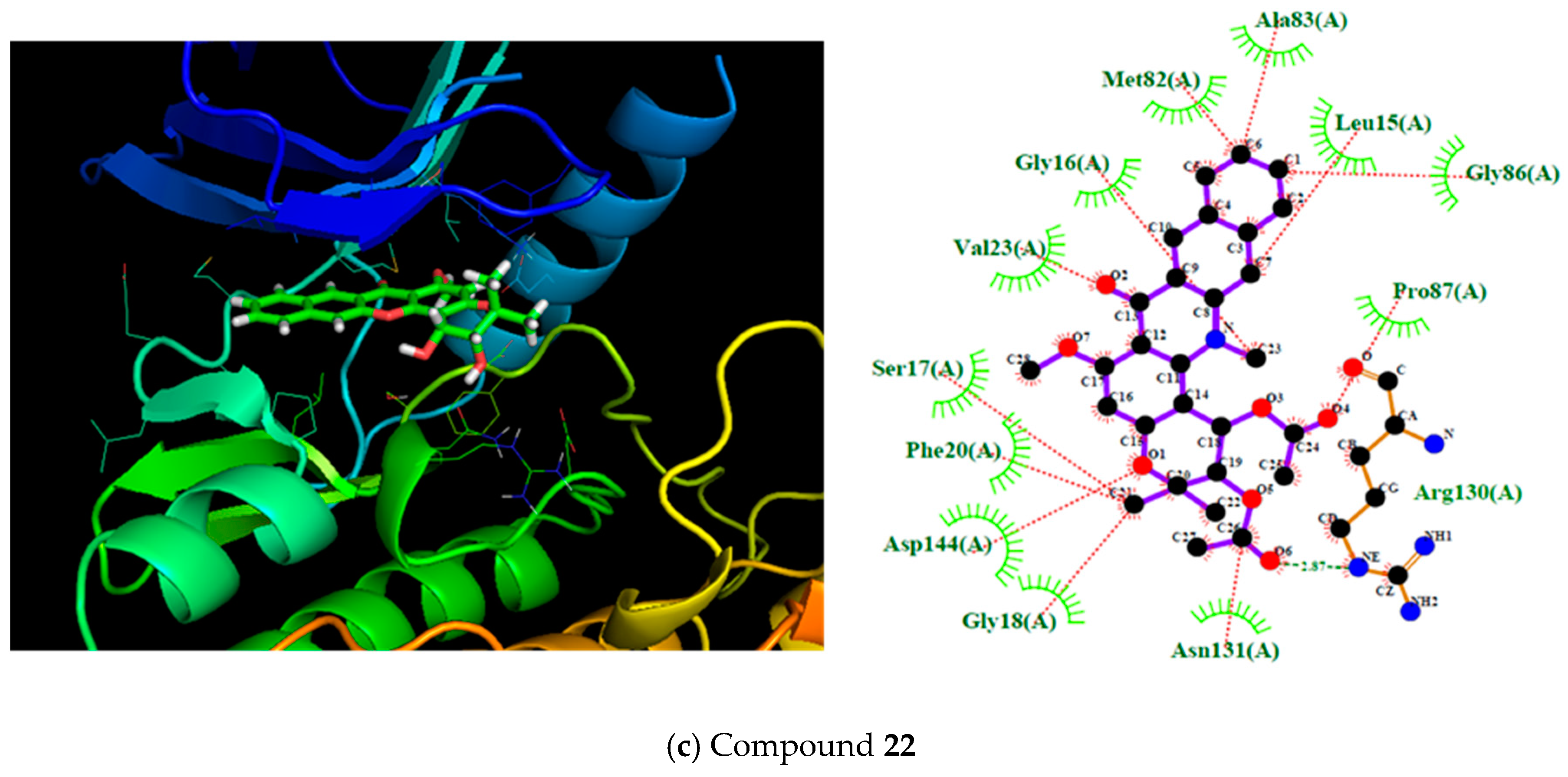
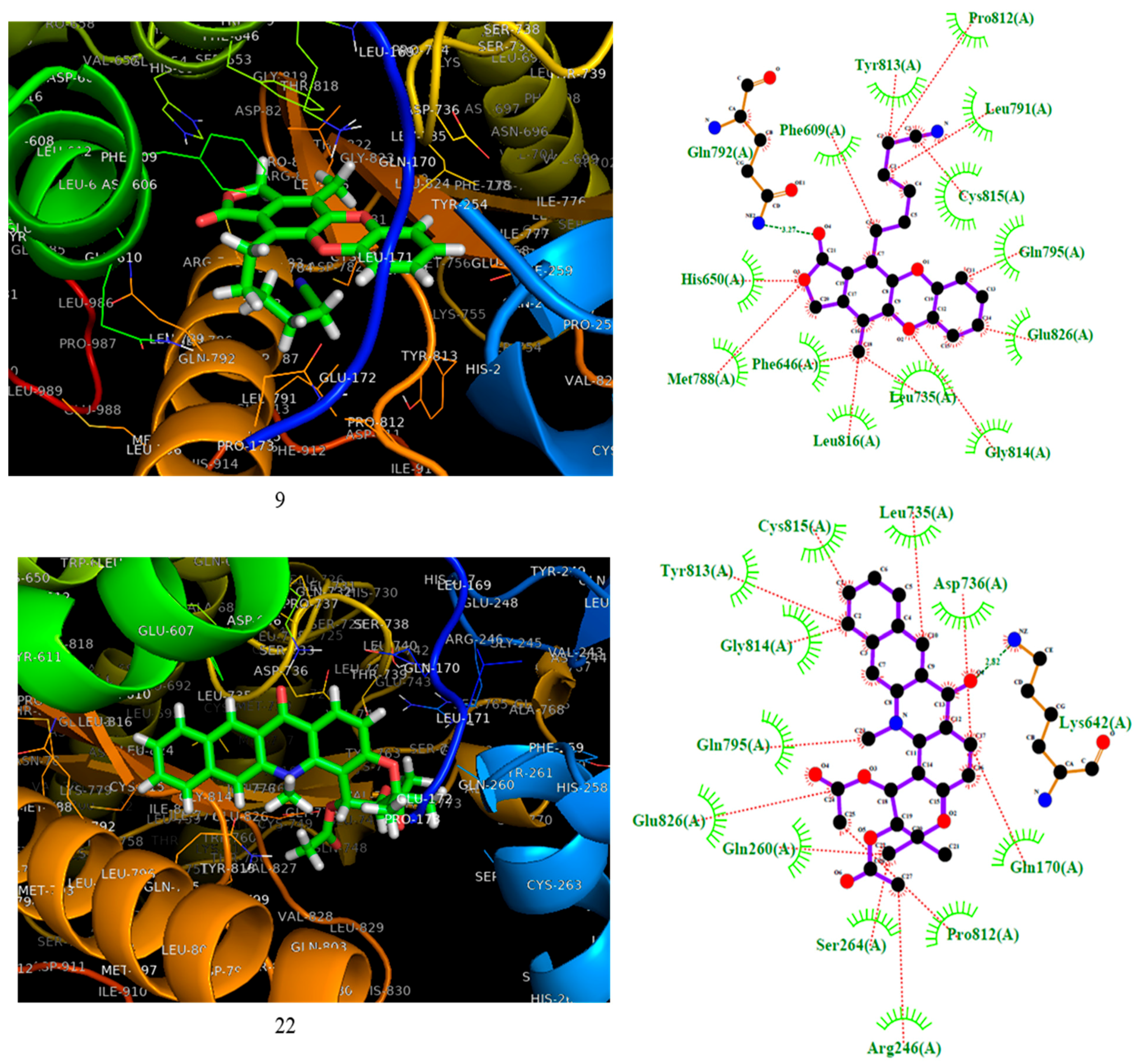
2.3. Molecular Docking
3. Materials and Methods
3.1. Pharmacological Data Collected
3.2. Molecular Modeling
3.3. Statistical Analysis
3.3.1. Quantitative Structure–Activity Relationship (QSAR) and Statistical Validation
3.3.2. Molecular Docking
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Song, M.; Giovannucci, E.L. Cancer risk: Many factors contribute. Science 2015, 347, 728–729. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Facts and Statistics|Leukemia and Lymphoma Society. Available online: https://www.lls.org/facts-and-statistics/facts-and-statistics-overview/facts-and-statistics (accessed on 18 December 2019).
- Hafez, H.A.; Soliaman, R.M.; Bilal, D.; Hashem, M.; Shalaby, L.M. Early Deaths in Pediatric Acute Leukemia: A Major Challenge in Developing Countries. J. Pediatr. Hematol. Oncol. 2019, 41, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Jain, N.; Ayer, T.; Wierda, W.G.; Flowers, C.R.; O’Brien, S.M.; Keating, M.J.; Kantarjian, H.M.; Chhatwal, J. Economic burden of chronic lymphocytic leukemia in the era of oral targeted therapies in the United States. J. Clin. Oncol. 2017, 35, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Vedi, A.; Mitchell, R.; Shanmuganathan, S.; Oswald, C.; Marshall, G.M.; Trahair, T.; Sivarajasingam, S.; Ziegler, D.S. Increased Survival for Children With Acute Myeloid Leukemia Results From Improved Postrelapse Treatment. J. Pediatric Hematol. Oncol. 2018, 40, 541–547. [Google Scholar] [CrossRef]
- Lins, M.M.; Santos, M.O.; Albuquerque, M.F.P.M.; de Castro, C.C.L.; Mello, M.J.G.; de Camargo, B. Incidence and survival of childhood leukemia in Recife, Brazil: A population-based analysis. Pediatric Blood Cancer 2017, 64, e26391. [Google Scholar] [CrossRef]
- Corella Aznar, E.G.; Ayerza Casas, A.; Carboné Bañeres, A.; Calvo Escribano, M.Á.C.; Labarta Aizpún, J.I.; Samper Villagrasa, P. Quality of life and chronic health conditions in childhood acute leukaemia survivors. Med. Clínica 2019, 152, 167–173. [Google Scholar] [CrossRef]
- Giannopoulos, K. Targeting immune signaling checkpoints in acute myeloid leukemia. J. Clin. Med. 2019, 8, 236. [Google Scholar] [CrossRef]
- Hoseini, S.S.; Cheung, N.K. Acute myeloid leukemia targets for bispecific antibodies. Blood Cancer J. 2017, 7, e522. [Google Scholar] [CrossRef]
- Charmsaz, S.; Al-Ejeh, F.; Yeadon, T.M.; Miller, K.J.; Smith, F.M.; Stringer, B.W.; Moore, A.S.; Lee, F.-T.; Cooper, L.T.; Stylianou, C.; et al. EphA3 as a target for antibody immunotherapy in acute lymphoblastic leukemia. Leukemia 2017, 31, 1779–1787. [Google Scholar] [CrossRef]
- Patrussi, L.; Capitani, N.; Baldari, C.T. Abnormalities in chemokine receptor recycling in chronic lymphocytic leukemia. Cell. Mol. Life Sci. 2019, 76, 3249–3261. [Google Scholar] [CrossRef] [PubMed]
- Flores-Sumoza, M.; Alcázar, J.J.; Márquez, E.; Mora, J.R.; Lezama, J.; Puello, E. Classical QSAR and docking simulation of 4-pyridone derivatives for their antimalarial activity. Molecules 2018, 23, 3166. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.R.; Márquez, E.A.; Calle, L. Computational molecular modelling of N-cinnamoyl and hydroxycinnamoyl amides as potential α-glucosidase inhibitors. Med. Chem. Res. 2018, 27, 2214–2223. [Google Scholar] [CrossRef]
- Cabrera, N.; Mora, J.R.; Marquez, E.A. Computational Molecular Modeling of Pin1 Inhibition Activity of Quinazoline, Benzophenone, and Pyrimidine Derivatives. J. Chem. 2019, 2019, 11. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Das, R.N. Chapter 3—Classical QSAR. In Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Roy, K., Kar, S., Das, R.N., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 81–102. [Google Scholar]
- Verma, J.; Khedkar, V.; Coutinho, E. 3D-QSAR in Drug Design—A Review. Curr. Top. Med. Chem. 2010, 10, 95–115. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar]
- Gupta, M.; Sharma, R.; Kumar, A. Docking techniques in pharmacology: How much promising? Comput. Biol. Chem. 2018, 76, 210–217. [Google Scholar] [CrossRef]
- Lushington, G.H.; Guo, J.X.; Wang, J.L. Whither Combine? New Opportunities for Receptor-Based QSAR. Curr. Med. Chem. 2007, 14, 1863–1877. [Google Scholar] [CrossRef]
- Talevi, A.; Gavernet, L.; Bruno-Blanch, L. Combined Virtual Screening Strategies. Curr. Comput. Aided-Drug Des. 2009, 5, 23–37. [Google Scholar] [CrossRef]
- Rasulev, B. Recent Developments in 3D QSAR and Molecular Docking Studies of Organic and Nanostructures. In Handbook of Computational Chemistry; Leszczynski, J., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2016; pp. 1–29. [Google Scholar]
- Danielson, M.L.; Hu, B.; Shen, J.; Desai, P.V. In Silico ADME Techniques Used in Early-Phase Drug Discovery. In Translating Molecules into Medicines; Bhattachar, S.N., Morrison, J.S., Mudra, D.R., Bender, D.M., Eds.; Springer International Publishing: Cham, Bayern, Deutschland, 2017; Volumn 25, pp. 81–117. [Google Scholar]
- Halder, A.K.; Moura, A.S.; Cordeiro, M.N.D.S. QSAR modelling: A therapeutic patent review 2010–present. Expert Opin. Ther. Pat. 2018, 28, 467–476. [Google Scholar] [CrossRef]
- Chen, Y.C. Beware of docking! Trends Pharmacol. Sci. 2015, 36, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Caflisch, A. Protein structure-based drug design: from docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018, 48, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Letis, A.S.; Seo, E.J.; Nikolaropoulos, S.S.; Efferth, T.; Giannis, A.; Fousteris, M.A. Synthesis and cytotoxic activity of new artemisinin hybrid molecules against human leukemia cells. Bioorganic Med. Chem. 2017, 25, 3357–3367. [Google Scholar] [CrossRef] [PubMed]
- Arthur, D.E.; Uzairu, A.; Mamza, P.; Abechi, S.E.; Shallangwa, G. Activity and toxicity modelling of some NCI selected compounds against leukemia P388ADR cell line using genetic algorithm-multiple linear regressions. J. King Saud Univ.-Sci. 2018. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Y.; Liu, N.; Li, L.; Xiao, S.; Li, X.; Chen, K.; Luo, C.; Chen, S.; Chen, H. Design, synthesis and anti leukemia cells proliferation activities of pyrimidylaminoquinoline derivatives as DOT1L inhibitors. Bioorg. Chem. 2018, 80, 649–654. [Google Scholar] [CrossRef]
- Melge, A.R.; Kumar, L.G.; Pavithran, K.; Nair, S.V.; Manzoor, K.; Gopi Mohan, C. Predictive models for designing potent tyrosine kinase inhibitors in chronic myeloid leukemia for understanding its molecular mechanism of resistance by molecular docking and dynamics simulations. J. Biomol. Struct. Dyn. 2019, 37, 4747–4766. [Google Scholar] [CrossRef]
- Cheng, G.; Wang, Z.; Yang, J.; Bao, Y.; Xu, Q.; Zhao, L.; Liu, D. Design, synthesis and biological evaluation of novel indole derivatives as potential HDAC/BRD4 dual inhibitors and anti-leukemia agents. Bioorg. Chem. 2019, 84, 410–417. [Google Scholar] [CrossRef]
- Furlan, V.; Konc, J.; Bren, U. Inverse molecular docking as a novel approach to study anticarcinogenic and anti-neuroinflammatory effects of curcumin. Molecules 2018, 23, 3351. [Google Scholar] [CrossRef]
- Canals, A.; Purciolas, M.; Aymamí, J.; Coll, M. The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 1009–1012. [Google Scholar] [CrossRef]
- Stiborová, M.; Poljaková, J.; Martínková, E.; Bořek-Dohalská, L.; Eckschlager, T.; Kizek, R.; Frei, E. Ellipticine cytotoxicity to cancer cell lines-a comparative study. Interdiscip. Toxicol. 2011, 4, 98–105. [Google Scholar] [CrossRef]
- Miller, C.M.; O’sullivan, E.C.; McCarthy, F.O. Novel 11-substituted ellipticines as potent anticancer agents with divergent activity against cancer cells. Pharmaceuticals 2019, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Bramhananda Reddy, N.; Burra, V.R.; Ravindranath, L.K.; Naresh Kumar, V.; Sreenivasulu, R.; Sadanandam, P. Synthesis and biological evaluation of benzimidazole fused ellipticine derivatives as anticancer agents. Mon. Fur Chem. 2016, 147, 599–604. [Google Scholar] [CrossRef]
- Vann, K.R.; Ergün, Y.; Zencir, S.; Oncuoglu, S.; Osheroff, N.; Topcu, Z. Inhibition of human DNA topoisomerase IIα by two novel ellipticine derivatives. Bioorganic. Med. Chem. Lett. 2016, 26, 1809–1812. [Google Scholar] [CrossRef] [PubMed]
- Russell, E.G.; Guo, J.; O’Sullivan, E.C.; O’Driscoll, C.M.; McCarthy, F.O.; Cotter, T.G. 7-formyl-10-methylisoellipticine, a novel ellipticine derivative, induces mitochondrial reactive oxygen species (ROS) and shows anti-leukaemic activity in mice. Invest. New Drugs 2016, 34, 15–23. [Google Scholar] [CrossRef]
- Madhavi, S.; Sreenivasulu, R.; Raju, R.R. Synthesis and biological evaluation of oxadiazole incorporated ellipticine derivatives as anticancer agents. Mon. Fur Chem. 2017, 148, 933–938. [Google Scholar] [CrossRef]
- Pujol, M.; Romero, M.; Sanchez, I. Synthesis and Biological Activity of New Class of Dioxygenated Anticancer Agents. Curr. Med. Chem. Agents 2005, 5, 215–237. [Google Scholar] [CrossRef]
- Romero, M.; Renard, P.; Caignard, D.-H.; Atassi, G.; Solans, X.; Constans, P.; Bailly, C.; Pujol, M.D. Synthesis and Structure–Activity Relationships of New Benzodioxinic Lactones as Potential Anticancer Drugs. J. Med. Chem. 2007, 50, 294–307. [Google Scholar] [CrossRef]
- Walters, W.P. Going further than Lipinski’s rule in drug design. Expert Opin. Drug Discov. 2012, 7, 99–107. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian16 Revision A.03; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Luiggi, M.; Mora, J.R.; Loroño, M.; Marquez, E.; Lezama, J.; Cordova, T.; Chuchani, G. Theoretical calculations on the gas-phase thermal decomposition kinetics of selected thiomethyl chloroalkanes: A new insight of the mechanism. Comput. Theor. Chem. 2014, 1027, 165–172. [Google Scholar] [CrossRef]
- Mora, J.R.; Cervantes, C.; Marquez, E. New insight into the chloroacetanilide herbicide degradation mechanism through a nucleophilic attack of hydrogen sulfide. Int. J. Mol. Sci. 2018, 19, 2864. [Google Scholar] [CrossRef] [PubMed]
- Lezama, J.; Márquez, E.; Mora, J.R.; Córdova, T.; Chuchani, G. Theoretical calculations on the mechanisms of the gas phase elimination kinetics of chlorocyclohexane, 3-chlorocyclohexene and 4-chlorocyclohexene. J. Mol. Struct. THEOCHEM 2009, 916, 17–22. [Google Scholar] [CrossRef]
- Marquez, E.; Domínguez, R.M.; Mora, J.R.; Córdova, T.; Chuchani, G. Experimental and theoretical studies of the homogeneous, unimolecular gas-phase elimination kinetics of trimethyl orthovalerate and trimethyl orthochloroacetate. J. Phys. Chem. A 2010, 114, 4203–4209. [Google Scholar] [CrossRef] [PubMed]
- McQuarrie, D.A. Statistical Mechanics; University Science Books: Sausalito, CA, USA, 2000; ISBN 978-1-891389-15-3. [Google Scholar]
- ChemAxon-Software Solutions and Services for Chemistry & Biology. Available online: https://chemaxon.com/ (accessed on 18 December 2019).
- García-Jacas, C.R.; Marrero-Ponce, Y.; Acevedo-Martínez, L.; Barigye, S.J.; Valdés-Martiní, J.R.; Contreras-Torres, E. QuBiLS-MIDAS: A parallel free-software for molecular descriptors computation based on multilinear algebraic maps. J. Comput. Chem. 2014, 35, 1395–1409. [Google Scholar] [CrossRef]
- Jungwirth, U.; Kowol, C.R.; Keppler, B.K.; Hartinger, C.G.; Berger, W.; Heffeter, P. Anticancer activity of metal complexes: Involvement of redox processes. Antioxid. Redox Signal. 2011, 15, 1085–1127. [Google Scholar] [CrossRef]
- Zhang, P.; Sadler, P.J. Redox-Active Metal Complexes for Anticancer Therapy. Eur. J. Inorg. Chem. 2017, 2017, 1541–1548. [Google Scholar] [CrossRef]
- Blunt, C.E.; Torcuk, C.; Liu, Y.; Lewis, W.; Siegel, D.; Ross, D.; Moody, C.J. Synthesis and Intracellular Redox Cycling of Natural Quinones and Their Analogues and Identification of Indoleamine-2,3-dioxygenase (IDO) as Potential Target for Anticancer Activity. Angew. Chem. Int. Ed. 2015, 54, 8740–8745. [Google Scholar] [CrossRef]
- Novak Jovanović, I.; Jadreško, D.; Miličević, A.; Hranjec, M.; Perin, N. An electrochemical study on the redox chemistry of cyclic benzimidazole derivatives with potent anticancer activity. Electrochim. Acta 2019, 297, 452–462. [Google Scholar] [CrossRef]
- Geerlings, P.; De Proft, F.; Langenaeker, W. Conceptual Density Functional Theory. Chem. Rev. 2003, 103, 1793–1874. [Google Scholar] [CrossRef]
- Sandoval-Yañez, C.; Mascayano, C.; Martínez-Araya, J.I. A theoretical assessment of antioxidant capacity of flavonoids by means of local hyper–softness. Arab. J. Chem. 2018, 11, 554–563. [Google Scholar] [CrossRef]
- Perea-Ramírez, L.I.; Guevara-García, A.; Galván, M. Using local softness to reveal oxygen participation in redox processes in cathode materials. J. Mol. Model. 2018, 24, 227. [Google Scholar] [CrossRef] [PubMed]
- Willems, R.E.M.; Weijtens, C.H.L.; de Vries, X.; Coehoorn, R.; Janssen, R.A.J. Relating Frontier Orbital Energies from Voltammetry and Photoelectron Spectroscopy to the Open-Circuit Voltage of Organic Solar Cells. Adv. Energy Mater. 2019, 9, 1803677. [Google Scholar] [CrossRef]
- Tagade, P.M.; Adiga, S.P.; Park, M.S.; Pandian, S.; Hariharan, K.S.; Kolake, S.M. Empirical Relationship between Chemical Structure and Redox Properties: Mathematical Expressions Connecting Structural Features to Energies of Frontier Orbitals and Redox Potentials for Organic Molecules. J. Phys. Chem. C 2018, 122, 11322–11333. [Google Scholar] [CrossRef]
- Meneses-Marcel, A.; Marrero-Ponce, Y.; Machado-Tugores, Y.; Montero-Torres, A.; Pereira, D.M.; Escario, J.A.; Nogal-Ruiz, J.J.; Ochoa, C.; Arán, V.J.; Martínez-Fernández, A.R.; et al. A linear discrimination analysis based virtual screening of trichomonacidal lead-like compounds: Outcomes of in silico studies supported by experimental results. Bioorganic Med. Chem. Lett. 2005, 15, 3838–3843. [Google Scholar] [CrossRef] [PubMed]
- Davids, M.S.; Brown, J.R. Targeting the B cell receptor pathway in chronic lymphocytic leukemia. Leuk. Lymphoma 2012, 53, 2362–2370. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Bojnik, E.; Ruppert, A.S.; Stefanovski, M.R.; Goettl, V.M.; Smucker, K.A.; Smith, L.L.; Dubovsky, J.A.; Towns, W.H.; MacMurray, J.; et al. Bruton’s tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL). Blood 2014, 123, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Bruton’s tyrosine kinase (BTK) inhibitors in clinical trials. Curr. Hematol. Malig. Rep. 2014, 9, 44–49. [Google Scholar] [CrossRef]
- Weisberg, E.L.; Puissant, A.; Stone, R.; Sattler, M.; Buhrlage, S.J.; Yang, J.; Manley, P.W.; Meng, C.; Buonopane, M.; Daley, J.F.; et al. Characterization of midostaurin as a dual inhibitor of FLT3 and SYK and potentiation of FLT3 inhibition against FLT3-ITD-driven leukemia harboring activated SYK kinase. Oncotarget 2017, 8, 52026–52044. [Google Scholar] [CrossRef]
- Thi Mai, H.D.; Gaslonde, T.; Michel, S.; Tillequin, F.; Koch, M.; Bongui, J.B.; Elomri, A.; Seguin, E.; Pfeiffer, B.; Renard, P.; et al. Structure-activity relationships and mechanism of action of antitumor benzo[b]pyrano[3,2-h]acridin-7-one acronycine analogues. J. Med. Chem. 2003, 46, 3072–3082. [Google Scholar] [CrossRef]
- Liu, L.; Shi, B.; Li, X.; Wang, X.; Lu, X.; Cai, X.; Huang, A.; Luo, G.; You, Q.; Xiang, H. Design and synthesis of benzofuro[3,2-b]pyridin-2(1H)-one derivatives as anti-leukemia agents by inhibiting Btk and PI3Kδ. Bioorganic Med. Chem. 2018, 26, 4537–4543. [Google Scholar] [CrossRef]
- Seeliger, D.; De Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.C.; Márquez, E.A.; Mora, J.R. Molecular modeling studies of bromopyrrole alkaloids as potential antimalarial compounds: A DFT approach. Med. Chem. Res. 2018, 27, 844–856. [Google Scholar] [CrossRef]
- Zhang, Z. Variable selection with stepwise and best subset approaches. Ann. Transl. Med. 2016, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Heinze, G.; Wallisch, C.; Dunkler, D. Variable selection—A review and recommendations for the practicing statistician. Biometrical. J. 2018, 60, 431–449. [Google Scholar] [CrossRef]
- Webb, G.I.; Sammut, C.; Perlich, C.; Horváth, T.; Wrobel, S.; Korb, K.B.; Noble, W.S.; Leslie, C.; Lagoudakis, M.G.; Quadrianto, N.; et al. Leave-One-Out Cross-Validation. In Encyclopedia of Machine Learning; Springer: Jersey City, NJ, USA, 2011; pp. 600–601. [Google Scholar]
- Cramer, R.D.; Bunce, J.D.; Patterson, D.E.; Frank, I.E. Crossvalidation, Bootstrapping, and Partial Least Squares Compared with Multiple Regression in Conventional QSAR Studies. Quant. Struct. Relatsh. 1988, 7, 18–25. [Google Scholar] [CrossRef]
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.P.; Agrawal, R.K. Validation of QSAR Models-Strategies and Importance. Int. J. Drug Des. Discov. 2011, 3, 511–519. [Google Scholar]
- Majumdar, S.; Basak, S.C. Beware of External Validation!—A Comparative Study of Several Validation Techniques used in QSAR Modelling. Curr. Comput. Aided. Drug Des. 2018, 14, 284–291. [Google Scholar] [CrossRef]
- Wong, T.T. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit. 2015, 48, 2839–2846. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB: Homepage. Available online: https://www.rcsb.org/ (accessed on 18 December 2019).

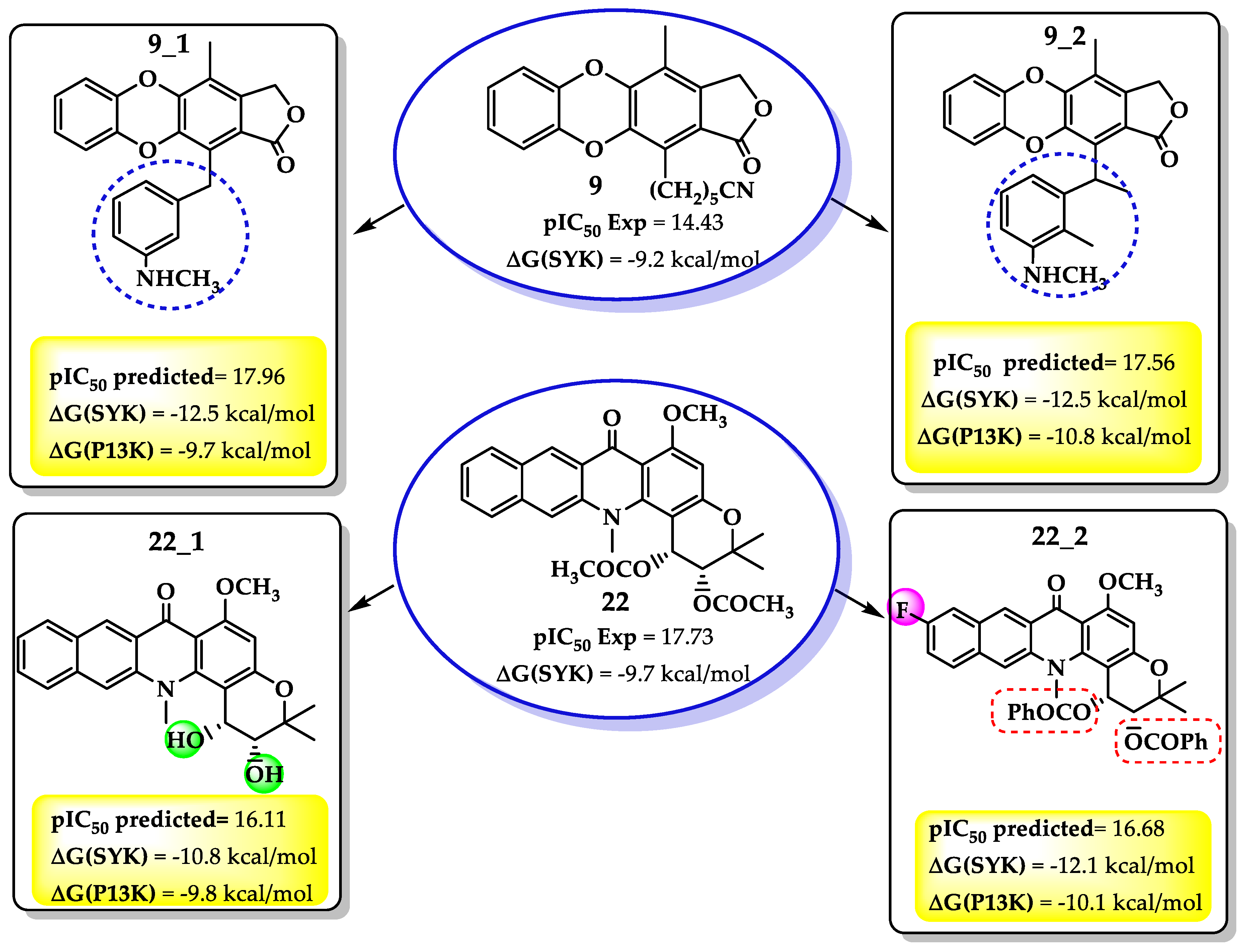
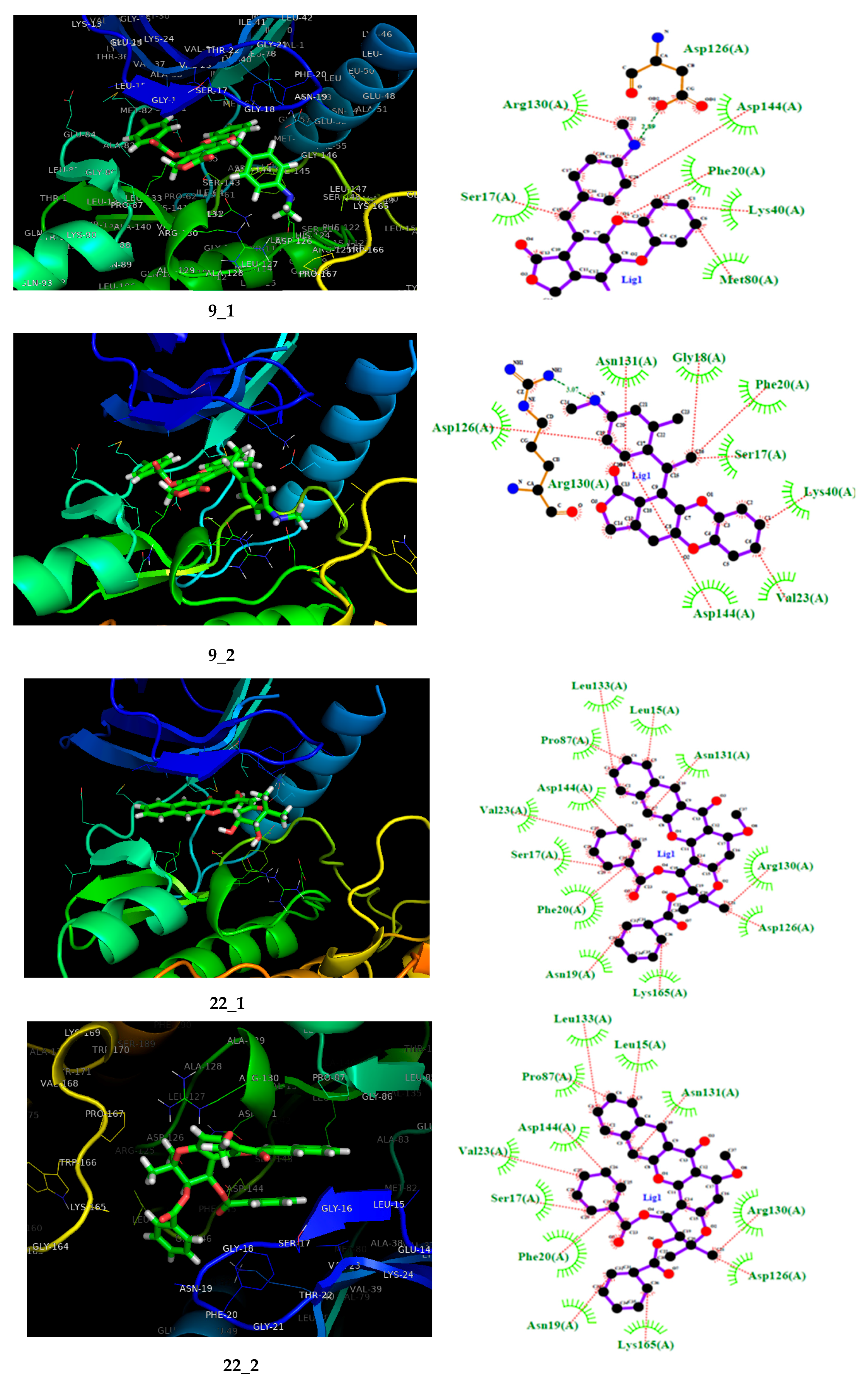
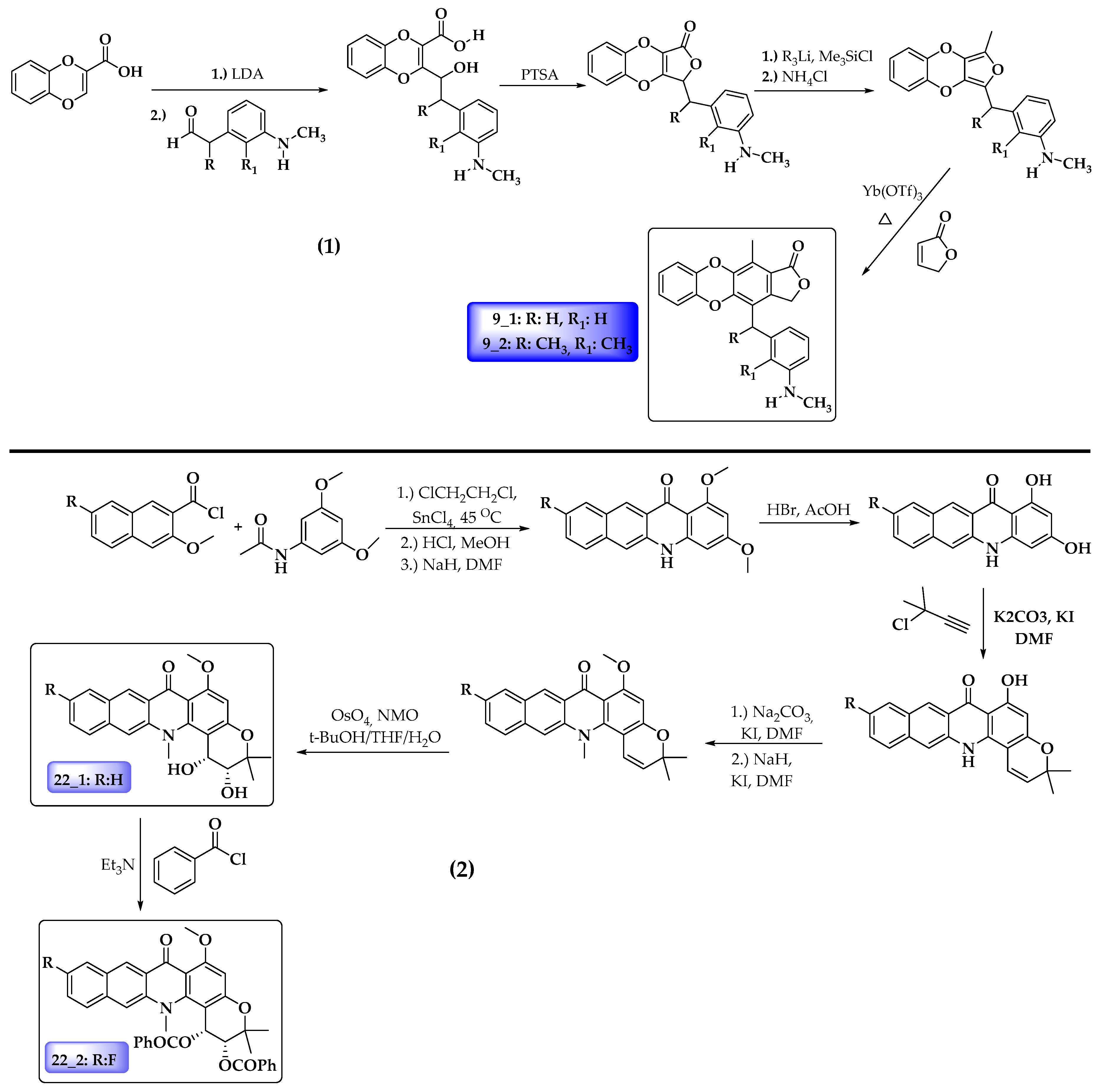
| Compound | pIC50 | HOMO | AC1RABABMID | TS1KFABMID | Softness (s) | LUMO | μ | PSA (A) |
|---|---|---|---|---|---|---|---|---|
| 1 | 12.94 | −0.288 | 0.015 | 0.762 | 3.615 | 0.011 | 5.636 | 87.38 |
| 2 | 13.23 | −0.288 | 0.007 | 0.187 | 4.613 | 0.011 | 5.646 | 91.98 |
| 3 | 13.12 | −0.288 | 0.038 | 0.142 | 3.614 | 0.011 | 5.610 | 96.58 |
| 4 | 12.45 | −0.291 | 0.011 | 0.197 | 3.576 | 0.011 | 5.575 | 101.18 |
| 5 | 11.21 | −0.294 | 0.010 | 0.767 | 3.535 | 0.011 | 5.475 | 105.55 |
| 6 | 10.87 | −0.288 | 0.015 | 0.342 | 3.502 | 0.003 | 5.099 | 102.75 |
| 7 | 9.47 | −0.295 | 0.010 | −0.029 | 3.427 | 0.003 | 9.575 | 73.94 |
| 8 | 14.43 | −0.291 | 0.214 | −0.199 | 4.528 | 0.007 | 6.154 | 73.94 |
| 9 | 9.33 | −0.289 | 0.022 | −0.303 | 3.612 | 0.012 | 6.567 | 88.38 |
| 10 | 11.27 | −0.289 | 0.061 | 0.036 | 3.685 | 0.011 | 4.510 | 73.94 |
| 11 | 12.94 | −0.281 | 0.022 | 0.169 | 4.011 | 0.015 | 6.952 | 69.18 |
| ELLIPTICINE | 15.48 | −0.281 | 0.095 | −1.020 | 6.100 | 0.000 | 3.545 | 24.06 |
| 12 | 13.63 | −0.286 | 0.169 | −1.020 | 3.495 | 13.630 | −0.286 | 23.59 |
| 13 | 13.63 | −0.286 | 0.169 | −1.020 | 3.495 | 0.007 | 3.274 | 30.49 |
| 14 | 10.59 | −0.274 | 0.007 | −0.193 | 3.914 | 0.016 | 5.912 | 59.95 |
| 15 | 11.01 | −0.262 | 0.055 | −0.093 | 2.046 | 0.016 | 6.150 | 69.18 |
| 16 | 10.82 | −0.270 | 0.014 | −0.453 | 3.835 | 0.007 | 5.121 | 41.93 |
| 17 | 11.62 | −0.287 | 0.027 | 0.382 | 3.369 | −0.010 | 3.638 | 52.93 |
| 18 | 11.17 | −0.285 | 0.046 | −0.018 | 3.410 | −0.009 | 2.842 | 41.93 |
| 19 | 14.51 | −0.274 | 0.019 | 1.636 | 3.571 | −0.008 | 7.458 | 100.85 |
| 20 | 15.42 | −0.274 | 0.022 | 2.150 | 3.567 | −0.008 | 7.597 | 100.85 |
| 21 | 10.11 | −0.278 | 0.026 | −0.660 | 3.560 | −0.003 | 6.429 | 82.39 |
| 22 | 17.73 | −0.288 | 0.201 | 2.337 | 3.292 | −0.015 | 7.236 | 100.85 |
| pIC50 | HOMO | AC1RABABMID | TS1KFABMID | Softness | |
|---|---|---|---|---|---|
| pIC50 | 1 | 0.012 | 0.497 | 0.512 | 0.210 |
| HOMO | 1 | −0.151 | 0.412 | 0.285 | |
| AC1RABABMID | 1 | −0.010 | 0.043 | ||
| TS1KFABMID | 1 | −0.400 | |||
| Softness | 1 |
| Compound | pIC50 | SYK | PI3K | BTK |
|---|---|---|---|---|
| 1 | 12.94 | −8.1 | −8.3 | −9.0 |
| 2 | 13.23 | −8.9 | −9.5 | −8.9 |
| 3 | 11.84 | −8.3 | −8.2 | −8.9 |
| 4 | 13.12 | −8.8 | −9.1 | −9.1 |
| 5 | 12.45 | −7.6 | −8.1 | −9.0 |
| 6 | 11.21 | −7.6 | −8.6 | −9.7 |
| 7 | 10.87 | −7.3 | −7.5 | −7.9 |
| 8 | 13.00 | −9.5 | −9.2 | −9.1 |
| 9 | 14.43 | −9.8 | −9.0 | −9.8 |
| 10 | 9.32 | −7.7 | −8.4 | −8.9 |
| 11 | 11.27 | −8.8 | −8.9 | −8.9 |
| 12 | 12.94 | −8.4 | −8.4 | −8.8 |
| Ellipticine | 15.48 | −9.5 | −9.9 | −9.5 |
| 13 | 13.63 | −9.1 | −9.0 | −8.4 |
| 14 | 10.58 | −7.5 | −7.4 | −10.1 |
| 15 | 11.06 | −9.0 | −10.0 | −10.5 |
| 16 | 10.83 | −7.7 | −6.9 | −6.8 |
| 17 | 11.61 | −7.7 | −7.9 | −7.4 |
| 18 | 11.16 | −7.5 | −9.8 | −10.1 |
| 19 | 14.51 | −8.9 | −9.3 | −10.1 |
| 20 | 15.42 | −8.9 | −8.7 | −8.9 |
| 21 | 10.11 | −7.7 | −7.3 | −6.7 |
| 22 | 17.73 | −9.7 | −8.7 | −9.7 |
| R788 | 13.23 | −9.1 | −9.5 | −9.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Márquez, E.; Mora, J.R.; Flores-Morales, V.; Insuasty, D.; Calle, L. Modeling the Antileukemia Activity of Ellipticine-Related Compounds: QSAR and Molecular Docking Study. Molecules 2020, 25, 24. https://doi.org/10.3390/molecules25010024
Márquez E, Mora JR, Flores-Morales V, Insuasty D, Calle L. Modeling the Antileukemia Activity of Ellipticine-Related Compounds: QSAR and Molecular Docking Study. Molecules. 2020; 25(1):24. https://doi.org/10.3390/molecules25010024
Chicago/Turabian StyleMárquez, Edgar, José R. Mora, Virginia Flores-Morales, Daniel Insuasty, and Luis Calle. 2020. "Modeling the Antileukemia Activity of Ellipticine-Related Compounds: QSAR and Molecular Docking Study" Molecules 25, no. 1: 24. https://doi.org/10.3390/molecules25010024
APA StyleMárquez, E., Mora, J. R., Flores-Morales, V., Insuasty, D., & Calle, L. (2020). Modeling the Antileukemia Activity of Ellipticine-Related Compounds: QSAR and Molecular Docking Study. Molecules, 25(1), 24. https://doi.org/10.3390/molecules25010024






